Abstract
The mammalian target of rapamycin (mTOR) is an evolutionarily conserved serine/threonine protein kinase that regulates numerous cellular processes including cell growth, proliferation, cell cycle, and autophagy. mTOR forms two different multi-protein complexes referred to as mTOR complex 1 (mTORC1) and mTORC2, and each complex exerts distinct functions exclusively. mTORC1 activity is sensitive to the selective inhibitor rapamycin, whereas mTORC2 is resistant. mTORC1 is regulated by many intra- and extra-cellular cues such as growth factors, nutrients, and energy-sensing signals, while mTORC2 senses ribosome maturation and growth factor signaling. This review focuses on current understandings by which mTORC1 pathway senses cellular nutrient availability for its activation.
Keywords: mTORC1, amino acids, rapamycin, Rag GTPase
Introduction
Mammalian target of rapamycin (mTOR) is an evolutionarily conserved large serine/threonine protein kinase and a member of the phosphoinositide-3-kinase (PI3K) related kinase family [1]. mTOR possesses multiple conserved domains including HEAT (Huntignton, elongation factor-3, a subunit of protein phoshpatase-2A, TOR1) repeats, a FAT (FRAP, ATM, and TRRAP) domain, an FATC domain, a FRB [FKBP12 (FK506-binding protein 12)-rapamycin-binding] domain, and a kinase domain [2].
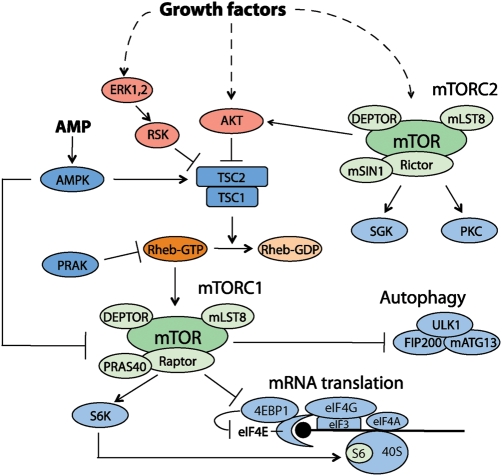
It is well known that mTOR forms two different functional complexes termed as mTOR complex 1 (mTORC1) and mTORC2 [3]. mTORC1 exists as a multi-protein complex containing mTOR, Raptor, mLST8 (GβL), and PRAS40 [4–8], while mTORC2 consists of mTOR, Rictor, mSin1 (MAPKAP1), PRR5, and mLST8 [3,9–15]. Importantly, the configuration of each complex is also conserved across species [3]. Raptor is an essential component for the mTORC1 signaling and functions as a scaffolding protein to recruit mTORC1 substrates such as S6 kinase (S6K) and eIF4E (eukaryotic translation initiation factor 4E) binding proteins (4EBPs) through their TOR signaling (TOS) motifs [16–18]. Interestingly, recent studies have shown that Raptor also plays a significant role for intra-cellular localization of mTORC1 in sensing amino acid availability, an essential cellular cue for mTORC1 activation (summarized below) [19]. PRAS40 (Proline-rich AKT substrate of 40 kDa) is originally identified as a substrate of Akt kinase [20], but recent studies have found that it may function as a negative regulator of mTORC1 [7,8]. It has also been shown that the interaction of PRAS40 with mTOR is disrupted due to phosphorylation by either mTOR or Akt, thereby inducing the ability of mTORC1 to phosphorylate other substrates such as S6K [21]. This module may be one of the mechanisms by which the PI3K–Akt pathway induces mTORC1 activity. The mechanism by which rapamycin suppresses mTORC1 function has been elucidated by the discoveries of these components and their structures. Rapamycin, an antifungal agent originally purified from Streptomyces hygroscopicus, binds to FKBP12 and this drug–protein complex directly binds to the FRB domain of mTOR [22]. Recent studies have revealed that mTORC1 forms a dimer and that this dimerization is important for mTORC1-mediated phosphorylation of 4EBP1 but not S6K [23–25]. The binding of Rapamycin-FKBP12 to the FRB domain of mTOR weakens the mTOR–Raptor interaction and ultimately disrupts the dimeric formation, thereby inhibiting mTORC1-dependent 4EBP1 phosphorylation [25]. It is important to note that Rapamycin–FKBP12 may also block access to the mTOR active site for phosphorylation of larger substrates such as S6K.
mTORC2 activity is known to be rapamycin resistant during short-term treatment [9,10,26]. Rictor is a core protein for mTORC2 catalytic activity, and it appears to recruit substrates to mTORC2 in a manner similar to that of Raptor in mTORC1 [9,27]. mSin1 is also an essential component for mTORC2, and its depletion abolishes mTORC2 activity while also disrupting the interaction between Rictor and mTOR [11–13]. Interestingly, ablation of mLST8, the common component of both mTORC1 and mTORC2, results in disruption of the Rictor–mTOR but not of the Raptor–mTOR interaction [28]. Consistent with these findings, mLST8 is crucial for mTORC2 activity but not for mTORC1 function in vivo [28]. More recently, it has been identified that the DEP domain containing mTOR interacting protein (DEPTOR) is the common component for both mTORC1 and mTORC2 [29]. DEPTOR negatively regulates both mTORC1 and mTORC2 functions, and loss of DEPTOR activates the mTOR kinase activity in both complexes.
