Abstract
Quantum dots (QDs) are attracting intense interest as fluorescence labeling agents for biomedical imaging because biocompatible coatings and relatively non-toxic rare earth metal QDs have emerged as possible options. QD photoemissions are bright, of narrow wavelength range, and very stable. We sought to encapsulate QDs within targeted PEGylated liposomes to reduce their propensity for liver uptake and to amplify the already strong QD emission signal. A novel lipid-QD conjugate initialized a process by which lipids in solution coalesced around the QDs. The liposomal structure was confirmed with size measurements, SEM, and IR spectroscopy. PEGylated QD liposomes injected into a xenograft tumor model largely cleared from the body within 24 hours. Residual liver labeling was low. Targeted QD liposomes exhibited robust tumor labeling compared with controls. This study highlights the potential of these near IR emitting QD liposomes for preclinical/clinical applications.
INTRODUCTION
Nanoparticles are increasingly being explored as labeling agents in fluorescence-based biomedical imaging. Fluorescence label-based laparoscopic/endoscopic imaging of the blood vessels, bronchi, esophagus, stomach, intestines, skin, and oral cavity, along with intra-operative fluorescence imaging, have the potential to facilitate the detection and assessment of pathological structures such as tumors, inflamed and/or infected tissue, ulcers and fibrotic lesions1–5. Quantum dot (QD) nanoparticles in particular are attracting considerable interest as imaging labels, as they produce a very intense, narrow wavelength band photoemission, and they are two-orders of magnitude more photostable than conventional fluorophores6.
Newer QD variants such as the rare earth metal compositions are relatively non-toxic, while the toxic cadmium-based QDs can be coated with polyethylene glycol (PEG) to possibly render them sufficiently inert for uneventful clearance from the body, if they are less than 15 nm in size and not too densely PEGylated7–11. In the absence of UV irradiation QDs with a stable coating have been found to be essentially nontoxic, so IR fluorescence may be preferable for cadmium-based QDs12. However, when they are intravenously injected both non-targeted, targeted and micelle-encapsulated QDs concentrate in the liver, spleen and bone marrow and may clear slowly if at all, and this accumulation is undesirable from the standpoint of toxicological and pharmacokinetic considerations7–12.
Liposomes offer an attractive potential solution to coating QDs to prevent liver accumulation. Liposomal nanoparticles are stable and long circulating when PEGylated13. They are comparatively large (≈100 nm) and have favorable charge characteristics even with a payload. They can be made multifunctional by at the same time accommodating any combination of QDs, a drug payload, a MRI or PET labeling agent, and a targeting ligand14–16. Gel core liposomes have favorable loading of all drug chemotypes17. Liposomes are already FDA approved for human use and there are currently two liposome-based products, Doxil® and Depocyte® that are used clinically, making liposomes an appealing and readily translatable potential platform for QD delivery in patients.
The goal of the present study was to address two key issues to support further investigations of QD liposomes as labeling agents for preclinical and clinical fluorescence imaging. First, various strategies of loading QDs into liposomes can be problematic for various reasons; (1) the approach of simply entrapping particles such as QDs and iron oxide within the liposomal bilayer may lead to leaching of these hydrophobic molecules in vivo18, (2) conjugation of QDs to the external liposome surface only via PEG would not be expected to result in the most extensive QD loading, (4) the use of long PEG chains (PEG2000) to bind the QDs to the external liposomal surface may expose the QDs and result in heavy liver concentrations even >24 hours after intravenous injection, and (3) additional moieties used to encapsulate the QDs can affect their emission spectrum19,20. The second major issue is that poor signal-to-noise in vivo is a limitation of virtually all labeling fluorophores21. The strong emission generated by QDs helps overcome this signal degradation by tissue autofluorescence, but further enhancement of signal by concentrating the QDs at the target is clearly an important objective.
Liposomal QD encapsulation was achieved using a novel and efficient linking of the QDs to a lipid anchor by covalent binding, so that initiation of the liposomal outer shell began at the lipid-QD conjugate and extended in all directions to effect complete QD encapsulation (Figure 1). This allows a higher density of QDs to be loaded, and this specific approach has been attempted previously but for micelles which have drug loading limitations and stability issues22,23. The signal-to-background issue was addressed by preparing QD liposomes targeted to the αvβ3 integrin expressed exclusively on angiogenic microvessels24,25,26. Our hypothesis was that unbound QD liposomes would wash out after several hours while targeted liposomes would remain bound to the target to generate a readily discriminated signal27.
Figure 1.
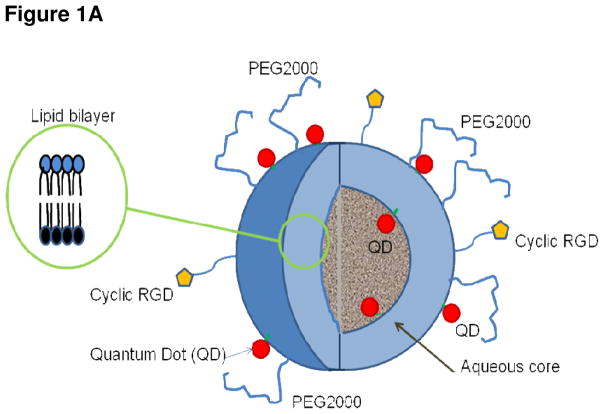
Figure 1A Cutaway drawing showing liposomal nanoparticle lipid bilayer (inset) and cyclic RGD peptides bound to the exterior liposomal surface. The QDs denoted by filled red circles are conjugated to a lipid moiety and reside inside and external to the bilayer. The liposome is in effect initiated at these QD-lipid conjugates and forms around them. The interior volume of this liposome is aqueous, but can be filled with a hydrogel to facilitate the loading of a broad range of drug chemotypes.
