Abstract
We investigated the identity and genetic diversity of more than 100 isolates belonging to Phyllosticta (teleomorph Guignardia), with particular emphasis on Phyllosticta citricarpa and Guignardia mangiferae s.l. occurring on Citrus. Phyllosticta citricarpa is the causal agent of Citrus Black Spot and is subject to phytosanitary legislation in the EU. This species is frequently confused with a taxon generally referred to as G. mangiferae, the presumed teleomorph of P. capitalensis, which is a non-pathogenic endophyte, commonly isolated from citrus leaves and fruits and a wide range of other hosts. DNA sequence analysis of the nrDNA internal transcribed spacer region (ITS1, 5.8S nrDNA, ITS2) and partial translation elongation factor 1-alpha (TEF1), actin and glyceraldehyde-3-phosphate dehydrogenase (GPDH) genes resolved nine clades correlating to seven known, and two apparently undescribed species. Phyllosticta citribraziliensis is newly described as an endophytic species occurring on Citrus in Brazil. An epitype is designated for P. citricarpa from material newly collected in Australia, which is distinct from P. citriasiana, presently only known on C. maxima from Asia. Phyllosticta bifrenariae is newly described for a species causing leaf and bulb spots on Bifrenaria harrisoniae (Orchidaceae) in Brazil. It is morphologically distinct from P. capitalensis, which was originally described from Stanhopea (Orchidaceae) in Brazil; an epitype is designated here. Guignardia mangiferae, which was originally described from Mangifera indica (Anacardiaceae) in India, is distinguished from the non-pathogenic endophyte, P. brazilianiae sp. nov., which is common on M. indica in Brazil. Furthermore, a combined phylogenetic tree revealed the P. capitalensis s.l. clade to be genetically distinct from the reference isolate of G. mangiferae. Several names are available for this clade, the oldest being P. capitalensis. These results suggest that endophytic, non-pathogenic isolates occurring on a wide host range would be more correctly referred to as P. capitalensis. However, more genes need to be analysed to fully resolve the morphological variation still observed within this clade.
Keywords: Guignardia endophyllicola, Guignardia mangiferae, Phyllosticta bifrenariae, Phyllosticta brazilianiae, Phyllosticta capitalensis, Phyllosticta citriasiana, Phyllosticta citribraziliensis, Phyllosticta citricarpa, taxonomy
INTRODUCTION
Phyllosticta species have often been reported as endophytes, plant pathogens or saprobes (Baayen et al. 2002, Glienke-Blanco et al. 2002, Okane et al. 2003, Silva et al. 2008, Huang et al. 2009, Wulandari et al. 2009). Many Phyllosticta species cause leaf blotch, leaf blight and black spots on fruits of various plants (Glienke-Blanco et al. 2002, Silva & Pereira 2007). Species of Phyllosticta s.str. represent anamorphs of Guignardia (Botryosphaeriaceae) (van der Aa & Vanev 2002, Crous et al. 2006, Schoch et al. 2009). Few studies have to date, however, elucidated the phylogenetic relationships among Phyllosticta species and their Guignardia teleomorphs. The generic concept of Phyllosticta was refined by van der Aa & Vanev (2002) who relocated 2 733 taxa to other coelomycetous genera. However, species concepts within Phyllosticta remain problematic.
Phyllosticta capitalensis was originally described on Stanhopea (Orchidaceae) from Brazil by Hennings (1908). Okane et al. (2001) reported an endophytic Phyllosticta in ericaceous plants from Japan, to which they attributed the name Phyllosticta capitalensis, describing the teleomorph as a new species, G. endophyllicola. Based on DNA sequence data of the ITS gene, Baayen et al. (2002) concluded that there was a common endophytic species associated with a wide host range of plants, which was similar to G. endophyllicola in morphology. Although several names were available for this species, they attributed the species to G. mangiferae (pathogenic on Mangifera indica (Anacardiaceae) in India), while the anamorph was referred to as P. capitalensis. Although no clear argument was presented for choosing the name G. mangiferae for this fungus, the choice of the anamorph name was based on the fact that two isolates from Orchidaceae (CBS 398.80, CBS 226.77) clustered in this clade. Uncertainty remains, therefore, as to which name applies to this species.
To determine the identity of the Phyllosticta species associated with several hosts including Citrus, Mangifera indica and the Orchidaceae, and to study the phylogenetic relationships among them, fungal isolates were subjected to DNA sequence analysis of the rDNA internal transcribed spacer (ITS1, 5.8S, ITS2) region, and partial translation elongation factor 1-alpha (TEF1), actin (ACT) and glyceraldehyde-3-phosphate dehydrogenase (GPDH) genes.
MATERIAL AND METHODS
Isolates
A total of 109 Phyllosticta / Guignardia isolates were investigated in the present study (Table 1). Single monosporic isolates were obtained from each culture prior to DNA sequence analysis. Isolates were obtained from several sources including the CBS Fungal Biodiversity Centre (CBS-KNAW), Utrecht, The Netherlands, the working collection of Pedro Crous housed at CBS (CPC), the LabGeM/UFPR collection, Curitiba, Brazil, the Dutch Quarantine Service (PD), and the Department of Primary Industries (BRIP), Brisbane, Australia. Two isolates (VIC30428 and VIC30556) were obtained from UFG collection, Viçosa, Brazil, and two isolates from the UNESP collection, Jaboticabal, Brazil (G22, Guig1). One strain of G. mangiferae was obtained from CABI Bioscience, UK (IMI 260576).
Table 1.
Guignardia and Phyllosticta isolates investigated in this study.
| Species | Strain no. 1 | Substrate | Country 2 | Collector(s) | GenBank Accession number |
|||
|---|---|---|---|---|---|---|---|---|
| ITS | TEF1 | ACT | GPDH 3 | |||||
| Guignardia mangiferae | IMI 260576 | Mangifera indica (Anacardiaceae), leaf endophyte | India | M.V. Leksshmi | JF261459 | JF261501 | JF343641 | JF343748 |
| Phyllosticta bifrenariae | VIC30556; CBS 128855 | Bifrenaria harrisoniae (Orchidaceae), living leaves | Brazil: MG | O. Pereira | JF343565 | JF343586 | JF343649 | JF343744 |
| Phyllosticta brazilianiae | LGMF330 | Mangifera indica (Anacardiaceae), leaf endophyte | Brazil: SP | C. Glienke | JF343572 | JF343593 | JF343656 | JF343758 |
| LGMF333 | Mangifera indica (Anacardiaceae), leaf endophyte | Brazil: SP | C. Glienke | JF343574 | JF343595 | JF343658 | JF343760 | |
| LGMF334 | Mangifera indica (Anacardiaceae), leaf endophyte | Brazil: SP | C. Glienke | JF343566 | JF343587 | JF343650 | JF343752 | |
| LGMF335 | Mangifera indica (Anacardiaceae), leaf endophyte | Brazil: SP | C. Glienke | JF343577 | JF343598 | JF343661 | JF343763 | |
| LGMF338 | Mangifera indica (Anacardiaceae), leaf endophyte | Brazil: SP | C. Glienke | JF343569 | JF343590 | JF343653 | JF343755 | |