Downstream of mTOR Complexes
mTOR regulates multiple cellular processes such as cell growth, cell cycle, cell survival, and autophagy. The most well-characterized substrates for mTORC1 are S6K and 4EBP1 [30–32]. mTORC1 phosphorylates S6K at multiple sites, particularly Thr389 and Ser371, the major mTORC1-targeting phosphorylation sites [33,34]. T389 phosphorylation on S6K1 creates the binding motif for phosphoinositide-dependent kinase-1 (PDK1) and the associated PDK1 phosphorylates T229 in the activation loop of S6K1, a process essential for the activation of S6K [35–37]. S6K plays important roles in the regulation of initiation, mRNA processing, and cell size control. In vivo experiments have shown that S6K depletion in Drosophila results in reductions in body size and decreases in cell size [38]. Similarly, S6K1 knockout mice have a body size that is ∼80% of that of their littermates [39].
4EBPs (4EBP1 and 4EBP2), which are known to function as translational repressors, are also well-known mTORC1 substrates [40]. 4EBPs have an eIF4E binding domain, which is shared by eIF4G, an essential scaffolding protein that forms the eIF4F complex [32]. Hypophosphorylated 4EBPs bind to eIF4E, which in turn recognizes the 5′-end cap of the majority of eukaryotic mRNAs. Phosphorylation of 4EBPs by mTORC1 promotes the dissociation of 4EBPs from eIF4E, thereby relieving the inhibitory effect of 4EBPs on eIF4E-dependent translation initiation [41]. Recent studies have shown that 4EBPs are crucial elements of the mTORC1 pathway that regulate cell number and proliferation [42].
mTORC1 is also an important player in the regulation of autophagy, a catabolic process involving the degradation of the cell's own components through the lysosomal machinery [43,44]. ULK1, a mammalian homolog of Atg1 in yeast, forms a complex with FIP200 and mAtg13 in mammals, and has been shown to have an essential role in autophagosomal formation [45,46]. Recent studies have shown that mTORC1 interacts with the ULK1 complex via Raptor and negatively phosphorylates mAtg13 and ULK1 to suppress ULK1 kinase activity, thereby inhibiting autophagosome formation [47,48]. ULK1 is also phosphorylated by AMP-activated protein kinase (AMPK). Recent studies have shown that AMPK-dependent ULK1 phosphorylation activates its kinase activity and plays a role in the induction of autophagy. These studies demonstrate that ULK1 phosphorylation by AMPK protects mTORC1-mediated ULK1 inhibition [49,50].
Due to its resistance to rapamcyin, knowledge of targets downstream of mTORC2 has lagged behind that of mTORC1. In contrast to the mTORC1 that is regulated by an array of extra and intra-cellular cues, mTORC2 is primarily regulated by growth factor-mediated PI3K signaling and plays important roles in cell survival and actin reorganization [9,12,26]. Recent studies have revealed that S6K-related kinases belonging to the AGC kinase family can be phosphorylated by mTORC2. These kinases include AKT, protein kinase Cα(PKC), and serum/glucocorticoid regulated kinase 1 (SGK1) [9,27,51]. AKT is the most well-characterized substrate of mTORC2 [27], and phosphorylation of the AKT hydrophobic motif (Ser473) as well as the turn motif (Ser450) is abolished in cells lacking an essential mTORC2 component such as Rictor or mSin1, indicative of an essential role of mTORC2 in the regulation of these phosphorylation sites [12,28]. However, mTORC2 catalyzes these phosphorylation sites through different mechanisms [52,53]. Recent studies have demonstrated that the turn motif phosphorylation of AKT is catalyzed by mTORC2 during the translation of newly synthesized AKT polypeptides, whereas the hydrophobic site is phosphorylated after AKT is matured [54]. Interestingly, Zinzalla et al. have demonstrated that PI3K activity induces an association of mTORC2 with the ribosome, which enhances mTORC2 kinase activity for the phosphorylation of both the hydrophobic and turn motifs of AKT [55]. Although it remains elusive as to whether all the substrates of mTORC2 can be catalyzed via ribosome/mTORC2-dependent manner, these observations indicate that mTORC2 functions with ribosomes to facilitate not only the maturation of AKT kinase through the turn motif phosphorylation but also the activation of AKT through the hydrophobic phosphorylation.
Upstream of mTORC1
mTORC1 is regulated by multiple upstream factors including growth factors, nutrients such as glucose and amino acids (Fig. 1) [56]. The molecular mechanisms by which these extra- and intra-cellular signals regulate mTORC1 have been extensively investigated. The most proximal molecule having a key role in the regulation of mTORC1 activity is the small GTPase Rheb. Rheb is a Ras-related GTPase protein originally identified as a gene that is rapidly induced in brain neurons by synaptic activity [57,58]. As is the case of other small GTPases, the GTP-bound form of Rheb is active while the GDP-bound form is inactive. Genetic and biochemical studies have unequivocally demonstrated that Rheb functions upstream of mTORC1 [59–65]. Biochemical studies have demonstrated that Rheb may associate with mTOR, although it remains unclear as to whether endogenous active Rheb preferentially interacts with mTOR in vivo [66,67]. However, importantly, purified active Rheb sufficiently stimulates mTORC1 kinase activity in vitro, suggesting that Rheb is a critical proximal activator of mTORC1 [8,68]. The activity of Rheb is tightly regulated by the tuberous sclerosis gene products, TSC1 (hamartin) and TSC2 (tuberin) [60,61,63]. Mutations of either the TSC1 or TSC2 genes causes tuberous sclerosis complex (TSC), a disease characterized by the formation of benign tumors (hamartomas) in multiple tissues [69,70]. TSC1 and TSC2 form a functional complex in vivo and this complex functions as a tumor suppressor [71]. TSC1 stabilizes TSC2 that possesses a GAP (GTPase-activating protein) domain in its carboxyl terminus. The identification of TSC2 as a negative regulator for the Rheb–mTORC1 pathway significantly advances our understanding as to how growth signals activate mTORC1. It has been shown that multiple growth-related kinases such as AKT, ERK, and RSK phosphorylate TSC2 and inhibit the function of the TSC complex, thereby activating the Rheb–mTORC1 pathway [72–74]. In TSC1 as well as TSC2 deficient cell lines, mTORC1 activity is constitutively active and deprivation of growth factors fails to inhibit mTORC1 activity in these cells [75]. However, it remains unclear as to how the phosphorylation of TSC2 by the above kinases affects TSC2 GAP activity.