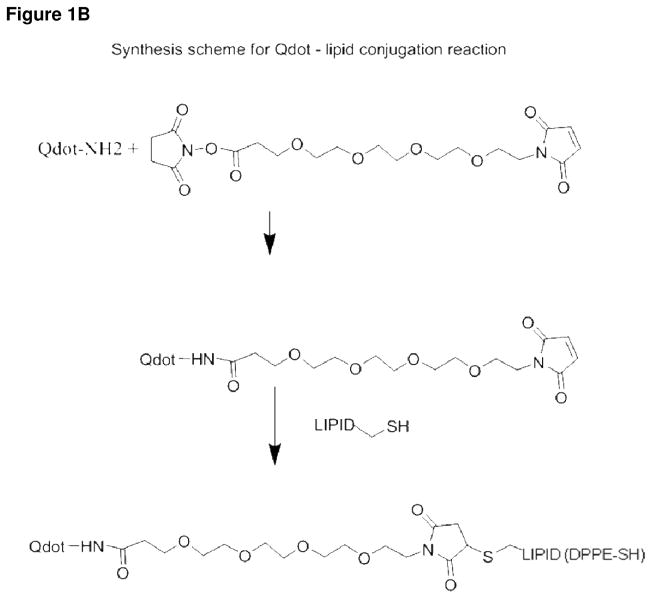
Figure 1B Schematic diagram depicting the conjugation of the QD and the DPPE-SH lipid prior to liposomal nanoparticle assembly. The first step is to couple the QD to the linker. The maleamide group of the linker reacts with the lipid-thiol to create a stable thioether bond.
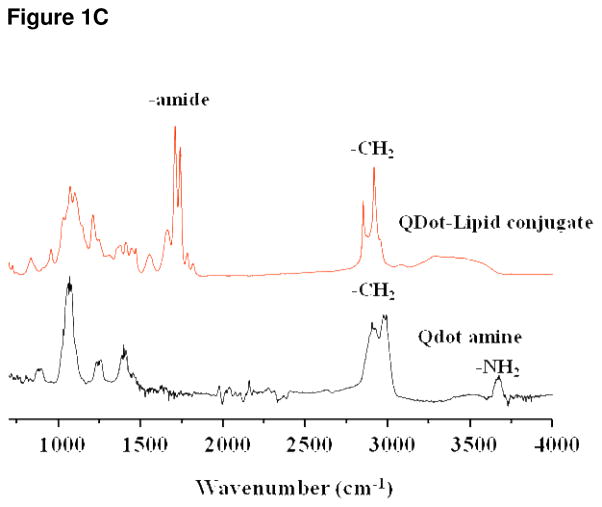
Figure 1C IR spectroscopy, the lower tracing shows the spectrum for QDs alone. The top tracing shows the spectrum of the putative lipid-QD conjugate. The doublet resulting from stretch of the amide bond is clearly indicated and is not present within the QD alone tracing. This data provides strong evidence that QD-lipid conjugation was successful. Fluorescence intensity of the QD800 nanocrystals was not diminished by encapsulation (data not shown).
We present physical measurements confirming the predicted structure of the QD liposomes with our assembly approach. Furthermore in a xenograft tumor model targeted QD liposomes (near IR 800 nm; non-UV emission) 24 hours after intravenous injection robustly labeled the tumor but not the liver, as they showed significant clearance. These results highlight the potential of QD liposomes as biomedical labeling agents and support subsequent, more comprehensive investigations of QD-based multifunctional particles for clinical diagnostic-therapeutic applications.
EXPERIMENTAL PROCEDURES
Preparation of QD-lipid Conjugates
Synthesis of QD-Lipid Conjugate and Assembly of Lipsomes
The final structure of the liposomal nanoparticle is shown in Figure 1A. Note the QD – lipid conjugates adorning the exterior and interior. To accomplish this, initially the amino-QD800 (1 μM; Invitrogen) in 300 μl water was conjugated to a short linker, succinimidyl ester-(PEO)4-maleimide (2.5 μM) in DMSO, as shown in the reaction scheme in Figure 1B. The reaction mixture was stirred at room temperature for one hour. Then DOPE-SH (2.5 μM; Avanti Polar Lipids, Inc, Alabaster, Alabama)) containing the free thiol group in 200 μl chloroform was reacted with the QD-(PEO)4-maleimide for one hour at room temperature in a biphasic solution to produce the QD-lipid conjugate. We performed thin layer chromatography of the QD-lipid conjugate, and then acquired the IR spectroscopic profile in order to prove the presence of the QD to lipid amide bond thus confirming conjugation.
The liposomal formulation comprised cholesterol/DOPE/DSPC/DOPE-(PEO)4-QD/DSPE-mPEG2000 (3:3:3:0.5:0.5 molar ratio) dissolved in chloroform and was evaporated under argon gas. The dried lipid film was hydrated in sterile water to create large multilamellar vesicles (LMVs). Liposomes were vortexed for 2–3 minutes to remove any adhering lipid film and sonicated in a bath sonicator (Ultrasonik 28X) for 2–3 minutes at room temperature. MLVs were then sonicated with a Ti-probe (Branson 450 sonicator) for 1–2 minutes to produce small unilamellar vesicles (SUVs) as indicated by the formation of a translucent solution. To reduce the size of the SUVs, stepwise extrusion was performed with the final step being extrusion through a polycarbonate filter with 100-nm pore size (Whatman). Unencapsulated QDs were separated from the QD liposomes by size exclusion chromatography using Sepharose CL-4B column, with PBS as an eluent.