| LGMF341 | Mangifera indica (Anacardiaceae), leaf endophyte | Brazil: SP | C. Glienke | JF343575 | JF343596 | JF343659 | JF343761 | |
| LGMF342 | Mangifera indica (Anacardiaceae), leaf endophyte | Brazil: SP | C. Glienke | JF343576 | JF343597 | JF343660 | JF343762 | |
| LGMF343 | Mangifera indica (Anacardiaceae), leaf endophyte | Brazil: SP | C. Glienke | JF343571 | JF343592 | JF343655 | JF343757 | |
| LGMF347 | Mangifera indica (Anacardiaceae), leaf endophyte | Brazil: SP | C. Glienke | JF343567 | JF343588 | JF343651 | JF343753 | |
| LGMF350 | Mangifera indica (Anacardiaceae), leaf endophyte | Brazil: SP | C. Glienke | JF343573 | JF343594 | JF343657 | JF343759 | |
| LGMF357 | Mangifera indica (Anacardiaceae), leaf endophyte | Brazil: PR | C. Glienke | JF343570 | JF343591 | JF343654 | JF343756 | |
| LGMF372 | Mangifera indica (Anacardiaceae), leaf endophyte | Brazil: PR | C. Glienke | JF343568 | JF343589 | JF343652 | JF343754 | |
| Phyllosticta capitalensis | 16 | Citrus paradisii (Rutaceae), fruit | Florida | – | JF261456 | JF261498 | JF343638 | JF343745 |
| 90 | Smilax kraussiana (Smilacaceae), leaf | South Africa | G.C. Carroll | JF261457 | JF261499 | JF343639 | JF343746 | |
| 106 | Encephalartos ferox (Zamiaceae), healthy leaves | South Africa | G.C. Carroll | JF261458 | JF261500 | JF343640 | JF343747 | |
| CBS 100175 | Citrus sp. (Rutaceae), healthy leaves | Brazil: SP | C. Glienke | FJ538320 | FJ538378 | FJ538436 | JF343699 | |
| CBS 100176 | Citrus sp. (Rutaceae), healthy leaves | Brazil: SP | C. Glienke | FJ538321 | FJ538379 | FJ538437 | JF343704 | |
| CBS 100250 | Psidium guajava (Myrtaceae), fruits | Brazil | C. Glienke | FJ538351 | FJ538409 | FJ538467 | JF343710 | |
| CBS 101228 | Nephelium lappaceum (Sapindaceae), discoloured spinters | USA: Hawaii | K.A. Nishijima | FJ538319 | FJ538377 | FJ538435 | JF343697 | |
| CBS 111638 | Capsicum sp. (Solanaceae), fruit | Dominican Republic | G.C. Carroll | FJ538345 | FJ538403 | FJ538461 | JF343709 | |
| CBS 114751 | Vaccinium sp. (Ericaceae), leaf | New Zealand | T. Fluher | FJ538349 | FJ538407 | FJ538465 | JF343722 | |
| CBS 115046 | Myracrodruon urundeuva (Anacardiaceae), leaf or bark | Brazil | K.F. Rodrigues | FJ538322 | FJ538380 | FJ538438 | JF343711 | |
| CBS 115047 | Aspidosperma polyneuron (Apocynaceae), leaf or bark | Brazil | K.F. Rodrigues | FJ538323 | FJ538381 | FJ538439 | JF343705 | |
| CBS 115049 | Bowdichia nitida (Fabaceae), leaf or bark | Brazil | K.F. Rodrigues | FJ538324 | FJ538382 | FJ538440 | JF343706 | |
| CBS 115051 | Spondias mombin (Anacardiaceae), leaf or bark | Brazil | K.F. Rodrigues | FJ538325 | FJ538383 | FJ538441 | JF343715 | |
| CBS 115052 | Spondias mombin (Anacardiaceae), leaf or bark | Brazil | K.F. Rodrigues | FJ538326 | FJ538384 | FJ538442 | JF343712 | |
| CBS 115053 | Myracrodruon urundeuva (Anacardiaceae), leaf or bark | Brazil | K.F. Rodrigues | FJ538327 | FJ538385 | FJ538443 | JF343717 | |
| CBS 115056 | Anacardium giganteum (Anacardiaceae), leaf or bark | Brazil | K.F. Rodrigues | FJ538328 | FJ538386 | FJ538444 | JF343720 | |
| CBS 115057 | Anacardium giganteum (Anacardiaceae), leaf or bark | Brazil | K.F. Rodrigues | FJ538329 | FJ538387 | FJ538445 | JF343716 | |
| CBS 115313 | Myracrodruon urundeuva (Anacardiaceae), leaf or bark | Brazil | K.F. Rodrigues | FJ538330 | FJ538388 | FJ538446 | JF343713 | |
| CBS 115345 | Bowdichia nitida (Fabaceae), leaf or bark | Brazil | K.F. Rodrigues | FJ538331 | FJ538389 | FJ538447 | JF343707 | |
| CBS 117118 | Musa acuminata (Musaceae) | Indonesia | I. Buddenhagen | FJ538339 | FJ538397 | FJ538455 | JF343723 | |
| CBS 119720 | Musa sp. (Musaceae) | USA: Hawaii | I. Buddenhagen | FJ538340 | FJ538398 | FJ538456 | JF343708 | |
| CBS 123373 | Musa paradisiaca (Musaceae) | Thailand | N.F. Wulandari | FJ538341 | FJ538399 | FJ538457 | JF343703 | |
| CBS 123374 | Citrus aurantium (Rutaceae) | Thailand | N.F. Wulandari | FJ538332 | FJ538390 | FJ538448 | JF343702 | |
| CBS 123404 | Musa paradisiaca (Musaceae) | Thailand | N.F. Wulandari | FJ538333 | FJ538391 | FJ538449 | JF343701 | |
| CBS 123405 | Musa acuminata (Musaceae) | Thailand | N.F. Wulandari | FJ538334 | FJ538392 | FJ538450 | JF343726 | |
| CBS 173.77 | Citrus aurantiifolia (Rutaceae) | New Zealand | – | FJ538335 | FJ538393 | FJ538451 | JF343725 | |
| CBS 226.77 | Paphiopedilum callosum (Orchidaceae), leaf spot | Germany | – | FJ538336 | FJ538394 | FJ538452 | JF343718 | |
| CBS 356.52; ATCC 11368 | Ilex sp. (Aquifoliaceae) | – | – | FJ538342 | FJ538400 | FJ538458 | JF343721 | |
| CBS 373.54 | Ilex sp. (Aquifoliaceae) | – | – | FJ538343 | FJ538401 | FJ538459 | JF343698 | |
| CMU131 | Magnolia liliifera (Magnoliaceae), leaf endophyte | Thailand | L.M. Duong | FJ538346 | FJ538404 | FJ538462 | JF343724 | |
| CMU139 | Magnolia liliifera (Magnoliaceae), leaf endophyte | Thailand | L.M. Duong | FJ538347 | FJ538405 | FJ538463 | JF343714 | |
| CMU142 | Magnolia liliifera (Magnoliaceae), leaf endophyte | Thailand | L.M. Duong | FJ538348 | FJ538406 | FJ538464 | JF343719 | |
| CPC 18845 | Stanhopea graveolens (Orchidaceae) | Brazil | O.L. Pereira | JF261463 | JF261505 | JF343645 | JF343774 | |
| CPC 18847 | Stanhopea graveolens (Orchidaceae) | Brazil | O.L. Pereira | JF261464 | JF261506 | JF343646 | JF343775 | |
| CPC 18848; CBS 128856 | Stanhopea graveolens (Orchidaceae) | Brazil | O.L. Pereira | JF261465 | JF261507 | JF343647 | JF343776 | |
| CPC 18849 | Stanhopea graveolens (Orchidaceae) | Brazil | O.L. Pereira | JF261466 | JF261508 | JF343648 | JF343777 | |
| G22 | Mangifera indica (Anacardiaceae), leaf endophyte | Brazil: SP | A. de Goes | JF261437 | JF261479 | JF343619 | JF343700 | |
| LGMF02 | Citrus latifolia (Rutaceae), healthy leaves | Brazil: SP | A. de Goes | JF261452 | JF261494 | JF343634 | JF343741 | |
| LGMF03 | Citrus latifolia (Rutaceae), healthy leaves | Brazil: SP | A. de Goes | JF261453 | JF261495 | JF343635 | JF343749 | |
| LGMF181 | Citrus reticulata (Rutaceae), black spot on fruit | Brazil: PR | C. Glienke | JF261447 | JF261489 | JF343629 | JF343736 | |
| LGMF217 | Citrus sinensis (Rutaceae), leaf endophyte | Brazil: PR | C. Glienke | JF261451 | JF261493 | JF343633 | JF343740 | |
| LGMF219 | Citrus sinensis (Rutaceae), leaf endophyte | Brazil: PR | C. Glienke | JF261448 | JF261490 | JF343630 | JF343737 | |
| LGMF220 | Citrus sinensis (Rutaceae), leaf endophyte | Brazil: PR | C. Glienke | JF261446 | JF261488 | JF343628 | JF343735 | |
| LGMF222 | Citrus sinensis (Rutaceae), leaf endophyte | Brazil: PR | C. Glienke | JF261450 | JF261492 | JF343632 | JF343739 | |
| LGMF231 | Citrus sinensis (Rutaceae), leaf endophyte | Brazil: SP | C. Glienke | JF261441 | JF261483 | JF343623 | JF343730 | |
| LGMF240 | Citrus sinensis (Rutaceae), leaf endophyte | Brazil: SP | C. Glienke | JF261443 | JF261485 | JF343625 | JF343732 | |
| LGMF244 | Citrus limonia (Rutaceae), leaf endophyte | Brazil: PR | C. Glienke | JF261442 | JF261484 | JF343624 | JF343731 | |