Figure 1.
mTOR signaling pathway The scheme depicts mTOR signaling cascade in the regulation of energy and growth factor-dependent translation and autophagy.
Recent studies have also revealed that mTORC1 activity is modulated by intra-cellular energy levels via multiple mechanisms. AMPK, an essential energy sensor, plays a key role in energy-dependent mTORC1 regulation. On nutritional crisis such as glucose deprivation, intra-cellular concentration of AMP is increased and results in the activation of AMPK. AMPK phosphorylates a wide array of substrates that inhibits anabolism but activates catabolism [76]. Protein synthesis is a process consuming large amounts of energy, and this anabolic process becomes an important target of AMPK [77]. Previous reports have demonstrated that AMPK also phosphorylates multiple proteins in the mTORC1 signaling including TSC2 and Raptor [78,79]. AMPK-dependent phosphorylation of these targets leads to suppression of mTORC1 activity, thereby inhibiting major anabolic processes under energy-deficient conditions. More recently, Zheng et al. have demonstrated another mechanism of down-regulation of the mTORC1 pathway in response to glucose deprivation [80]. This study has revealed that the p38β MAPK–PRAK cascade can be activated in cells under glucose starvation conditions. Interestingly, PRAK phosphorylates Rheb, and PRAK-dependent Rheb phosphorylation reduces the nucleotide-binding ability of Rheb. This may lead to destabilization of Rheb, thereby inhibiting the mTORC1 pathway.
Although a body of evidence indicates an essential role of Rheb for the activation of mTORC1, the molecular mechanisms by which Rheb activates mTORC1 have not been satisfactory addressed. The phosphatidic acid (PA) biosynthetic pathway has been postulated as an important element downstream of Rheb to activate mTORC1 pathway. Previous studies have demonstrated that PA directly interacts with mTOR and has a positive role for the mTORC1 pathway, because an inhibition of PA accumulation using 1-butanol or knockdown of PL-D1, an enzyme essential for PA production, diminishes mTORC1 activity in vivo [81,82]. Interestingly, recent study has shown that active Rheb (GTP-bound form) binds and activates PL-D1 in vitro, raising the possibility that Rheb also indirectly activates mTORC1 via PA production [83]. It is intriguing to test the effect of PA on mTORC1 activity in vitro as well as mTOR localization in cells to dissect the role of Rheb-PL-D1-PA axis in the activation of mTORC1 pathway.
Amino Acids Sensing
Amino acids, fundamental nutrients for all cells, are also essential for mTORC1 signaling. In response to amino acid depletion, mTORC1 activity is rapidly abolished [84]. In contrast, exposure to amino acids enhances mTORC1 activity in the presence of growth factors. It is clear that both amino acids and growth factors are two indispensable factors for mTORC1 activity. Among amino acids, leucine and glutamine are two most important for mTORC1 activity [84,85]. Leucine deprivation rapidly inhibits both S6K and 4EBP1 phosphorylation. Although it remains unclear as to how a single amino acid or its metabolite is proximally important for mTORC1 activation, a recent study has revealed an unexpected role of glutamine uptake for leucine-induced mTORC1 activation. Glutamine, another essential amino acid for mTORC1 activation, is incorporated into cells via the SLC1A5 transporter in a Na+-dependent manner. Interestingly, the incorporated intra-cellular glutamine becomes a key substance to trigger leucine uptake via the bidirectional transporter SLC7A5–SLC3A2. These observations indicate that glutamine is an essential source for the uptake of leucine. Consistently, loss of SLC1A5 suppresses mTORC1 activity and cell growth while inducing autophagy [86]. The mechanism by which amino acids such as leucine stimulate mTORC1 activity has been a major point of focus in this pathway.