Physical Characterization of Bioconjugates
Particle size and net charge (Zeta potential)
The liposome suspension was diluted in 1/10 in MilliQ water, and 100 μl of the dilution was sized using light backscattering (Malvern Zetasizer, ZEN 3600). The same instrument was used to measure the particle net charge which was expressed in mV. The QD liposome size and surface ζ-potential were obtained from three repeat measurements with a backscattering angle of 173°. Liposome morphology and size were further characterized using scanning electron microscopy (SEM). Samples were prepared by dropping 5 μl of QD liposome suspension onto a polished silicon wafer. After drying the droplet at room temperature overnight, the sample was coated with chromium and then imaged by SEM.
Electron microscopy (SEM) for structure confirmation
Further confirmation of liposome size and structure was obtained with SEM. The particle samples were diluted 1/10 in milliQ water, drying 5 μl onto polished silicon wafers overnight, coating with chromium, and then imaged on a Philips XL-30 electron microscope to 30,000X. Particular emphasis was placed on confirming that the liposomes were spherical.
Fluorescent emission peak and IR spectrum
For the emission peak QDs alone and QD800-lipsomes were prepared using known volumes and concentrations, followed by purification with size exclusion chromatography. A Tecan fluorescence spectrophotometer was used to measure the peak emission of the intact nanoparticles at an excitation wavelength of 405 nm, and to confirm that the emission peak (780 nm) was that expected for the QD800 nanocrystals. For the IR spectrum the QD-lipid conjugate and the QDs alone were prepared in solution and the emitted IR was measured using a Thermo Scientific Nicolet 6700 FT-IR spectrometer.
Particle Stability
Liposomal nanoparticles encapsulating QDs were prepared and were characterized in terms of size and charge using a Malvern Zetasizer. The particles were stored at room temperature for 7 days and then were rescanned, and changes quantified in terms of percent change from the pre-storage data.
Biological Characterization
Cell viability study
U87 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% Fetal bovine serum (FBS), cells were cultured at 37°C in a humidified atmosphere containing 5% CO2. For cell viability study 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction assay was performed. For MTT assay 2000 cells (U87) per well were seeded into a 96 well plate and cultured overnight, then incubated with 10 different concentrations of the QD liposomes starting from 0.2 uM to 0.39 nM. 10 ul of MTT was added 72 hr after addition of the QD particles to the cells. Results were expressed as percent viability = {[A540(treated cells) − background]/[A540(untreated cells) − background]} × 100.
In vitro imaging of targeted QD liposomes
U87 cells were seeded on to a 6 well culture plate at a density of 200K cell per well and incubated in DMEM with 10% FBS for 24 hrs, QD encapsulated particles or native QDs were added to the wells at a final concentration of 0.5 nM. Cells were incubated for 4 hr, washed with PBS twice and observed with a laser scanning confocal microscope. QD conjugates in the cytoplasm excited with 450 nm and fluorescence emission was observed at 780 nm.
In vivo Imaging of native QDs, and targeted vs non-targeted QD-lipsomes
Athymic nude mice were anesthetized with ketamine/medetomidine and subcutaneously inoculated in the left flank with 4×106 cells/100 μl PBS of the U87 glioblastoma cell line. When the flank tumors grew to a volume of 60 mm3, the animals were anesthetized with isofluorane administered via a nose cone, and imaged in an Advanced Research Technologies (ART) Optix-MX2® imaging system. This apparatus includes four pulsed lasers (picoseconds pulse, 80 MHz duty cycle) of different wavelengths to excite a variety of fluorophores with corresponding emission filters and a time-correlated single photon counting (TCSPC) photomultiplier tube for fluorescence time domain detection which can provide both fluorescence intensity and fluorescence lifetime imaging. Here 470 nm/780 nm excitation/emission wavelengths were employed to image the nanoparticles. The nanoparticles were intravenously injected via the tail vein and the mice were imaged before injection, and at 2 and 24 hours after injection. A brightfield image was acquired first, followed by fluorescence intensity images based on single photon counting. The image data were saved directly on a PC for later processing. Images were acquired both before and after nanoparticle injection.
Data Processing
The data was processed using ART Optix analysis software and the counts represented as heat maps superimposed on the brightfield grayscale image. Identity of the fluorophore (QD800) was confirmed by fluorescence lifetime determination.
RESULTS
Physical Characterization of QD-lipid Conjugates
Confirmation of conjugation
Thin layer chromatography revealed the presence both a QD and QD conjugate in the mobile phase (shown in supporting data). IR spectroscopy clearly indicated the amide bond linking the QD with the lipid (DPPE-SH) and therefore confirmed the existence of the QD-lipid conjugate (Figure 1C). The QD emission peak wavelength was unchanged by the lipid conjugation (shown in supporting data).