| LGMF253 | Citrus limonia (Rutaceae), leaf endophyte | Brazil: PR | C. Glienke | JF261460 | JF261502 | JF343642 | JF343750 | |
| LGMF259 | Citrus latifolia (Rutaceae), leaf endophyte | Brazil: PR | C. Glienke | JF261461 | JF261503 | JF343643 | JF343751 | |
| LGMF317 | Citrus reticulata (Rutaceae), leaf endophyte | Brazil: PR | C. Glienke | JF261440 | JF261482 | JF343622 | JF343729 | |
| LGMF318 | Citrus reticulata (Rutaceae), leaf endophyte | Brazil: PR | C. Glienke | JF261454 | JF261496 | JF343636 | JF343742 | |
| LGMF319 | Citrus reticulata (Rutaceae), leaf endophyte | Brazil: PR | C. Glienke | JF261445 | JF261487 | JF343627 | JF343734 | |
| LGMF326 | Citrus reticulata (Rutaceae), leaf endophyte | Brazil: PR | C. Glienke | JF261444 | JF261486 | JF343626 | JF343733 | |
| LGMF332 | Mangifera indica (Anacardiaceae), leaf endophyte | Brazil: SP | C. Glienke | JF261439 | JF261481 | JF343621 | JF343728 | |
| LGMF358 | Mangifera indica (Anacardiaceae), leaf endophyte | Brazil: PR | C. Glienke | JF261449 | JF261491 | JF343631 | JF343738 | |
| LGMF366 | Mangifera indica (Anacardiaceae), leaf endophyte | Brazil: PR | C. Glienke | JF261438 | JF261480 | JF343620 | JF343727 | |
| VIC30428 | Cymbidium sp. (Orchidaceae), leaf blight | Brazil: MG | M. Silva & O.L. Pereira | JF261455 | JF261497 | JF343637 | JF343743 | |
| Phyllosticta citriasiana | CBS 120486; PD 05/01969753 | Citrus maxima (Rutaceae) | Thailand | J. de Gruyter | FJ538360 | FJ538418 | FJ538476 | JF343686 |
| CBS 120487; PD 05/03081053 | Citrus maxima (Rutaceae) | China | K. Rosendahl-Peters | FJ538361 | FJ538419 | FJ538477 | JF343687 | |
| CBS 123370; PD 08/04453736 | Citrus maxima (Rutaceae) | Vietnam | J. de Gruyter | FJ538355 | FJ538413 | FJ538471 | JF343689 | |
| CBS 123371; PD 08/04454173 | Citrus maxima (Rutaceae) | Vietnam | J. de Gruyter | FJ538356 | FJ538414 | FJ538472 | JF343690 | |
| CBS 123393; PD 08/04453728 | Citrus maxima (Rutaceae) | Vietnam | J. de Gruyter | FJ538358 | FJ538416 | FJ538474 | JF343688 | |
| Phyllosticta citribraziliensis | CBS 100098 | Citrus sp. (Rutaceae), healthy leaves | Brazil: PR | C. Glienke | FJ538352 | FJ538410 | FJ538468 | JF343691 |
| LGMF08 | Citrus sp. (Rutaceae), healthy leaves | Brazil: PR | C. Glienke | JF261435 | JF261477 | JF343617 | JF343692 | |
| LGMF09 | Citrus sp. (Rutaceae), healthy leaves | Brazil: PR | C. Glienke | JF261436 | JF261478 | JF343618 | JF343693 | |
| Phyllosticta citricarpa | 29 | Citrus sinensis (Rutaceae), black spot on fruit | South Africa | G.C. Carroll | JF261433 | JF261475 | JF343615 | JF343683 |
| 71 | Citrus sinensis (Rutaceae), black spot on fruit | South Africa | G.C. Schutte | JF261432 | JF261474 | JF343614 | JF343682 | |
| CBS 102373 | Citrus aurantium (Rutaceae), black spot on fruit | Brazil | – | FJ538312 | FJ538370 | FJ538428 | JF343678 | |
| CBS 102374 | Citrus aurantium (Rutaceae), black spot on fruit | Brazil | – | FJ538313 | FJ538371 | FJ538429 | JF343679 | |
| CBS 111.20 | – | – | – | FJ538314 | FJ538372 | FJ538430 | JF343681 | |
| CBS 120489 | Citrus limon (Rutaceae) | Brazil | J. de Gruyter | FJ538315 | FJ538373 | FJ538431 | JF343685 | |
| CBS 122384 | Citrus limon (Rutaceae) | South Africa | M. Truter | FJ538316 | FJ538374 | FJ538432 | JF343680 | |
| CBS 122482 | Citrus sinensis (Rutaceae), lesions on fruit | Zimbabwe | L. Huisman | FJ538317 | FJ538375 | FJ538433 | JF343677 | |
| CBS 127451; CPC 18173 | Citrus reticulata (Rutaceae) | Australia | S.L. Willingham | JF343580 | JF343601 | JF343664 | JF343768 | |
| CBS 127452; CPC 18174 | Citrus reticulata (Rutaceae) | Australia | S.L. Willingham | JF343581 | JF343602 | JF343665 | JF343769 | |
| CBS 127453; CPC 18175 | Citrus reticulata (Rutaceae) | Australia | S.L. Willingham | JF343582 | JF343603 | JF343666 | JF343770 | |
| CBS 127454; CPC 18176 | Citrus limon (Rutaceae) | Australia | S.L. Willingham | JF343583 | JF343604 | JF343667 | JF343771 | |
| CBS 127455; CPC 18177 | Citrus sinensis (Rutaceae) | Australia | S.L. Willingham | JF343584 | JF343605 | JF343668 | JF343772 | |
| Guig1 | Citrus maxima (Rutaceae), black spot on fruit | Brazil: SP | A. de Goes | JF261429 | JF261471 | JF343611 | JF343674 | |
| LGMF06 | Citrus sinensis (Rutaceae), black spot on fruit | Brazil: SP | A. de Goes | JF261431 | JF261473 | JF343613 | JF343676 | |
| LGMF20 | Citrus sinensis (Rutaceae), black spot on fruit | Brazil: PR | C. Glienke | JF261430 | JF261472 | JF343612 | JF343675 | |
| LGMF25 | Citrus sinensis (Rutaceae), black spot on fruit | Brazil: PR | C. Glienke | JF261428 | JF261470 | JF343610 | JF343673 | |
| LGMF27 | Citrus sinensis (Rutaceae), black spot on fruit | Brazil: PR | C. Glienke | JF261427 | JF261469 | JF343609 | JF343672 | |
| LGMF45 | Citrus reticulata (Rutaceae), black spot on fruit | Brazil: PR | C. Glienke | JF261426 | JF261468 | JF343608 | JF343671 | |
| LGMF63 | Citrus reticulata (Rutaceae), black spot on fruit | Brazil: PR | C. Glienke | JF261425 | JF261467 | JF343607 | JF343670 | |
| LGMF247 | Citrus limonia (Rutaceae), on leaves | Brazil: PR | C. Glienke | JF261434 | JF261476 | JF343616 | JF343684 | |
| Phyllosticta cussonia | CPC 14873 | Cussonia sp. | South Africa | P.W. Crous | JF343578 | JF343599 | JF343662 | JF343764 |
| CPC 14875 | Cussonia sp. | South Africa | P.W. Crous | JF343579 | JF343600 | JF343663 | JF343765 | |
| Phyllosticta hypoglossi | CBS 101.72; IFO 32916 | Ruscus aculeatus (Ruscaceae), living leaves | Italy | W. Gams | FJ538365 | FJ538423 | FJ538481 | JF343694 |
| CBS 167.85 | Ruscus hypoglosum (Ruscaceae) | Italy | W. Gams | FJ538366 | FJ538424 | FJ538482 | JF343696 | |
| CBS 434.92 | Ruscus aculeatus (Ruscaceae), dead cladodes | Italy | W. Gams | FJ538367 | FJ538425 | FJ538483 | JF343695 | |
| Phyllosticta owaniana | CBS 776.97 | Brabejum stellatifolium (Proteaceae), leaf spot | South Africa | A. den Breeÿen | FJ538368 | FJ538426 | FJ538484 | JF343767 |
| CPC 14901 | Brabeijum stellatifolium (Proteaceae), leaf spot | South Africa | P.W. Crous | JF261462 | JF261504 | JF343644 | JF343766 | |
| Phyllosticta spinarum | CBS 292.90 | Chamaecyparis pisifera (Cupressaceae) | France | M. Morelet | JF343585 | JF343606 | JF343669 | JF343773 |
| CBS 937.70 | Hedera helix (Araliaceae), leaf litter | Italy | W. Gams | FJ538350 | FJ538408 | FJ538466 | JF411745 | |
1 ATCC: American Type Culture Collection, Virginia, USA; CBS: CBS Fungal Biodiversity Centre, Utrecht, The Netherlands; CMU: Microbiology Section, Chiang Mai University (MSCMU), Department of Biology, Faculty of Science, Chang Mai University, Thailand; CPC: Culture collection of P.W. Crous, housed at CBS; IFO: Institute for Fermentation, Osaka, Japan; IMI: International Mycological Institute, CABI-Bioscience, Egham, Bakeham Lane, U.K.; LGMF: Culture collection of Laboratory of Genetics of Microorganisms, Federal University of Parana, Curitiba, Brazil; PD: Plant Protection Service, Wageningen, The Netherlands; VIC: Culture collection of Federal University of Viçosa, Viçosa, Brazil.