Recent studies using genetic as well as a biochemical approaches have started to uncover this black box. Using RNAi screening in Drosophila cells, CG7097, a mammalian ortholog of MAP4K3 [also known as GLK (germinal centre-like kinase)] has been identified as an amino acids-sensitive regulator of the mTORC1 [87]. Loss of function of MAP4K3 in mammalian cells causes reduction of mTORC1 activity, while gain of function of the MAP4K3 enhances mTOR1 activity and renders the mTORC1 pathway insensitive to amino acids. Importantly, the study demonstrates that the kinase activity of MAPK4 is regulated by amino acids. Consistent with these biochemical observations in mammalian cells, lack of CG7097 in Drosophila contributes to small body size that is a phenocopy with reduced dTOR function or growth under low-nutrients condition [88,89]. More recent study has shown that PR61 epsilon, a PP2A-targeting subunit, plays an important role in the regulation of MAP3K3 phosphorylation in response to amino acids [90]. Phosphorylation of Ser170 in the activation segment of MAP3K3 is required for MAP4K3 activity and its activation of mTORC1 signaling. Importantly, on amino acid deprivation, PP2A/PR61 interacts with MAP4K3, thereby promoting dephosphorylation of Ser170 and inhibiting mTOR signaling. These studies have demonstrated an important role of MAP4K3 in amino acid-sensitive mTORC1 regulation. However, the mechanism by which MAP4K3 activates the mTORC1 pathway remains unclear.
Class III phosphatidylinositol-3-kinase, Vps34 has also been proposed as a factor implicated in the regulation of amino acids-induced mTORC1 activation. Overexpression of Vps34 increases S6K phosphorylation in the presence of amino acids, while amino acids fails to enhance S6K phosphorylation in Vps34 deficient mammalian cells [91,92]. These observations suggest that Vps34 functions upstream of mTORC1 in amino acid sensing. Intriguingly, neither ablation of dVps34 nor overexpression of the kinase-inactive dVps34 has impact on dTORC1-mediated cell growth in Drosophila fat body [93]. Furthermore, ubiquitous overexpression of wild type or kinase-inactive dVps34 has no effect on the phosphorylation of dS6K in larval extracts. However, loss of dVps34 function does suppress dTOR-Atg1-mediated autophagy induction under starvation condition. These observations indicates that dVps34 functions downstream of dTORC1-dependent nutrient signaling in Drosophila. The discrepancy of above observations could be due to the difference of primary role of PI(3)P generated by Vps34 in cultured mammalian cells and Drosophila organisms, since Vps34 has multiple functions not only in autophagy but also endocytosis and membrane as well as protein sorting/trafficking [93,94]. It is possible that disruption of some of those Vps34 functions in cultured mammalian cells may affect amino acid sensing mechanisms of mTORC1. Nevertheless, future studies using in vivo mammalian system will be necessary to clarify physiological role of Vps34 in amino acids-sensitive mTORC1 regulation.
Rag-Ragulator
Another important development elucidating the molecular mechanism by which amino acids enhance the mTORC1 pathway is the identification of the Rag small GTPase family in the spatial regulation of mTORC1 localization. Two biochemical and genetic studies have complementarily demonstrated that Rag plays a crucial role in amino acid-sensitive mTORC1 regulation [19,95]. The mammalian Rag subfamily of GTPase consists of Rag A, B, C, and D, and belongs to the super-family of Ras-related GTPases [96]. RagA and RagB are homologous to yeast Gtr1p, while the yeast Gtr2p is a homolog of RagC and RagD [97]. As seen in the yeast Gtr1p/Gtr2p complex, mammalian RagA or RagB forms a heterodimer with RagC or RagD and functions as a hetrodimeric complex. A unique feature of this Rag complex is that RagA/B is the GTP form whereas RagC/D is the GDP form in the active complex [98]. It has previously been demonstrated that the Gtr1p–Gtr2p heterodimer forms a complex with EGO1 (GSE2) as well as EGO3 (GSE1) and localizes at endosomes where the complex may exert a crucial role for GAP1 (general amino acid transpoter) translocalization to the plasma membrane in response to amino acid insufficiency [98,99].
Sancak et al. have identified that the Rag heterodimer interacts with Raptor, an essential component of mTORC1 [19]. Importantly, the interaction between Raptor and the Rag heterodimer is dependent on the levels of GTP-bound RagA or RagB in the heterodimer. Furthermore, amino acid deprivation reduces the levels of GTP-charged RagB. More importantly, two studies have demonstrated that knockdown of RagA/B or dominant negative RagA/B expression significantly inhibits the mTORC1 pathway [19,95]. In contrast, constitutively active RagA/B renders mTORC1 resistant to amino acid deprivation. Consistent with these biochemical data in the mammalian system, loss or gain of function of Drosophila Gtr1 (dRagA) causes an increase or a decrease in wing cell size, respectively [95]. Furthermore, constitutively active dRagAQ61L inhibits starvation-induced autophagy in fat body cells, thereby suggesting that dRags (dRagA/dRagC) function upstream of dTORC1. Importantly, the epistatic studies in Drosophila show that dRags functions upstream of dRheb in the regulation of dTORC1 activation [95].