Particle size and net charge (Zeta potential)
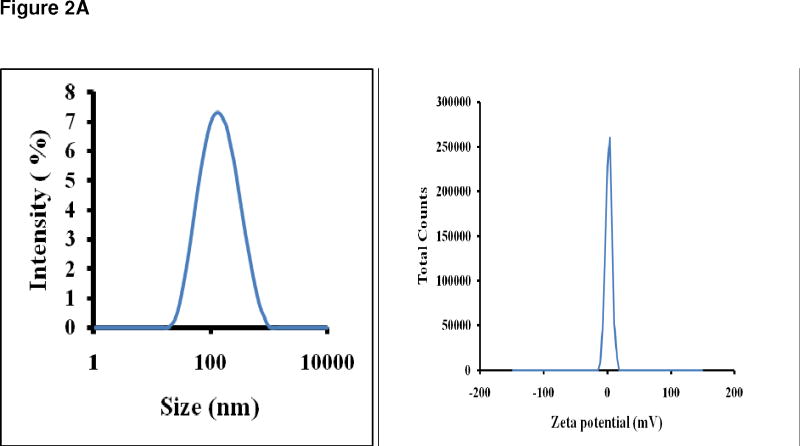
Figure 2A, left panel, shows that the average particle size was 100 nm indicating that fully formed liposomes were present in the suspension mixture, since this was the final extrusion pore size. The right panel of Figure 2A indicates the zeta potential, or net charge, which was close to the desired neutrality, consistent with complete QD encapsulation. We found that the zeta potential and average particle size were not significantly changed after storage in PBS at 4°C for 7 days (not shown).
Figure 2.
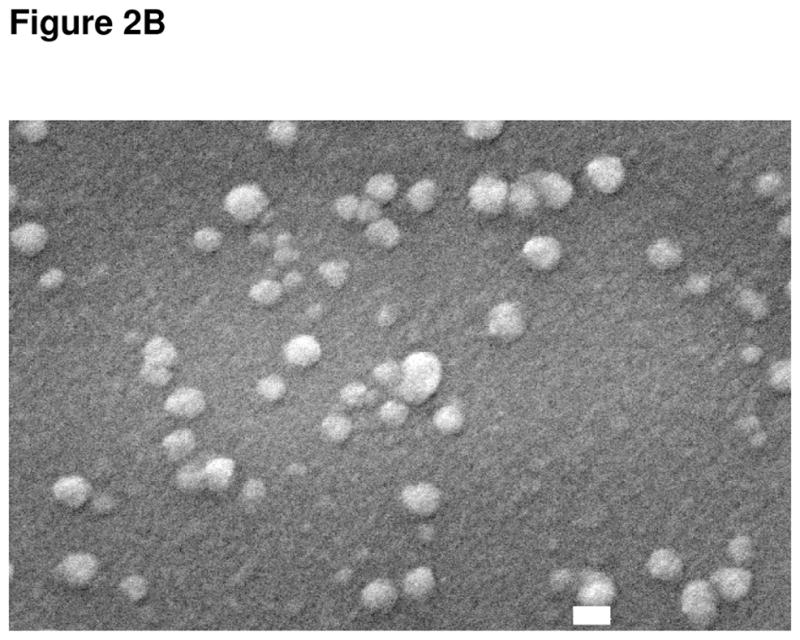
Figure 2A Left panel is the Zetasizer plot of a QD liposome suspension and reveals an average QD liposome size of 109 nm and a symmetrical size distribution. This data confirms that the nanoparticles containing QDs are of the desired size, which indicates that the encapsulation of the relatively large QDs did occur. The right panel is the plot of charge distribution indicating that the liposomal nanoparticles containing QDs were neutral, indicating that encapsulation of the QDs was complete. The lack of charge allows for a much longer in vivo circulation time.
Figure 2B Electron microscope image of liposomal nanoparticles containing QDs. The spherical shape of the nanoparticles is clearly visible and they are the correct size, approximately 100 nm, as indicated by the white bar which denotes 100 nm.
Electron microscopy (SEM) for structure confirmation
Figure 2B is the SEM of the QD liposomes. The white bar indicates 100 nm, and the SEM confirmed that the particles were 100 or greater in size, that they had a spherical shape, and were intact.
Biological Characterization
Cell viability study
We found that the QD liposomes and the QDs were non-toxic in the concentrations tested (0.01 to 0.20 μM) when applied to U87 cells in culture (shown in supporting data). This data shows that encapsulating the QDs within liposomes did not make them toxic at physiologically relevant doses.
In vitro Imaging
The QD targeted QD liposomes were taken up more avidly by U87 cells than non-targeted QD liposomes. In Figure 3 panels A – C depict the uptake of particles in cultured U87 glioblastoma cells. Panel A is the phase contrast image, while B is the fluorescence acquisition. Panel C shows the merge and it can be seen that a significant proportion of the cells have internalized the targeted QD liposomes. In comparison panels D – F which are arranged according to the same format, reveal the lower uptake of non-targeted QD liposomes into U87 cells. The merge of D and E, denoted by panel F, reveals that endocytosis alone of the non-targeted QDs resulted in considerably less internalization than was the case with targeted particles. This data suggests that that targeting αvβ3 integrins, which are expressed by U87 cells, confers a delivery advantage with tumor cells. The targeted QD liposomes are most probably being taken up via the αvβ3 receptor and internalized, while the non-targeted liposomes are mostly being taken up via endocytosis.
Figure 3.