2 Abbreviations used with Brazil: MG: State of Minas Gerais; PR: State of Paraná; SP: State of São Paulo.
3 ITS: Internal transcribed spacers 1 and 2 together with 5.8S nrDNA; TEF1: partial translation elongation factor 1-alpha gene; ACT: partial actin gene; GPDH: partial glyceraldehyde-3-phosphate dehydrogenase gene.
DNA isolation, amplification and analyses
Genomic DNA extraction was done using the UltraClean™ Microbial DNA Kit (MO Bio, Carlsbad, CA, USA) according to manufacturer’s protocol or according to Glienke-Blanco et al. (2002). The primers V9G (de Hoog & Gerrits van den Ende 1998) and ITS4 (White et al. 1990) were used to amplify the internal transcribed spacer region (ITS) of the nuclear ribosomal RNA operon, including the 3′ end of the 18S rRNA, the first internal transcribed spacer region, the 5.8S rRNA gene; the second internal transcribed spacer region and the 5′ end of the 28S rRNA gene. The primers EF1-728F (Carbone & Kohn 1999) and EF2 (O’Donnell et al. 1998) were used to amplify part of the translation elongation factor 1-α gene (TEF1) and the primers ACT-512F and ACT-783R (Carbone & Kohn 1999) were used to amplify part of the actin gene (ACT). Amplification conditions followed Arzanlou et al. (2008). The primers GDF1 (Guerber et al. 2003) and Gpd2-LM (Myllys et al. 2002) or GDR1 (Guerber et al. 2003) were used to amplify part of the glyceraldehyde-3-phosphate dehydrogenase (GPDH) gene of G. mangiferae s.l. isolates. Amplification reactions were performed under two different conditions, depending on the laboratory in which those specific reactions were performed. The first condition had a total reaction volume of 15.5 μL, which was composed of 1× PCR Buffer (Applied Biosystems, Foster City, USA), 2 mM MgCl2, 40 μM dNTPs, 0.08 μM of each forward and reverse primer, 0.5 U of Taq DNA polymerase (Roche Diagnostics, Indianapolis, USA) and 1–10 ng of genomic DNA. The PCR cycle conditions were 4 min of 94 °C, followed by 13 cycles of 94 °C for 30 s, the annealing temperature was decreased in 0.7 for every subsequent set of cycles, 72 °C for 60 s, followed by 23 cycles of 94 °C for 30 s, 56 °C for 30 s, 72 °C for 60 s and a final elongation at 72 °C for 7 min. The second condition had a total reaction volume of 12.5 μL, which was composed of 1× PCR Buffer (Bioline GmbH, Luckenwalde, Germany), 5.6 % DMSO (v/v), 2 mM MgCl2, 20 μM dNTPs, 0.2 μM of each forward and reverse primer, 0.25 U of BioTaq Taq DNA polymerase (Bioline GmbH, Luckenwalde, Germany) and 1–10 ng of genomic DNA. The PCR cycle conditions were 5 min of 94 °C, followed by 40 cycles of 94 °C for 30 s, 52 °C for 30 s, 72 °C for 30 s and a final elongation step at 72 °C for 7 min. The partial GPDH gene of G. citricarpa isolates was amplified with the primers GDF1 (Guerber et al. 2003) and a primer developed in the present study, GPDHR2 (5′-CTCRGMRGCRGCCTTGATGG-3′). A 1 000 bp fragment was obtained with this primer combination. Amplification reactions were performed in a final reaction volume of 12.5 μL, which was composed of 1× PCR Buffer (Applied Biosystems, Foster City, USA), 2.5 mM MgCl2, 40 μM dNTPs, 0.12 μM of each forward and reverse primer, 0.5 U of Taq DNA polymerase (Roche Diagnostics, Indianapolis, USA) and 1–10 ng of genomic DNA. The PCR cycle conditions were 5 min of 95 °C, followed by 35 cycles of 95 °C for 30 s, 50 °C for 45 s, 72 °C for 90 s, and a final elongation at 72 °C for 7 min. Amplicons were sequenced using both PCR primers with a BigDye Terminator Cycle Sequencing Kit v3.1 (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions, and sequences were analyzed on an ABI Prism 3700 DNA Sequencer (Perkin-Elmer, Norwalk, Foster City, CA, USA).
Consensus sequences were manually aligned using MEGA v4 software (Kumar et al. 2008) by inserting gaps. Phylogenetic analyses of the aligned sequence data (no nucleotides were excluded) were performed with PAUP (Phylogenetic Analysis Using Parsimony) v4.0b10 (Swofford 2003) as described previously (Cheewangkoon et al. 2008). Based on previous phylogenetic studies (e.g. Wulandari et al. 2009), Phyllosticta owaniana was used as outgroup in the phylogenetic analyses. Statistical parameters calculated by PAUP included Tree length (TL), consistency index (CI), retention index (RI) and rescaled consistency index (RC). Novel sequence data were deposited in GenBank (Table 1) and alignments in TreeBASE (www.treebase.org).
Morphology
Isolates were established on 2 % malt extract agar (MEA), 2 % potato-dextrose agar (PDA), pine-needle agar (PNA; tap water agar with autoclaved pine needles; Crous et al. 2006) and oatmeal agar (OA; Crous et al. 2009c), and incubated at 25 °C under near-ultraviolet light to promote sporulation. Fungal structures were mounted on glass slides in clear lactic acid for microscopic examination after 14 d of incubation. Thirty measurements were determined per structure, where possible, from colonies sporulating on PNA. Colony colours (surface and reverse) were determined using the colour charts of Rayner (1970) after 1 mo at 25 °C in the dark. Nomenclatural novelties and descriptions were deposited in MycoBank (www.MycoBank.org; Crous et al. 2004).
RESULTS
Phylogenetic analysis
The manually adjusted combined (ITS, TEF1, ACT and GPDH) alignment contained 105 isolates (including two outgroup sequences) and, of all 1 580 characters used in the phylogenetic analysis, 442 were parsimony-informative, 61 were variable and parsimony-uninformative, and 1 077 were conserved. Distance analyses using the three substitution models on the sequence data yielded trees with identical topology and similar bootstrap values. Only the first 1 000 equally most parsimonious trees were retained, the first of which is shown in Fig. 1 (TL = 932, CI = 0.790, RI = 0.982, RC = 0.776). These trees only differed with regard to the order of the small terminal branches within the well-supported clades (see the thickened strict consensus branches in Fig. 1).
Fig. 1.
The first of 1 000 equally most parsimonious trees obtained from a heuristic search with 100 random taxon additions of the combined sequence alignment. The scale bar shows 10 changes, and bootstrap support values from 1 000 replicates are shown at the nodes. Branches present in the strict consensus tree are thickened and original species names are indicated next to the strain number for clade 10. The tree was rooted to sequences of two Phyllosticta owaniana strains.
Ten well-supported clades could be resolved (Fig. 1). The first clade consists of the strain VIC30556, which was isolated from leaf and pseudobulb lesions on Bifrenaria harrisoniae (Orchidaceae) in Brazil (Silva et al. 2008) and was morphologically identified as Phyllosticta capitalensis by the authors. This isolate, described here as P. bifrenariae sp. nov., caused dark, large spots on orchid leaves, in contrast to the symptoms associated with endophytic isolates (Silva et al. 2008).
The second clade consists of two isolates of Phyllosticta cussonia from South Africa, while the third clade consists of three isolates from Ruscus hypoglossum in Italy, representing a species complex presently treated as P. hypoglossi. The fourth clade consists of two isolates identified as P. spinarum from Chamaecyparis pisifera in France and Hedera helix in Italy, respectively, and probably also represents a species complex. Three Citrus (Rutaceae) endophytic isolates from Brazil, described here as P. citribraziliensis, make up clade 5.
The sixth clade is represented by isolates of P. citriasiana (Wulandari et al. 2009), associated with tan spot on Citrus maxima fruits. Clade 7 represents isolates of P. citricarpa from Australia, Brazil, South Africa and Zimbabwe. Clade 8 consists of 12 endophytic isolates of Mangifera indica (Anacardiaceae) from Brazil. These isolates are morphologically distinct, and exhibited insignificant homology to any sequence found in the GenBank nucleotide database, and these are described below as P. brazilianiae sp. nov. Clade 9 consists of a single isolate (IMI 260576), which was isolated in India from Mangifera indica, and is considered authentic for the name G. mangiferae.