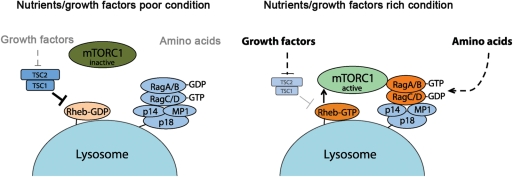
How do Rag GTPases regulate mTORC1 activity in response to amino acids? In in vitro studies, purified active Rag GTPases, unlike the Rheb small GTPase, are not able to stimulate mTORC1 kinase activity, therefore suggesting that Rags may stimulate mTORC1 pathway indirectly in vivo [8,19,68]. Strikingly, immunofluorescence studies demonstrate that amino acid treatment induces co-localization of mTOR with LAMP2 (late endosome or lysosome marker) [19]. Furthermore, Rag small GTPases are constitutively expressed at LAMP2-positive compartments. In addition, mTOR or Raptor localization at the LAMP2-positive compartments is dependent on the levels of GTP-bound form of RagA/B. These data suggest that mTORC1 may translocate to the lysosomal membrane in a manner dependent on Rag, whose activity is regulated by amino acid availability. Consistent with these models, knockdown of RagA/B, or dominant inactive RagB mutants block amino acid-induced mTOR localization at the lysosome. The active Rag heterodimer, however, sufficiently translocates mTOR to the lysosomal membrane with concomitant mTORC1 activation under amino acid starvation conditions. Given the fact that exogenous Rheb can be expressed in the LAMP2-positive compartment, these observations together indicate that the active Rag heterodimer allows mTORC1 to translocate on the lysosome or late endosome where mTORC1 interacts with Rheb, thereby enhancing mTORC1 activity in response to amino acids. These spatial and temporal regulations of mTORC1 by Rag and Rheb logically explain how the signals from amino acids and growth factors are integrated and activate the mTORC1 pathway [19] (Fig. 2).
Figure 2.
Amino acids sensing of mTORC1 Rag-Ragulator-dependent mTORC1 activation in response to amino acid. On amino acids stimulation, Rag anchored by Ragulators (MP1/p14/p18) on the lysosome membrane can be activated by an unknown mechanism, and the active Rag complex subsequently recruits mTORC1. On the lysosomal membrane, the mTORC1 is activated by Rheb, whose activity is enhanced by growth factor-mediated suppression of TSC complex. This model nicely explains how amino acids and growth factor signals are coordinated to produce maximal activation of mTORC1 at the lysosome by active Rag and Rheb small GTPases. The dotted or solid line indicates indirect or direct regulation, respectively.
Lack of lipid modification motifs such as those for farnesylation or myristoylation represents a unique feature of the Rag small GTPases within the Ras-related GTPase family [96]. However, studies suggest that Rags constitutively express and function on the lysosomal membrane in the regulation of mTORC1 [19]. Using biochemical approaches, Sancak et al. have identified that mammalian Rag forms a complex with the MAPK scaffolding protein 1 (MP1), p14, and p18 encoded by MAKSP1, ROBLD3, and c11orf59, respectively [100]. This is similar to the EGO complex in Saccharomyces cerevisiae, which acts as a binding protein of Rag GTPases [99]. Previous studies have reported that MP1, p14, and p18 form a functional hetero-trimer [101]. These three proteins are conserved from Drosophila to mammals [100]. Interestingly, it has previously been demonstrated that the MP1/p14/p18 complex localizes endosomal and lysosomal membrane via the myristoylation and the palmitoylation of p18 [101]. p18 acts as a scaffolding protein in the complex, and for the endosomal MEK1–ERK signaling. p18 knockout mice show an embryonic lethal phenotype and display severe defects in endosome/lysosome organization. Lack of one of the members of the MP1/p14/p18 complex displays similar phenotypes to those in Rag deficient cells. For instance, amino acid treatment fails to stimulate mTORC1 translocalization as well as activity. Importantly, lack of the functional MP1/p14/p18 complex causes a mis-localization of Rag GTPases [100]. These studies suggest that the MP1/p14/p18 complex functions as an anchor for Rag GTPases to recruit mTORC1 at the lysosomal membrane for its activity. Given the fact that both the MP1/p14/p18 complex and the EGO complex play an important role for mTORC1 and TORC1 function, respectively [102], they should be evolutionarily conserved functional complexes in the nutrient-sensitive TORC1 regulation. Consistently, recent studies have demonstrated structural similarities between p18 and the EGO1. Furthermore, structural analyses have revealed a significant similarity between the fold of the MP1/p14 hetrodimeric complex and EGO3 [103]. Thus, the RagA/B–RagC/D–MP1/p14/p18 complex may be a structurally as well as functionally conserved homolog of the yeast Gtr1p–Gtr2p–EGO3/EGO1 complex.
Considering an important role of organelles such as late endosome/lysosome as essential working places in amino acids-sensitive mTORC1 activation, it is conceivable that many other functional proteins in the late endosome/lysosome as well as the proteins that have a role in membrane dynamics may directly or indirectly implicate in the regulation of amino acids-sensitive mTORC1 pathway. Consistently, recent studies have demonstrated that other small GTPases such as RalA, Rab5, Rab7, and Arf1 that regulate protein trafficking and membrane dynamics involves in the regulation of mTORC1 activation [68,104–106].
Concluding Remarks
With recent important findings, current understandings of the molecular mechanisms underlying nutrient-sensitive mTORC1 regulation have significantly advanced. Spatial and temporal activation of mTORC1 by two distinct small GTPases have highlighted the delicate regulation of the mTORC1 pathway. However, many questions remain unanswered. For instance, given the new players such as the Rag small GTPase hetrodimeric complex in the amino acid-sensitive mTORC1 regulation, how do amino acids regulate RagA/B as well as RagC/D activities (GTP–GDP ratio)? There should be guanine exchange factors (GEF) and GAPs for Rag GTPases, which might be regulated by amino acids; however, they have not been identified yet in mammals. It is conceivable that multiple regulators may be involved in the regulatory processes in which GDP is replaced by GTP in RagA/B, while GTP is exchanged by GDP in RagC/D on amino acid stimulation. Recent studies have shown that Vam6 functions as a GEF for Gtr1p and promotes GTP-loading of Gtr1p, which results in activation of TORC1 in yeast. Vps39, however, a mammalian homologue of yeast Vam6, has little effect on the amino acid-induced mTOR translocalization to the lysosomal membrane, although ablation of Vps39 in mammalian cells does show inhibitory effects on mTORC1 activity [102, 105]. Another question is where the accumulated mTORC1 on the lysosome or late endosome membrane under nutrient rich condition quickly disappears in response to amino acid starvation. Is there any special mechanism to transfer active mTORC1 to another organelle without using consumable energy? Furthermore, where exactly is endogenous Rheb expressed, and do any additional Rheb GEFs exist? Although it seems that MAP4K3 functions upstream of the Rag system in amino acid-sensitive mTORC1 regulation, what is the precise relationship between the Rag system and MAP4K3? Although there are still many questions in this pathway, we anticipate that these answers will be uncovered soon.