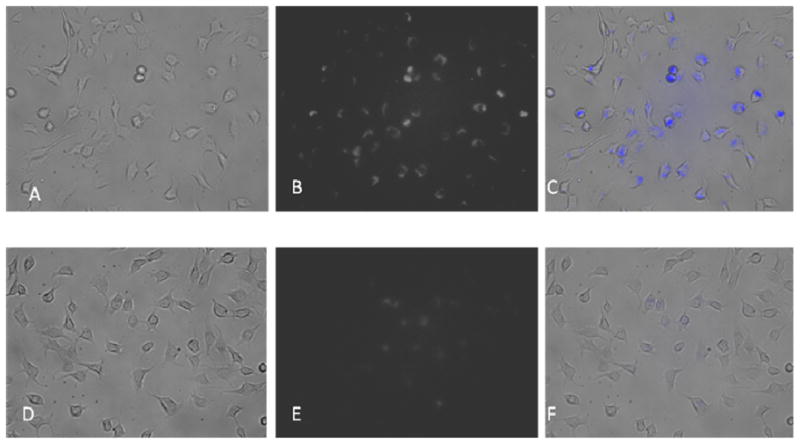
Panels A – C depict the uptake of particles in cultured U87 glioblastoma cells. Panel A is the phase contrast image, while B is the fluorescence acquisition. Panel C shows the merge and it can be seen that significant proportion of the cells have internalized the targeted QD liposomes. In comparison panels D – F show the uptake of non-targeted QD liposomes by U87 cells. The merge of D and E, denoted by panel F, reveals that endocytosis of the non-targeted QDs resulted in considerably less internalization than was the case with targeted particles.
In vivo Imaging of native QDs, and targeted vs non-targeted QD-liposomes
Targeted and non-targeted QD liposomes were widely distributed at two hours after injection of the mice as shown in Figures 4A–D. The photon counts were high in images taken with the animal in both the prone and supine positions. Peaks counts for both targeted and non-targeted QD liposomes were measured in the liver, kidney and bone marrow. After 24 hours the unbound QD liposomes had washed out of most body areas and the liver labeling has substantially declined (Figures 4E–F). However these PEGylated QD liposomes had evidently cleared the liver. In view of the extensive clearance and the finding that QD liposomes were not toxic in cell culture, there exists the possibility that even cadmium-based QD liposomes may be practicable in terms of some cancer related clinical applications.
Figure 4.
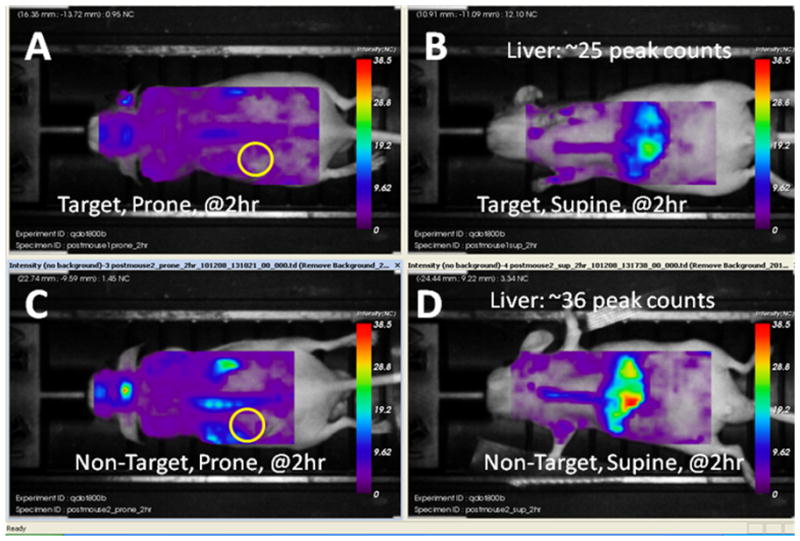
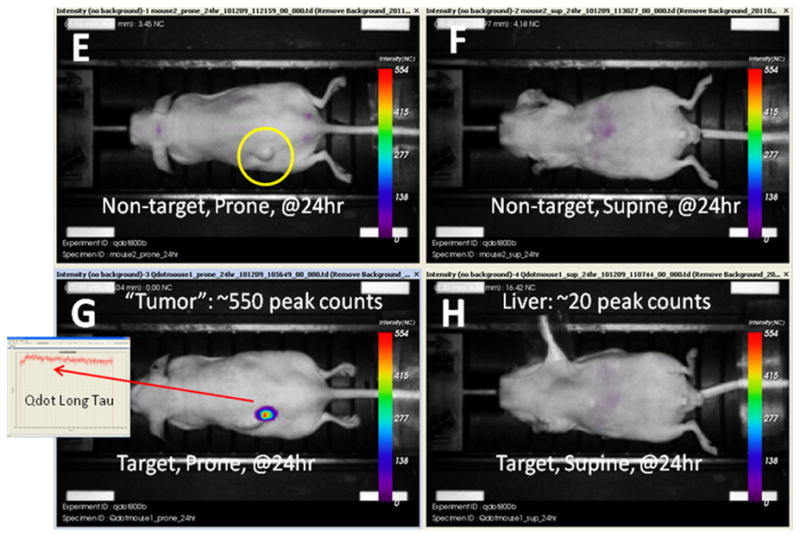
Panels A–D above and E–H below. Series of brightfield grayscale images fused with fluorescence intensity images. Panels A through D were obtained at 2 hours after the intravenous injection of either targeted or non-targeted QD liposomes. The tumor site is indicated by a yellow circle on the prone images. Note that there is considerable liver uptake of both targeted and non-targeted QD liposomes resulting in strong liver labeling. At 24 hours after injection, denoted by panels E though H the QD liposomes, or at least the QDs have cleared the body either from renal clearance or via the feces. However, in 4G the targeted QD liposomes have bound to the tumor and/or its blood vessels to generate robust, clearly defined labeling. The identity of this fluorescence is confirmed to be the nanoparticle by the characteristic long fluorescence lifetime. The tumor site in the mouse that received non-targeted liposomes is shown by the yellow circle in 4E. In 4F and 4H the decline in liver labeling is quite substantial. The images were not processed and are original.