Clade 10 represents several different hosts and countries (Fig. 1, Table 1). This clade included isolates from Rutaceae (Citrus spp.), Anacardiaceae (Mangifera indica, Spondias mombin, Myracrodruon urundeuva, Anacardium giganteum), Myrtaceae (Psidium guajava), Sapindaceae (Nephelium lappaceum), Solanaceae (Capsicum), Fabaceae (Bowdichia nitida), Apocynaceae (Aspidosperma polyneuron), Musaceae (Musa spp.), Orchidaceae (Cymbidium sp., Paphiopedilum callosum, Stanhopea graveolens), Aquifoliaceae (Ilex sp.), Magnoliaceae (Magnolia liliifera), Smilacaceae (Smilax kraussiana) and Zamiaceae (Encephalartos ferox). This clade contains isolates previously identified as G. mangiferae, G. endophylicolla, G. psidii, G. capsici, G. musae, G. vaccini, G. philoprina, G. musarum, Guignardia sp. and P. capitalensis. However, the low sequence homology found between the reference isolate of G. mangiferae (clade 9) (IMI 260576) and clade 10 isolates, strongly supports these as two distinct species (Fig. 1).
Morphology
Several new species were identified during this study, which are described below. Furthermore, an epitype could also be designated for P. citricarpa based on Citrus collections newly obtained from Australia. Similarly, an epitype could be designated for P. capitalensis, based on fresh collections obtained on Stanhopea from Brazil. Although isolates belonging to clade 10 are all treated as P. capitalensis, some morphological variation was observed in conidium morphology (sheath thickness, appendage length and conidium shape), and growth in culture. Most cultures produced conidia with sheaths more than 2 μm thick, as reported by Baayen et al. (2002) for P. capitalensis. Several isolates also produced a Guignardia state in culture. Additional genes need to be sequenced to determine if the observed variation in clade 10 is intra- or interspecific. Furthermore, in moving to a single nomenclature for species of Ascomycetes (Rossman & Samuels 2005, Crous et al. 2006, 2007, 2009a, b, Aveskamp et al. 2010, Lechat et al. 2010, Lombard et al. 2010a, b, c), the older generic name, Phyllosticta (1818), is chosen above the later Guignardia (1892), which should be regarded as synonym.
Guignardia mangiferae A.J. Roy, Indian Phytopathol. 20: 348. 1968
Type specimen. India, Shitlakhet in Almora, on leaves of Mangifera indica, 9 July 1963, B.S. Khati, holotype HFRS 1056 (could not be obtained for examination).
Colonies on OA. Pycnidia black, aggregated, erumpent, globose to ampulliform, exuding a colourless, glossy conidial mass; pycnidia up to 300 μm diam, 250 μm tall; pycnidial wall consisting of several layers, up to 40 μm thick, of textura angularis. Ostiole single, central, up to 30 μm wide, consisting of thickened, brown cells. Conidiophores subcylindrical to doliiform, frequently reduced to conidiogenous cells, coated in mucoid layer, 6–15 × 3–6 μm. Conidiogenous cells terminal, subcylindrical to doliiform, hyaline, smooth, 6–10 × 3–4 μm; proliferating 2–3 times percurrently near apex. Conidia (8–)10–12 × (5–)6–7 μm, solitary, hyaline, aseptate, thin- and smooth-walled, coarsely guttule, ellipsoid to obovoid, tapering toward a narrowly truncate base, enclosed in a mucilaginous sheath, 2–5 μm thick, and bearing a hyaline, mucoid apical appendage, 7–13 × 1–1.5 μm, straight to flexible, unbranched, tapering towards an acute apex. No teleomorph other than ascomatal initials developed in agar (OA, SNA, PDA, MEA, PNA), and the isolate sporulated poorly.
Specimen examined. India, on leaves of Mangifera indica (Anacardiaceae), 1981, M.V. Leksshmi, culture IMI 260576.
Notes — Two other species occurring on Mangifera indica in Brazil need to be discussed. Phyllosticta mangiferae has fusiform, 11–23 × 6–7 μm conidia, resembling the genus Fusicoccum (van der Aa & Vanev 2002). Phyllosticta anacardiacearum differs from G. mangiferae by having shorter conidiophores, and a narrower sheath, although the conidia are similar in size (van der Aa 1973). No cultures of P. anacardiacearum are, however, available for study. Because the name Phyllosticta mangiferae is occupied, a new name would have to be proposed for Guignardia mangiferae when it eventually is placed in Phyllosticta. However, because mango has been poorly studied, we choose to wait until more isolates become available.
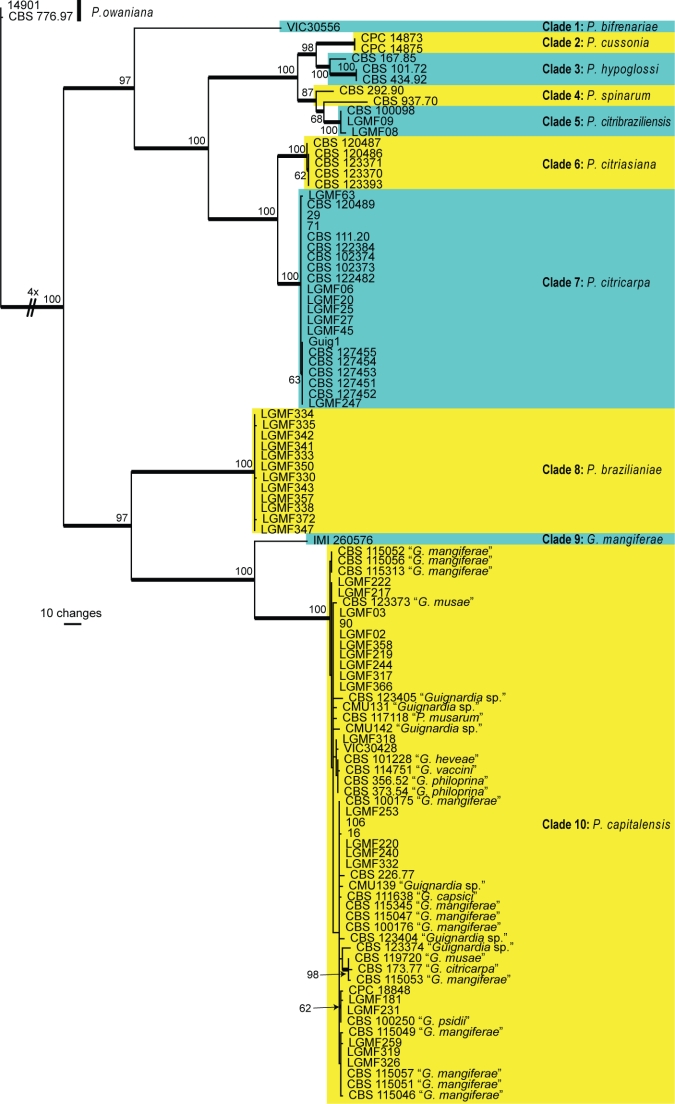
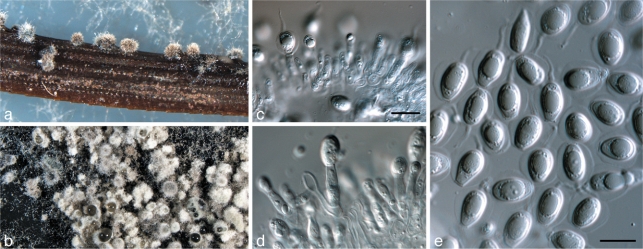
Phyllosticta bifrenariae O.L. Pereira, C. Glienke & Crous, sp. nov. — MycoBank MB517969; Fig. 2
Fig. 2.
Phyllosticta bifrenariae. a. Pycnidium forming on PNA; b. pycnidia forming on PDA; c, d. conidiophores giving rise to conidia; e, f. conidia; g. spermatia (all: CBS H-20520 holotype). — Scale bars = 10 μm.
Phyllostictae capitalensis similis, sed conidiis maioribus, 10–16 × 7–9 μm.
Etymology. Named after the host genus from which it was isolated, Bifrenaria.
Colonies on PNA. Pycnidia black, solitary, or arranged in clusters of up to 6, ampulliform, base ovoid, up to 250 μm diam, with elongated subcylindrical neck up to 1 100 μm long, and rounded apex, 180 μm diam; pycnidial wall consisting of several layers, up to 40 μm thick; outer region of dark brown textura angularis to globularis; inner region consisting of 1–2 pale cell layers, that become hyaline toward interior, textura angularis. Ostiole single, central, up to 40 μm wide. Conidiophores reduced to Conidiogenous cells, subcylindrical to ampulliform, hyaline, smooth, 7–10 × 4–5 μm; inconspicuously proliferating once or twice percurrently near apex. Conidia (10–)11–13(–16) × (7–)8–9 μm, solitary, hyaline, aseptate, thin- and smooth-walled, with large central guttule, ellipsoid to ovoid or obovoid, tapering toward a narrowly truncate base, 3–4 μm wide, enclosed in a thick mucilaginous sheath, 3–6 μm thick, and bearing a hyaline, mucoid apical appendage, 6–20 × 1–1.5 μm, straight to flexible, unbranched, tapering towards an acute tip. Spermatia at times forming in conidial conidiomata, hyaline, bacilliform, 5–10 × 1.5–2 μm.