Funding
This work was supported by the grant from the National Institutes of Health (DK083491).
Acknowledgements
We thank Aristotle Mannan for discussion.
References
- 1.Abraham RT. PI3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair (Amst) 2004;3:883–887. doi: 10.1016/j.dnarep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Cafferkey R, McLaughlin MM, Young PR, Johnson RK, Livi GP. Yeast TOR (DRR) proteins: amino-acid sequence alignment and identification of structural motifs. Gene. 1994;141:133–136. doi: 10.1016/0378-1119(94)90141-4. [DOI] [PubMed] [Google Scholar]
- 3.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 4.Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 5.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 6.Kim DH, Sarbassov dos D, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, Tempst P, et al. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- 7.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 8.Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 10.Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 11.Yang Q, Inoki K, Ikenoue T, Guan KL. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 2006;20:2820–2832. doi: 10.1101/gad.1461206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 13.Frias MA, Thoreen CC, Jaffe JD, Schroder W, Sculley T, Carr SA, Sabatini DM. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr Biol. 2006;16:1865–1870. doi: 10.1016/j.cub.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Thedieck K, Polak P, Kim ML, Molle KD, Cohen A, Jeno P, Arrieumerlou C, et al. PRAS40 and PRR5-like protein are new mTOR interactors that regulate apoptosis. PLoS One. 2007;2:e1217. doi: 10.1371/journal.pone.0001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woo SY, Kim DH, Jun CB, Kim YM, Haar EV, Lee SI, Hegg JW, et al. PRR5, a novel component of mTOR complex 2, regulates platelet-derived growth factor receptor beta expression and signaling. J Biol Chem. 2007;282:25604–25612. doi: 10.1074/jbc.M704343200. [DOI] [PubMed] [Google Scholar]
- 16.Schalm SS, Blenis J. Identification of a Conserved Motif Required for mTOR Signaling. Curr Biol. 2002;12:632–639. doi: 10.1016/s0960-9822(02)00762-5. [DOI] [PubMed] [Google Scholar]
- 17.Schalm SS, Fingar DC, Sabatini DM, Blenis J. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr Biol. 2003;13:797–806. doi: 10.1016/s0960-9822(03)00329-4. [DOI] [PubMed] [Google Scholar]
- 18.Nojima H, Tokunaga C, Eguchi S, Oshiro N, Hidayat S, Yoshino K, Hara K, et al. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J Biol Chem. 2003;278:15461–15464. doi: 10.1074/jbc.C200665200. [DOI] [PubMed] [Google Scholar]
- 19.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovacina KS, Park GY, Bae SS, Guzzetta AW, Schaefer E, Birnbaum MJ, Roth RA. Identification of a proline-rich Akt substrate as a 14-3-3 binding partner. J Biol Chem. 2003;278:10189–10194. doi: 10.1074/jbc.M210837200. [DOI] [PubMed] [Google Scholar]
- 21.Oshiro N, Takahashi R, Yoshino K, Tanimura K, Nakashima A, Eguchi S, Miyamoto T, et al. The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J Biol Chem. 2007;282:20329–20339. doi: 10.1074/jbc.M702636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi J, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 23.Takahara T, Hara K, Yonezawa K, Sorimachi H, Maeda T. Nutrient-dependent multimerization of the mammalian target of rapamycin through the N-terminal HEAT repeat region. J Biol Chem. 2006;281:28605–28614. doi: 10.1074/jbc.M606087200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Billington CJ, Jr, Pan D, Neufeld TP. Drosophila target of rapamycin kinase functions as a multimer. Genetics. 2006;172:355–362. doi: 10.1534/genetics.105.051979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yip CK, Murata K, Walz T, Sabatini DM, Kang SA. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol Cell. 2010;38:768–774. doi: 10.1016/j.molcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarbassov dos D, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 27.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 28.Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 29.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Avruch J, Belham C, Weng Q, Hara K, Yonezawa K. The p70 S6 kinase integrates nutrient and growth signals to control translational capacity. Prog Mol Subcell Biol. 2001;26:115–154. doi: 10.1007/978-3-642-56688-2_5. [DOI] [PubMed] [Google Scholar]
- 31.Thomas G. The S6 kinase signaling pathway in the control of development and growth. Biol Res. 2002;35:305–313. doi: 10.4067/s0716-97602002000200022. [DOI] [PubMed] [Google Scholar]
- 32.Gingras AC, Kennedy SG, O'Leary MA, Sonenberg N, Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev. 1998;12:502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearson RB, Dennis PB, Han JW, Williamson NA, Kozma SC, Wettenhall RE, Thomas G. The principal target of rapamycin-induced p70s6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. EMBO J. 1995;14:5279–5287. doi: 10.1002/j.1460-2075.1995.tb00212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saitoh M, Pullen N, Brennan P, Cantrell D, Dennis PB, Thomas G. Regulation of an activated S6 kinase 1 variant reveals a novel mammalian target of rapamycin phosphorylation site. J Biol Chem. 2002;277:20104–20112. doi: 10.1074/jbc.M201745200. [DOI] [PubMed] [Google Scholar]