The targeted particles remained bound to the tumor and produced strong labeling that could be cleanly discriminated from background. The identity of the QD800 liposomes was unambiguously confirmed by measuring the species specific fluorescence lifetime, which as expected was much longer than the surrounding tissue autofluorescence (inset in Figure 4G) 28–30. The mouse which received non-targeted QD liposomes had a low degree of liver labeling and no tumor labeling. None of the acquired images were filtered or modified from the original.
DISCUSSION
QDs have attracted attention owing to their photoemission qualities which include the generation of an intense narrow wavelength band of fluorescent light and resistance to photobleaching. Cadmium-based QDs unfortunately are toxic to cultured human cells and little is known about their clearance from the human body. Several reports have indicated that coating cadmium-based QDs with inert materials renders them essentially non-toxic as long as exposure to UV light is avoided11,12. Newer rare earth metal and organic QDs are much less toxic, but encapsulation is still desirable for purposes of targeting, stealth, and drug loading12. Micelles tend to be unstable and drug uptake into a micelle already containing a QD would be challenging23. Liposomes are ideal QD coating candidates but liposomal encapsulation is technically comlex.
The present study was designed to develop a practicable and reliable liposomal QD encapsulation protocol to create a fluorescence labeling platform that could be targeted and also used as a drug delivery vehicle. We sought to determine whether lipid encapsulation reduced toxicity in vitro, and facilitated targeting for amplification of the QD signal using an in vivo system. A novel chemistry was implemented to allow conjugation of the QD to a lipid anchor which acted as an initiation site for lipid encapsulation. Conjugation rather than simple encapsulation alone is required as otherwise the hydrophobic QDs will prematurely leach out from between the liposomal bilayer.
The PEG2000 is represents an optimized PEG variant in terms of toxicity and increasing circulation stability. PEG2000 has been chosen as it is not toxic and because it is used in many clinical liposomal formulations, such as Doxil®. It prevents opsonization and removal from the circulation and thereby enhancing liposomal circulation half-life. At the same time PEG2000 is non-toxic17,25,31.
A near IR emitting QD (QD800) was tested to demonstrate that near IR fluorescence imaging of QDs in vivo was feasible, since avoidance of UV light prevents coated QDs from becoming toxic12. The targeted QD liposomes largely cleared the test mice within 24 hours, suggesting that coated QDs may be cleared rapidly and thus represent a lower toxicity hazard. Since the QD liposomes were too large (100 nm plus) for kidney clearance it is possible that the route of excretion was through the liver and the intestines and then finally via the feces7,8. Importantly the targeting results suggest that targeting allowed for washout to reduce unbound, background QDs27. This may have facilitated an amplification of the target labeling with enhanced signal-to-noise.
The focus of future studies will be to more comprehensively characterize QD liposomal clearance and toxicity, and to combine target fluorescence labeling with drug delivery16,32–35. This ‘theranostic’ combination within a single nano-platform would potentially allow the tumor to be located, and drug delivery to be chronicled and related to the response of the tumor. Some reports describe the loading of doxorubicin into lipsomes, but this is an older compound and relatively straightforward to load into liposomes. Newer agents such as taxol are more challenging owing to their hydrophobicity36. Gel core liposomes allow drug compounds of virtually all chemotypes to be loaded effectively16,36–39.
In summary the present study has shown that QD encapsulation within a large (100 nm plus) liposome can be accomplished and that such a QD liposome may be targeted. Moreover the lipid coating and PEGylation renders the QD essentially non-toxic and coupled with rapid clearance may allow both cadmium-based and less toxic rare earth metal and organic QDs to be feasible for patient use. The data provide a basis for pursuing subsequent, comprehensive studies involving the targeted delivery of a therapeutic agent using a QD bearing liposomal platform, the chronicling of the therapeutic effect of such a nanoparticle on an in vivo implanted tumor, and the detailed characterization of particle biodistribution and clearance.
Supplementary Material
Acknowledgments
The authors wish to thank Dr. Santosh Aryal for the SEM image, Dr. Kersi Pestonjamasp and Mr. Joseph Aguilera for the in vitro fluorescence images, and Mr. Ying Chao for the in vivo tumor implantations. The authors gratefully acknowledge ICMIC NCI-P50-CA128346 and the UCSD In Vivo Small Animal Imaging Resource for the in vivo imaging.