Culture characteristics — Colonies after 14 d at 25 °C in the dark on OA flat, spreading, olivaceous-grey, with moderate aerial mycelium.
Specimen examined. Brazil, Gerdau Açominas RPPN, Serra de Ouro Branco, Ouro Branco, Minas Gerais, on Bifrenaria harrisoniae (Orchidaceae), 6 Nov. 2007, O.L. Pereira, CBS H-20520 holotype, culture ex-type VIC 30556 = CBS 128855.
Notes — Although the isolate now described as P. bifrenariae was originally considered to be representative of P. capitalensis, it is ecologically distinct in being a pathogen on Bifrenaria harrisoniae (Orchidaceae) (Silva et al. 2008), and is also phylogenetically distinct (Fig. 1). Morphologically P. capitalensis (conidia (10–)11–12(–14) × (5–)6–7 μm) is distinct by having smaller conidia than P. bifrenariae (10–16 × 7–9 μm). Phyllosticta aplectri, which occurs on Aplectrum hyemale (Orchidaceae, USA), has smaller conidia, 5–8 × 4–6 μm (van der Aa 1973).
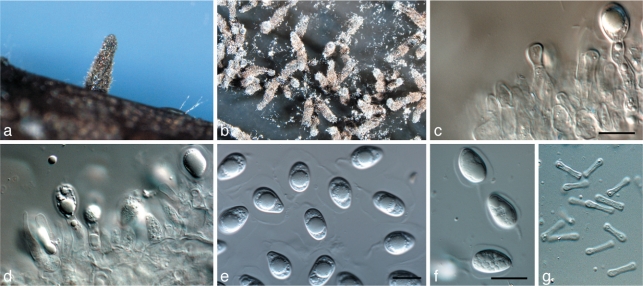
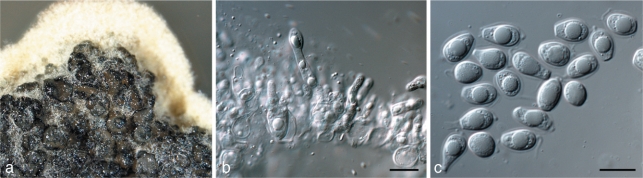
Phyllosticta brazilianiae D. Stringari, C. Glienke & Crous, sp. nov. — MycoBank MB517970; Fig. 3
Fig. 3.
Phyllosticta brazilianiae. a. Pycnidia forming on PDA; b, c. conidiophores giving rise to conidia; d. conidia (all: CBS H-20521 holotype). — Scale bars = 10 μm.
Phyllostictae anacardiacearum similis, sed endophytice, neque vero phytoparasitice crescenti.
Etymology. Named after the country from which it was collected, Brazil.
Colonies on PNA. Pycnidia black, aggregated, superficial to erumpent, globose to ampulliform, exuding a colourless, glossy conidial mass; pycnidia up to 300 μm diam; pycnidial wall consisting of several layers, up to 40 μm thick; outer region of dark brown, thickened, textura angularis to globularis; inner region up to 20 μm wide, consisting of 1–2 pale cell layers of textura angularis. Ostiole single, central, 5–10 μm wide, consisting of thickened, brown cells. Conidiophores subcylindrical to doliiform, reduced to conidiogenous cells, or with one supporting cell, coated in mucoid layer, 10–20 × 4–5 μm. Conidiogenous cells terminal, subcylindrical to doliiform, hyaline, smooth, 7–15 × 3–4 μm; proliferating 1–3 times percurrently near apex. Conidia (8–)10–11(–12.5) × (5–)6(–7) μm, solitary, hyaline, aseptate, thin- and smooth-walled, coarsely guttulate, ellipsoid to obovoid, tapering toward a narrowly truncate base, enclosed in a thin mucilaginous sheath, 1–2 μm thick, and bearing a hyaline, mucoid apical appendage, (5–)8–10(–15) × 1.5–2 μm, straight to flexible, unbranched, tapering towards an acute apex.
Culture characteristics — Colonies after 14 d at 25 °C in the dark on OA flat, spreading, olivaceous-grey, becoming pale olivaceous-grey towards the margin, with moderate aerial mycelium.
Specimen examined. Brazil, Pompéia, São Paulo, on Mangifera indica (Anacardiaceae), May 2007, D. Stringari, CBS H-20521 holotype, culture ex-type LGMF 330 = CBS 126270.
Notes — Van der Aa (1973) introduced the name Phyllosticta anacardiacearum as a nom. nov. for Phyllostictina mangiferae occurring on mango in Brazil. The name Phyllosticta mangiferae was found to be a species of Fusicoccum, while Phyllosticta mortonii, occurring on mango in Mexico, was thought to be a species of Phoma (van der Aa & Vanev 2002). While no authentic material could be located for Phyllosticta anacardiacearum, it was originally described from subcircular to angular leaf spots, reaching 1 cm diam, surrounded by a red-purple margin. The same was also found to be the case when van der Aa (1973) redescribed the fungus from a specimen collected on Mangifera indica in Miami. The species described here as P. brazilianae is ecologically distinct from P. anacardiacearum being an endophyte, and failing to induce leaf spots despite repeated inoculations on mango.
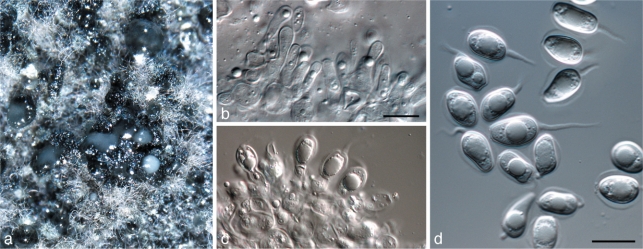
Phyllosticta capitalensis Henn., Hedwigia 48: 13. 1908 — Fig. 4
Fig. 4.
Phyllosticta capitalensis. a, b. Asci with ascospores; c, d. conidiogenous cells giving rise to conidia; e. conidia (all: CBS H-20522 epitype). — Scale bars = 10 μm.
Colonies on OA. Ascomata erumpent, in section globose to pyriform, often irregularly shaped, unilocular, central ostiole forming by dehiscence when mature, up to 250 μm diam. Peridium comprising three strata, an outer stratum of thick-walled, small-lumened, brown textura angularis, becoming thin-walled with larger lumina in the middle layer, inner layer of thin-walled, hyaline textura angularis, altogether 14–45 μm thick. Asci attached to the basal peridium, clavate, with a wide, slightly squared apex, tapering gradually to a small pedicel, bitunicate, with a well-developed ocular chamber, 8-spored, 58–80 × 11–15 μm. Ascospores limoniform, sometimes slightly elongated, aseptate, hyaline, thick-walled, refractive, with a large central guttule and large mucilaginous polar appendages, overlapping biseriate, 15–17 × 5–6 μm, 3.5 μm wide at each end. Pycnidia black, aggregated, erumpent, globose to ampulliform, exuding a colourless, glossy conidial mass; pycnidia up to 300 μm diam, 250 μm tall; pycnidial wall consisting of 6–8 layers, up to 40 μm thick, of textura angularis. Ostiole single, central, 5–15 μm diam. Conidiophores subcylindrical to ampulliform, frequently reduced to conidiogenous cells, or branching from a basal supporting cell, coated in mucoid layer, 7–20 × 3–7 μm. Conidiogenous cells terminal, subcylindrical to ampulliform to doliiform, hyaline, smooth, 7–10 × 3–5 μm; proliferating 1–2 times percurrently near apex. Conidia (10–)11–12(–14) × (5–)6–7 μm, solitary, hyaline, aseptate, thin- and smooth-walled, coarsely guttule, ellipsoid to obovoid, tapering toward a narrowly truncate base, enclosed in a mucilaginous sheath, 2–4 μm thick, and bearing a hyaline, mucoid apical appendage, 6–8 × 1–1.5 μm, straight to curved, unbranched, tapering towards a bluntly rounded apex.
Specimens examined. Brazil, São Paulo, on leaves of Stanhopea sp., Apr. 1903, B, holotype; São Paulo, Lindóia, on leaves of Stanhopea graveolens, 17 Oct. 2010, O.L. Pereira, epitype designated here CBS H-20522, culture ex-epitype CBS 128856 = CPC 18848, CPC 18849.