- 35.Pullen N, Dennis PB, Andjelkovic M, Dufner A, Kozma SC, Hemmings BA, Thomas G. Phosphorylation and activation of p70s6k by PDK1. Science. 1998;279:707–710. doi: 10.1126/science.279.5351.707. [DOI] [PubMed] [Google Scholar]
- 36.Williams MR, Arthur JS, Balendran A, van der Kaay J, Poli V, Cohen P, Alessi DR. The role of 3-phosphoinositide-dependent protein kinase 1 in activating AGC kinases defined in embryonic stem cells. Curr Biol. 2000;10:439–448. doi: 10.1016/s0960-9822(00)00441-3. [DOI] [PubMed] [Google Scholar]
- 37.Biondi RM, Kieloch A, Currie RA, Deak M, Alessi DR. The PIF-binding pocket in PDK1 is essential for activation of S6K and SGK, but not PKB. EMBO J. 2001;20:4380–4390. doi: 10.1093/emboj/20.16.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G. Drosophila S6 kinase: a regulator of cell size. Science. 1999;285:2126–2129. doi: 10.1126/science.285.5436.2126. [DOI] [PubMed] [Google Scholar]
- 39.Shima H, Pende M, Chen Y, Fumagalli S, Thomas G, Kozma SC. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 1998;17:6649–6659. doi: 10.1093/emboj/17.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, Lawrence JC, Jr., et al. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 41.Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 42.Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 44.Shigemitsu K, Tsujishita Y, Hara K, Nanahoshi M, Avruch J, Yonezawa K. Regulation of translational effectors by amino acid and mammalian target of rapamycin signaling pathways. Possible involvement of autophagy in cultured hepatoma cells. J Biol Chem. 1999;274:1058–1065. doi: 10.1074/jbc.274.2.1058. [DOI] [PubMed] [Google Scholar]
- 45.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, et al. ULK-Atg13–FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, et al. Nutrient-dependent mTORC1 association with the ULK1–Atg13–FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia-Martinez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) Biochem J. 2008;416:375–385. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- 52.Facchinetti V, Ouyang W, Wei H, Soto N, Lazorchak A, Gould C, Lowry C, et al. The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 2008;27:1932–1943. doi: 10.1038/emboj.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ikenoue T, Inoki K, Yang Q, Zhou X, Guan KL. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J. 2008;27:1919–1931. doi: 10.1038/emboj.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oh WJ, Wu CC, Kim SJ, Facchinetti V, Julien LA, Finlan M, Roux PP, et al. mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. EMBO J. 2010;29:3939–3951. doi: 10.1038/emboj.2010.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 2011;144:757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 56.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 57.Yamagata K, Sanders LK, Kaufmann WE, Yee W, Barnes CA, Nathans D, Worley PF. Rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel Ras-related protein. J Biol Chem. 1994;269:16333–16339. [PubMed] [Google Scholar]
- 58.Aspuria PJ, Tamanoi F. The Rheb family of GTP-binding proteins. Cell Signal. 2004;16:1105–1112. doi: 10.1016/j.cellsig.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 59.Gao X, Zhang Y, Arrazola P, Hino O, Kobayashi T, Yeung RS, Ru B, et al. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat Cell Biol. 2002;4:699–704. doi: 10.1038/ncb847. [DOI] [PubMed] [Google Scholar]
- 60.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, et al. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- 62.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein ocmplex toward Rheb. Curr Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol. 2003;5:578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- 64.Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol. 2003;5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- 65.Stocker H, Radimerski T, Schindelholz B, Wittwer F, Belawat P, Daram P, Breuer S, et al. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- 66.Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr Biol. 2005;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 67.Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 2005;280:18717–18727. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- 68.Sato T, Nakashima A, Guo L, Tamanoi F. Specific activation of mTORC1 by Rheb G-protein in vitro involves enhanced recruitment of its substrate protein. J Biol Chem. 2009;284:12783–12791. doi: 10.1074/jbc.M809207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kwiatkowski DJ. Tuberous sclerosis: from tubers to mTOR. Ann Hum Genet. 2003;67:87–96. doi: 10.1046/j.1469-1809.2003.00012.x. [DOI] [PubMed] [Google Scholar]
- 70.Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. N Engl J Med. 2006;355:1345–1356. doi: 10.1056/NEJMra055323. [DOI] [PubMed] [Google Scholar]
- 71.van Slegtenhorst M, Nellist M, Nagelkerken B, Cheadle J, Snell R, van den Ouweland A, Reuser A, et al. Interaction between hamartin and tuberin, the TSC1 and TSC2 gene products. Hum Mol Genet. 1998;7:1053–1057. doi: 10.1093/hmg/7.6.1053. [DOI] [PubMed] [Google Scholar]