Footnotes
We characterized the effects of conjugation in terms of emission wavelength and liposomal toxicity. We found that the QD emission peak wavelength was unchanged by the lipid conjugation. Moreover, encapsulating the QDs within liposomes did not make them more toxic at the doses typically used in vitro. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Roberts DW, Valdés PA, Harris BT, Fontaine KM, Hartov A, Fan X, Ji S, Lollis SS, Pogue BW, Leblond F, Tosteson TD, Wilson BC, Paulsen KD. Coregistered fluorescence-enhanced tumor resection of malignant glioma: relationships between delta-aminolevulinic acid-induced protoporphyrin IX fluorescence, magnetic resonance imaging enhancement, and neuropathological parameters. J Neurosurg. 2011;114(3):595–603. doi: 10.3171/2010.2.JNS091322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zou P, Xu S, Povoski SP, Wang A, Johnson MA, Martin EW, Jr, Subramaniam V, Xu R, Sun D. Near-infrared fluorescence labeled anti-TAG-72 monoclonal antibodies for tumor imaging in colorectal cancer xenograft mice. Mol Pharm. 2009;6:428–440. doi: 10.1021/mp9000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu RC, Traverso LW. Diagnostic laparoscopy improves staging of pancreatic cancer deemed locally unresectable by computed tomography. Surg Endosc. 2005;19:638–642. doi: 10.1007/s00464-004-8165-x. [DOI] [PubMed] [Google Scholar]
- 4.Loning M, Diddens H, Kupker W, Diedrich K, Huttmann G. Laparoscopic fluorescence detection of ovarian carcinoma metastases using 5-aminolevulinic acid-induced protoporphyrin IX. Cancer. 2004;100:1650–1656. doi: 10.1002/cncr.20155. [DOI] [PubMed] [Google Scholar]
- 5.He Y, Xu H, Chen C, Peng J, Tang H, Zhang Z, Li ZP. In Situ Spectral Imaging of Marker Proteins in Gastric Cancer with Near-infrared and Visible Quantum Dots Probes. Talanta epub. 2011 doi: 10.1016/j.talanta.2011.03.035. [DOI] [PubMed] [Google Scholar]
- 6.Smith AM, Dave S, Nie S, True L, Gao X. Multicolor quantum dots for molecular diagnostics of cancer. Expt Rev Mol Diagn. 2006;6(2):231–244. doi: 10.1586/14737159.6.2.231. [DOI] [PubMed] [Google Scholar]
- 7.Schipper ML, Iyer G, Koh AL, Cheng Z, Ebenstein Y, Aharoni A, Keren S, Bentolila LA, Li J, Rao J, Chen X, Banin U, Wu AM, Sinclair R, Weiss S, Gambhir SS. Particle size, surface coating, and PEGylation influence the biodistribution of quantum dots in living mice. Small. 2009;5(1):126–34. doi: 10.1002/smll.200800003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi HS, Liu W, Misra P, Tanaka E, Zimmer JP, Itty Ipe B, Bawendi MG, Frangioni JV. Renal clearance of quantum dots. Nat Biotechnol. 2007;25(10):1165–70. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pozzi-Mucelli S, Boschi F, Calderan L, Sbarbati A, Osculati F. Quantum Dots: proteomics characterization of the impact on biological systems. Journal of Physics: Conference Series. 2009;170:1–5. [Google Scholar]
- 10.Lewinski N, Zhu H, Drezek R. Evaluating strategies for risk assessment of nanomaterials. In: Sahu SC, Casciano D, editors. Nanotoxicity: from in vivo and in vitro models to health risks. Wiley; New Delhi: 2009. p. 476. [Google Scholar]
- 11.Das GK, Chan PP, Teo A, Loo JS, Anderson JM, Tan TT. In vitro cytotoxicity evaluation of biomedical nanoparticles and their extracts. J Biomed Mater Res A. 2010;93(1):337–46. doi: 10.1002/jbm.a.32533. [DOI] [PubMed] [Google Scholar]
- 12.Lee J, Ji K, Kim J, Park C, Lim KH, Yoon TH, Choi K. Acute toxicity of two CdSe/ZnSe quantum dots with different surface coating in Daphnia magna under various light conditions. Environ Toxicol. 2010;25(6):593–600. doi: 10.1002/tox.20520. [DOI] [PubMed] [Google Scholar]
- 13.Remaut K, Lucas B, Braeckmans K, Demeester J, De Smedt SC. Pegylation of liposomes favours the endosomal degradation of the delivered phosphodiester oligonucleotides. J Control Release. 2007;12;117(2):256–66. doi: 10.1016/j.jconrel.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 14.Hu R, Yong K-T, Roy I, Ding H, Law W-C, Cai H, Zhang X, Vathy LA, Bergey EJ, Prasad PN. Functionalized near-infrared quantumdots for in vivo tumor vasculature imaging. Nanotechnology. 2010;21:1–9. doi: 10.1088/0957-4484/21/14/145105. [DOI] [PubMed] [Google Scholar]
- 15.Garg A, Tisdale AW, Haidari E, Kokkoli E. Targeting colon cancer cells using PEGylated liposomes modified with a fibronectin-mimetic peptide. Int J Pharmaceutics. 2009;366:201–210. doi: 10.1016/j.ijpharm.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L, Granick S. How to Stabilize Phospholipid Liposomes (Using Nanoparticles) Nanoletters. 2006;6(4):694–698. doi: 10.1021/nl052455y. [DOI] [PubMed] [Google Scholar]
- 17.Erten A, Wrasidlo W, Scadeng M, Esener S, Hoffman R, Bouvet M, Makale M. MR and fluorescence imaging of doxorubicin loaded nanoparticles using a novel in vivo model. Nanomedicine. 2010;6(6):797–807. doi: 10.1016/j.nano.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bulte JWM, Modo HMJ. Nanoparticles in Biomedical Imaging - Fundamental Biomedical Technologies. 1. Vol. 102. Springer; New York: 2008. Introduction: The Emergence of Nanoparticles as an Imaging Platform in Biomedicine; pp. 2–3. [Google Scholar]