Notes — Phyllosticta capitalensis is the name proposed for the isolates in clade 10 (formerly incorrectly referred to as Guignardia mangiferae; Baayen et al. 2002), representing a taxon that is frequently isolated as endophyte, and has a wide host range and geographic distribution.
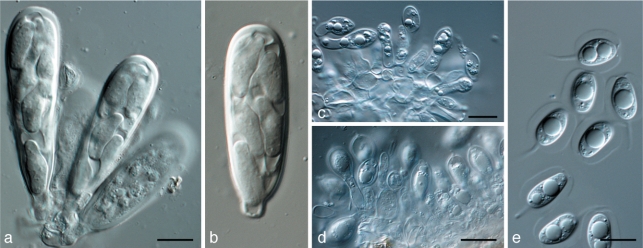
Phyllosticta citribraziliensis C. Glienke & Crous, sp. nov. — MycoBank MB517971; Fig. 5
Fig. 5.
Phyllosticta citribraziliensis. a. Pycnidia forming on PNA; b. pycnidia forming on PDA; c, d. conidiophores giving rise to conidia; e. conidia (all: CBS H-20523 holotype). — Scale bars = 10 μm.
Phyllostictae citricarpae similis, sed conidiis maioribus, 10–16 × 5–8 μm.
Etymology. Named after the host (Citrus) and country from which it was isolated, Brazil.
Colonies on PNA. Pycnidia black, solitary, erumpent, globose, exuding colourless to opague conidial masses; pycnidia up to 250 μm diam; pycnidial wall consisting of several layers, up to 40 μm thick; outer region of dark brown, thickened, textura angularis to globularis; inner region up to 25 μm wide, consisting of 1–2 pale cell layers, that become hyaline toward interior, textura angularis. Ostiole single, central, up to 30 μm wide. Conidiophores subcylindrical to ampulliform, reduced to conidiogenous cells, or with 1–2 supporting cells, at times branched at the base, 20–45 × 6–9 μm. Conidiogenous cells terminal, subcylindrical to doliiform, hyaline, smooth, coated in a mucoid layer, 7–20 × 3–4 μm; inconspicuously proliferating once or twice percurrently near apex. Conidia (8–)10–12(–13) × 6–7(–8) μm, solitary, hyaline, aseptate, thin- and smooth-walled, coarsely guttulate, ellipsoid to obovoid, tapering toward a narrowly truncate base, 2–3 μm wide, enclosed in a thick mucilaginous sheath, 2–4 μm thick, and bearing a hyaline, mucoid apical appendage, 7–15 × 1.5–2 μm, straight to flexible, unbranched, tapering towards an acutely rounded tip.
Culture characteristics — Colonies after 14 d at 25 °C in the dark on OA flat, spreading, olivaceous grey, with moderate aerial mycelium.
Specimen examined. Brazil, Rio Negro, Paraná, on Citrus limon, Mar. 1997, C. Glienke, CBS H-20523 holotype, culture ex-type CBS 100098.
Notes — Although isolates occurring on Citrus have in the past been treated as representative of P. spinarum (Stringari et al. 2009), they are phylogenetically distinct (Fig. 1), and can also be distinguished morphologically by having larger conidia (8–)10–12(–13) × 6–7(–8) μm than the type of P. spinarum (8–)9.8(–12) × (6–)6.6(–7) μm; Nag Raj & Morelet 1997). Furthermore, P. citribraziliensis also has branched conidiophores, a thick mucilaginous sheath surrounding its conidia (2–4 μm), whereas those in P. spinarum are reduced to conidiogenous cells, and the sheath is 1–2 μm thick (Nag Raj & Morelet 1997).
Phyllosticta citricarpa (McAlpine) Aa, Stud. Mycol. 5: 40. 1973. — Fig. 6
Fig. 6.
Phyllosticta citricarpa. a. Pycnidia forming on OA, with diffuse yellow pigment visible in agar; b. conidiophores giving rise to conidia; c. conidia (all: CBS H-20524 epitype). — Scale bars = 10 μm.
Basionym. Phoma citricarpa McAlpine, Fungus diseases of Citrus trees in Australia, and their treatment: 21. 1899.
Teleomorph. Guignardia citricarpa Kiely, Proc. Linn. Soc. New South Wales 73: 259. 1948.
Colonies on OA. Pycnidia black, aggregated, superficial to erumpent, globose to ampulliform, exuding a colourless, opaque conidial mass; pycnidia up to 250 μm diam; pycnidial wall consisting of several layers, 20–50 μm thick; outer region of dark brown, thickened, textura angularis to globularis; inner region consisting of 1–2 pale cell layers of textura angularis. Ostiole single, central, 10–15 μm wide, consisting of thickened, brown cells. Conidiophores subcylindrical to doliiform, reduced to conidiogenous cells, or branched from a supporting cell, coated in mucoid layer, 10–20 × 4–7 μm. Conidiogenous cells terminal, subcylindrical to somewhat doliiform, hyaline, smooth, 7–12 × 3–4 μm; proliferating 1–2 times percurrently near apex. Conidia (10–)11–12(–14) × (6–)7(–8) μm, solitary, hyaline, aseptate, thin- and smooth-walled, coarsely guttulate, ellipsoid to obovoid, tapering toward a narrowly truncate base, enclosed in a thin mucilaginous sheath, 1(–2) μm thick, and bearing a hyaline, mucoid apical appendage, 5–10(–17) × 1–1.5 μm, straight to flexible, unbranched, tapering towards an acute apex.
Culture characteristics — Colonies after 14 d at 25 °C in the dark on OA flat, spreading, olivaceous-grey, becoming pale olivaceous-grey towards the margin, with sparse to moderate aerial mycelium; surrounded by a diffuse yellow pigment in the agar medium.
Specimens examined. Australia, Sydney, on Citrus sinensis, 1898, D. McAlpine, VPRI 1536, Lectotype selected here; Queensland, Emerald, ex Citrus black spot on leaf of Citrus sinensis, anon., 16 Dec. 2004, BRIP 46098 = CBS 127455; Queensland, Mundubbera, ex Citrus black spot on fruit of C. reticulata cv. Imperial, 27 Mar. 2001, S.L. Willingham, BRIP 27890 = CBS 127453, BRIP 27889 = CBS 127452, BRIP 27888 = CBS 127451; Gayndah, Queensland, ex Citrus black spot on C. limon, 3 Mar. 2009, A.K. Miles, CBS H-20524 epitype designated here, culture ex-epitype BRIP 52614 = CBS 127454.
Notes — The most characteristic features of P. citricarpa are the narrower sheaths (1(–2) μm thick), compared to that of P. capitalensis (2–3 μm thick), and the yellow pigment that diffuses into the agar when isolates of P. citricarpa are cultivated on oatmeal agar.
DISCUSSION
The present study aimed to resolve the taxonomy of the Phyllosticta species occurring on Citrus, either as pathogens, or as harmless endophytes. In the process we also had to resolve the status of the common endophytic taxon with a known wide host range and geographic distribution. Several names have in the past been linked to this taxon, including Guignardia mangiferae and Phyllosticta capitalensis. By obtaining reference strains considered authentic for these names, we could show that G. mangiferae is a distinct taxon from P. capitalensis, and that P. capitalensis is the name to be used for this cosmopolitan endophyte (clade 10, Fig. 1). In the process we also designated epitypes for P. capitalensis and P. citricarpa, described a novel species on orchids in Brazil as P. bifrenariae, one on Citrus as P. citribraziliensis, and another on Mangifera indica as P. brazilianiae.
Several species of Phyllosticta are now known to occur on Citrus, namely P. citriasiana, which is a pathogen of C. maxima, causing tan spot in Asia (Wulandari et al. 2009), P. citricarpa, which causes Citrus Black Spot in many countries, and is of quarantine concern (Baayen et al. 2002), P. citribraziliensis, which is an endophyte on Citrus in Brazil, and P. capitalensis, which is a wide host range endophyte, that also occurs on Citrus.
Although the genus Phyllosticta has received much taxonomic attention of late (refs), very few phylogenetic studies have thus far been conducted, and hence the taxonomy of this group is still problematic. Due to the lack of reference strains, and the fact that few gene loci other than ITS have thus far been used for DNA analysis, most of the conclusions reached thus far have been incorrect, meaning that published literature will have to be interpreted with care. Furthermore, in spite of the multi-gene approach taken in the present study, some morphological variation is still present among isolates treated here as P. capitalensis (clade 10), and more gene loci need to be investigated to confirm whether this is indeed a single taxon. Further studies are presently underway to address this issue.