- 72.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 73.Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 74.Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, Blenis J. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc Natl Acad Sci USA. 2002;99:13571–13576. doi: 10.1073/pnas.202476899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kwiatkowski DJ, Zhang H, Bandura JL, Heiberger KM, Glogauer M, el-Hashemite N, Onda H. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum Mol Genet. 2002;11:525–534. doi: 10.1093/hmg/11.5.525. [DOI] [PubMed] [Google Scholar]
- 76.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 77.Buttgereit F, Brand MD. A hierarchy of ATP-consuming processes in mammalian cells. Biochem J. 1995;312(Pt 1):163–167. doi: 10.1042/bj3120163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 79.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zheng M, Wang YH, Wu XN, Wu SQ, Lu BJ, Dong MQ, Zhang H, et al. Inactivation of Rheb by PRAK-mediated phosphorylation is essential for energy-depletion-induced suppression of mTORC1. Nat Cell Biol. 2011;13:263–272. doi: 10.1038/ncb2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- 82.Fang Y, Park IH, Wu AL, Du G, Huang P, Frohman MA, Walker SJ, et al. PLD1 regulates mTOR signaling and mediates Cdc42 activation of S6K1. Curr Biol. 2003;13:2037–2044. doi: 10.1016/j.cub.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 83.Sun Y, Fang Y, Yoon MS, Zhang C, Roccio M, Zwartkruis FJ, Armstrong M, et al. Phospholipase D1 is an effector of Rheb in the mTOR pathway. Proc Natl Acad Sci USA. 2008;105:8286–8291. doi: 10.1073/pnas.0712268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 85.Nakajo T, Yamatsuji T, Ban H, Shigemitsu K, Haisa M, Motoki T, Noma K, et al. Glutamine is a key regulator for amino acid-controlled cell growth through the mTOR signaling pathway in rat intestinal epithelial cells. Biochem Biophys Res Commun. 2005;326:174–180. doi: 10.1016/j.bbrc.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 86.Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Findlay GM, Yan L, Procter J, Mieulet V, Lamb RF. A MAP4 kinase related to Ste20 is a nutrient-sensitive regulator of mTOR signalling. Biochem J. 2007;403:13–20. doi: 10.1042/BJ20061881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bryk B, Hahn K, Cohen SM, Teleman AA. MAP4K3 regulates body size and metabolism in Drosophila. Dev Biol. 2010;344:150–157. doi: 10.1016/j.ydbio.2010.04.027. [DOI] [PubMed] [Google Scholar]
- 89.Resnik-Docampo M, de Celis JF. MAP4K3 is a component of the TORC1 signalling complex that modulates cell growth and viability in Drosophila melanogaster. PLoS One. 2011;6:e14528. doi: 10.1371/journal.pone.0014528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yan L, Mieulet V, Burgess D, Findlay GM, Sully K, Procter J, Goris J, et al. PP2A T61 epsilon is an inhibitor of MAP4K3 in nutrient signaling to mTOR. Mol Cell. 2010;37:633–642. doi: 10.1016/j.molcel.2010.01.031. [DOI] [PubMed] [Google Scholar]
- 91.Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci USA. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem. 2005;280:33076–33082. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- 93.Juhasz G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM, Neufeld TP. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol. 2008;181:655–666. doi: 10.1083/jcb.200712051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 95.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J Biol Chem. 2001;276:7246–7257. doi: 10.1074/jbc.M004389200. [DOI] [PubMed] [Google Scholar]
- 97.Hirose E, Nakashima N, Sekiguchi T, Nishimoto T. RagA is a functional homologue of S. cerevisiae Gtr1p involved in the Ran/Gsp1-GTPase pathway. J Cell Sci. 1998;111:11–21. doi: 10.1242/jcs.111.1.11. [DOI] [PubMed] [Google Scholar]
- 98.Gao M, Kaiser CA. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat Cell Biol. 2006;8:657–667. doi: 10.1038/ncb1419. [DOI] [PubMed] [Google Scholar]
- 99.Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell. 2005;19:15–26. doi: 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 100.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nada S, Hondo A, Kasai A, Koike M, Saito K, Uchiyama Y, Okada M. The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK–ERK pathway to late endosomes. EMBO J. 2009;28:477–489. doi: 10.1038/emboj.2008.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Binda M, Peli-Gulli MP, Bonfils G, Panchaud N, Urban J, Sturgill TW, Loewith R, et al. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell. 2009;35:563–573. doi: 10.1016/j.molcel.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 103.Kogan K, Spear ED, Kaiser CA, Fass D. Structural conservation of components in the amino acid sensing branch of the TOR pathway in yeast and mammals. J Mol Biol. 2010;402:388–398. doi: 10.1016/j.jmb.2010.07.034. [DOI] [PubMed] [Google Scholar]
- 104.Maehama T, Tanaka M, Nishina H, Murakami M, Kanaho Y, Hanada K. RalA functions as an indispensable signal mediator for the nutrient-sensing system. J Biol Chem. 2008;283:35053–35059. doi: 10.1074/jbc.M805822200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Flinn RJ, Yan Y, Goswami S, Parker PJ, Backer JM. The late endosome is essential for mTORC1 signaling. Mol Biol Cell. 2010;21:833–841. doi: 10.1091/mbc.E09-09-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li L, Kim E, Yuan H, Inoki K, Goraksha-Hicks P, Schiesher RL, Neufeld TP, et al. Regulation of mTORC1 by the Rab and Arf GTPases. J Biol Chem. 2010;285:19705–19709. doi: 10.1074/jbc.C110.102483. [DOI] [PMC free article] [PubMed] [Google Scholar]