- 19.Weng KC, Noble CO, Papahadjopoulos-Sternberg B, Chen FF, Drummond DC, Kirpotin DB, Wang D, Hom YK, Hann B, Park JW. Targeted tumor cell internalization and imaging of multifunctional quantum dot-conjugated immunoliposomes in vitro and in vivo. . Nano Lett. 2008;8(9):2851–7. doi: 10.1021/nl801488u. [DOI] [PubMed] [Google Scholar]
- 20.Chen CS, Yao J, Durst RA. Liposome encapsulation of fluorescent nanoparticles: Quantum dots and silica Nanoparticles. Journal of Nanoparticle Research. 2006;8:1033–1038. [Google Scholar]
- 21.Hinterdorfer P, van Oijen A, editors. Handbook of Single Molecule Biophysics. Springer; New York: 2009. pp. 2–5.pp. 143 [Google Scholar]
- 22.Kim YH, Subramanyam E, Im JH, Huh KM, Choi H, Choi JS, Lee YK, Park SW. A new PEG-lipid conjugate micelle for encapsulation of CdSe/ZnS quantum dots. J Nanosci Nanotechnol. 2010;10(5):3275–9. doi: 10.1166/jnn.2010.2230. [DOI] [PubMed] [Google Scholar]
- 23.Kim KS, Hur W, Park SJ, Hong SW, Choi JE, Eun Goh EJ, Yoon SK, Hahn SK. Bioimaging for Targeted Delivery of Hyaluronic Acid Derivatives to the Livers in Cirrhotic Mice Using Quantum Dots. ACSNANO. 2010;4(6):3005–3014. doi: 10.1021/nn100589y. [DOI] [PubMed] [Google Scholar]
- 24.Cai W, Zhang X, Wu Y, Chen X. RGD peptide-labeled quantum dots for integrin αvβ3 targeting. In: Blondelle SE, editor. Understanding Biology Using Peptides. American Peptide Society; 2005. pp. 463–464. [Google Scholar]
- 25.Murphy EA, Majeti BK, Barnes LA, Makale M, Weis SM, Lutu-Fuga K, Wrasidlo W, Cheresh DA. Nanoparticle-mediated drug delivery to tumor vasculature suppresses metastasis. Proc Natl Acad Sci U S A. 2008;8(105)(27):9343–8. doi: 10.1073/pnas.0803728105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arap W, Pasqualini R, Ruoslahti E. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science. 1998;16(279)(5349):377–80. doi: 10.1126/science.279.5349.377. [DOI] [PubMed] [Google Scholar]
- 27.Papagiannaros A, Upponi J, Hartner W, Mongayt D, Levchenko T, Torchilin V. Quantum dot loaded immunomicelles for tumor imaging. BMC Medical Imaging. 2010;10:22. doi: 10.1186/1471-2342-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tadrous PJ. Methods for imaging the structure and function of living tissues and cells: 2. Fluorescence lifetime imaging. J Pathol. 2000;191:229–234. doi: 10.1002/1096-9896(200007)191:3<229::AID-PATH623>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 29.Elson D, et al. Time-domain fluorescence lifetime imaging applied to biological tissue. Photochem Photobiol Sci. 2004;3:795–801. doi: 10.1039/b316456j. [DOI] [PubMed] [Google Scholar]
- 30.Hall DJ, Sunar U, Farshchi-Heydari S, Han SH. In vivo simultaneous monitoring of two fluorophores with lifetime contrast using a full-field time domain system. Appl Opt. 2009;48:D74–D78. doi: 10.1364/ao.48.000d74. [DOI] [PubMed] [Google Scholar]
- 31.Liao D, Liu Z, Wrasidlo W, Chen T, Luo Y, Xiang R, Reisfeld RA. Synthetic enzyme inhibitor: a novel targeting ligand for nanotherapeutic drug delivery inhibiting tumor growth without systemic toxicity. Nanomedicine. 2011 Mar 17; doi: 10.1016/j.nano.2011.03.001. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 32.Kim SH, Tan JP, Nederberg F, Fukushima K, Colson J, Yang C, Nelson A, Yang YY, Hedrick JL. Hydrogen bonding-enhanced micelle assemblies for drug delivery. Biomaterials. 2010;31(31):8063–71. doi: 10.1016/j.biomaterials.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 33.May A, Bhaumik S, Gambhir SS, Zhan C, Yazdanfar S. Whole-body, real-time preclinical imaging of quantum dot fluorescence with time-gated detection. J Biomed Opt. 2009;14:060504. doi: 10.1117/1.3269675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Botchway SW, Charnley M, Haycock JW, Parker AW, Rochester DL, Weinstein JA, Williams JA. Time-resolved and two-photon emission imaging microscopy of live cells with inert platinum complexes. Proc Natl Acad Sci U S A. 2008;105:16071–16076. doi: 10.1073/pnas.0804071105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akers WJ, Berezin MY, Lee H, Achilefu S. Predicting in vivo fluorescence lifetime behavior of near-infrared fluorescent contrast agents using in vitro measurements. J Biomed Opt. 2008;13:054042. doi: 10.1117/1.2982535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kan P, Tsao C-W, Wang A-J, Su W-C, Liang H-F. A Liposomal Formulation Able to Incorporate a High Content of Paclitaxel and Exert Promising Anticancer Effect. Journal of Drug Delivery. 2011:1–9. doi: 10.1155/2011/629234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casadei MA, Cerreto F, Cesa S, Giannuzzo M, Feeney M, Marianecci C, Paolicelli P. Solid lipid nanoparticles incorporated dextran hydrogels. A new drug delivery system for oral formulations. Int J Pharmaceutics. 2006;325:140–146. doi: 10.1016/j.ijpharm.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 38.Kim I, Jeong Y, Kim S-ZH. Self-assembled hydrogel nanoparticles compoused of dextran and polyethyleneglycol macromer. Int J Pharmaceutics. 2000;205:109–116. doi: 10.1016/s0378-5173(00)00486-5. [DOI] [PubMed] [Google Scholar]
- 39.De Geest BG, Stubbe BG, Jonas AM, Van Thienen T, Hinrichs WL, Demeester J, De Smedt SC. Self-exploding lipid coated Microgels. Biomacromolecules. 2006;7:373–379. doi: 10.1021/bm0507296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.