Guignardia mangiferae was first described on Mangifera indica in India (Roy 1968), but the type specimen has not been available for study. In spite of the reference isolate (IMI 260576) being genetically distinct from others in the P. capitalensis clade (Fig. 1), this isolate proved to only form the anamorph in culture. Furthermore, no cultures are available for the plant pathogenic species, P. anacardiacearum, which we regard as distinct from the common endophyte for which the name P. brazilianiae has been introduced. This situation on mango is similar to the one on Citrus, where the plant pathogenic species are represented by P. citricarpa and P. citriasiana, and the endophytic strains by P. citribraziliensis and P. capitalensis. Despite the large production of mango in Brazil, the Phyllosticta leaf spot disease has not been found in commercial orchards, and it is possible that the species is either distinct, or vary rare, and not occurring on commercial cultivars. To help clarify the relationship of endophytic Phyllosticta spp. and their hosts, pathogenicity tests similar to those performed for endophytes of Musa acuminata (Photita et al. 2004), must be conducted on a range of different hosts in future studies.
Acknowledgments
We thank the Brazilian agency CNPq for financial support to C. Glienke. We are grateful to A. van Iperen, M. Vermaas, M. Starink (CBS, Utrecht) and J. Wolter-Sadlers (INRES, Bonn) for providing technical assistance.
REFERENCES
- Aa HA van der.1973. Studies in Phyllosticta I. Studies in Mycology 5: 1 – 110 [Google Scholar]
- Aa HA van der, Vanev S.2002. A revision of the species described in Phyllosticta. CBS, Utrecht, The Netherlands: [Google Scholar]
- Arzanlou M, Groenewald JZ, Fullerton RA, Abeln ECA, Carlier J, Zapater MF, Buddenhagen IW, Viljoen A, Crous PW.2008. Multiple gene genealogies and phenotypic characters differentiate several novel species of Mycosphaerella and related anamorphs on banana. Persoonia 20: 19 – 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aveskamp M, Gruyter H de, Woudenberg J, Verkley G, Crous PW.2010. Highlights of the Didymellaceae: A polyphasic approach to characterise Phoma and related pleosporalean genera. Studies in Mycology 65: 1 – 60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baayen RP, Bonants PJM, Verkley G, Carroll GC, Aa HA van der, et al. 2002. Nonpathogenic isolates of the citrus black spot fungus, Guignardia citricarpa, identified as a cosmopolitan endophyte of woody plants, Guignardia mangiferae (Phyllosticta capitalensis). Phytopathology 92: 464 – 477 [DOI] [PubMed] [Google Scholar]
- Carbone I, Kohn LM.1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553 – 556 [Google Scholar]
- Cheewangkoon R, Crous PW, Hyde KD, Groenewald JZ, Toanan C.2008. Species of Mycosphaerella and related anamorphs on Eucalyptus leaves from Thailand. Persoonia 21: 77 – 91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Braun U, Groenewald JZ.2007. Mycosphaerella is polyphyletic. Studies in Mycology 58: 1 – 32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G.2004. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19 – 22 [Google Scholar]
- Crous PW, Schoch CL, Hyde KD, Wood AR, Gueidan C, et al. 2009a. Phylogenetic lineages in the Capnodiales. Studies in Mycology 64: 17 – 47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Slippers B, Wingfield MJ, Rheeder J, Marasas WFO, et al. 2006. Phylogenetic lineages in the Botryosphaeriaceae. Studies in Mycology 55: 235 – 253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Carnegie AJ, Wingfield MJ, Hunter GC, et al. 2009b. Unravelling Mycosphaerella: do you believe in genera? Persoonia 23: 99 – 118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Verkleij GJM, Groenewald JZ, Samson RA. (eds). 2009c. Fungal Biodiversity. CBS Laboratory Manual Series 1. Centraalbureau voor Schimmelcultures, Utrecht, Netherlands: [Google Scholar]
- Glienke-Blanco C, Aguilar-Vildoso CI, Vieira MLC, Barroso PAV, Azevedo JL.2002. Genetic variability in the endophytic fungus Guignardia citricarpa isolated from citrus plants. Genetic and Molecular Biology 25: 251 – 255 [Google Scholar]
- Guerber JC, Liu B, Correll JC, Johnston PR.2003. Characterization of diversity in Colletotrichum acutatum sensu lato by sequence analysis of two gene introns, mtDNA and intron RFLPs, and mating compatibility. Mycologia 95: 872 – 895 [PubMed] [Google Scholar]
- Hennings P.1908. Fungi S. Paulenses IV a cl. Puttemans collecti. Hedwigia 48: 1 – 20 [Google Scholar]
- Hoog GS de, Gerrits van den Ende AHG.1998. Molecular diagnostics of clinical strains of filamentous basidiomycetes. Mycoses 41: 183 – 189 [DOI] [PubMed] [Google Scholar]
- Huang WY, Cai YZ, Surveswaran S, Hyde KD, Corke H, Sun M.2009. Molecular phylogenetic identification of endophytic fungi isolated from three Artemisia species. Fungal Diversity 36: 69 – 88 [Google Scholar]
- Kumar S, Dudley J, Nei M, Tamura K.2008. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in Bioinformatics 9: 299 – 306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechat C, Crous PW, Groenewald JZ.2010. The enigma of Calonectria species occurring on leaves of Ilex aquifolium in Europe. IMA Fungus 1: 101 – 108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, Crous PW, Wingfield BD, Wingfield MJ.2010a. Multigene phylogeny and mating tests reveal three cryptic species related to Calonectria pauciramosa. Studies in Mycology 66: 15 – 30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, Crous PW, Wingfield BD, Wingfield MJ.2010b. Phylogeny and systematics of the genus Calonectria. Studies in Mycology 66: 31 – 69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, Crous PW, Wingfield BD, Wingfield MJ.2010c. Species concepts in Calonectria (Cylindrocladium). Studies in Mycology 66: 1 – 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myllys L, Stenroos S, Thell A.2002. New genes for phylogenetic studies of lichenized fungi: glyceraldehyde-3-phosphate dehydrogenase and beta-tubulin genes. Lichenologist 34: 237 – 246 [Google Scholar]
- Nag Raj TR, Morelet M.1979. Observations on Mucosetospora (Coelomycetes). Canadian Journal of Botany 57: 1295 – 1297 [Google Scholar]
- O’Donnell K, Kistler HC, Cigelnik E, Ploetz RC.1998. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Science USA 95: 2044 – 2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okane I, Lumyong S, Nakagiri A, Ito T.2003. Extensive host range of an endophytic fungus, Guignardia endophyllicola (anamorph: Phyllosticta capitalensis). Mycoscience 44: 353 – 363 [Google Scholar]
- Okane I, Nakagiri A, Ito T.2001. Identity of Guignardia sp. inhabiting ericaceous plants. Canadian Journal of Botany 79: 101 – 109 [Google Scholar]
- Photita W, Lumyong S, Lumyong P, McKenzie EHC, Hyde KD.2004. Are some endophytes of Musa acuminata latent pathogens? Fungal Diversity 16: 131 – 140 [Google Scholar]
- Rayner RW.1970. A mycological colour chart. CMI and British Mycological Society, Kew, Surrey, England: [Google Scholar]
- Rossman AY, Samuels GJ.2005. Towards a single scientific name for species of fungi. Inoculum 56: 3 – 6 [Google Scholar]
- Roy AJ.1968. Some fungi from Almora. Indian Phytopathology 20: 340 – 348 [Google Scholar]
- Schoch CL, Crous PW, Groenewald JZS, Boehm EWA, Burgess TI, et al. 2009. A class-wide phylogenetic assessment of Dothideomycetes. Studies in Mycology 64: 1 – 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M, Pereira OL.2007. First report of Guignardia endophyllicola leaf blight on Cymbidium (Orchidaceae) in Brazil. Australasian Plant Disease Notes 2: 31 – 32 [Google Scholar]
- Silva M, Pereira OL, Braga IF, Leli SM.2008. Leaf and pseudobulb diseases on Bifrenaria harrisoniae (Orchidaceae) caused by Phyllosticta capitalensis in Brazil. Australasian Plant Disease Notes 3: 53 – 56 [Google Scholar]
- Stringari D, Glienke C, Christo D de, Maccheroni W, Jr, Azevedo JL de.2009. High molecular diversity of the fungus Guignardia citricarpa and Guignardia mangiferae and new primers for the diagnosis of the citrus black spot. Brazilian Archives of Biology and Technology 52: 1063 – 1073 [Google Scholar]
- Swofford DL.2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, Massachusetts: [Google Scholar]
- White TJ, Bruns T, Lee J, Taylor J.1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (eds), PCR protocols: a guide to methods and applications: 315 – 322 Academic Press, San Diego, California, USA: [Google Scholar]
- Wulandari NF, Toanun C, Hyde KD, Duong LM, Gruyter J de, Meffert JP, Groenewald JZ, Crous PW.2009. Phyllosticta citriasiana sp. nov., the cause of Citrus tan spot of Citrus maxima in Asia. Fungal Diversity 34: 23 – 39 [Google Scholar]