Abstract
In a taxonomic study of yeasts that have been isolated in French Guiana and Thailand, five yeast strains isolated from plants were found to belong to the Yamadazyma clade of Saccharomycotina. On the basis of morphology, physiology and the nucleotide divergence in the D1/D2 domain of the 26S nuclear ribosomal RNA (nrRNA) gene, as well as the internal transcribed spacers (ITS) domain of the nrRNA gene operon, these strains were identified to represent three novel species in this teleomorphic clade. An additional isolate, that is publicly available from the CBS yeast collection and isolated from Taiwan, was found to be similar to one of the novel species described from Thailand. Yeast species belonging to the Yamadazyma clade have previously been described as members of the Candida membranifaciens clade. These species are widely distributed and were isolated from diverse habitats, including water, plants, animals and guts of insects and termites. In the present study the ITS region is shown to be a valuable region for species identification within this clade, and the novel species proposed are Candida vaughaniae (ex-type strain CBS 8583), Candida khao-thaluensis (ex-type strain CBS 8535) and Candida tallmaniae (ex-type strain CBS 8575).
Keywords: French Guiana, systematics, Thailand, Yamadazyma
INTRODUCTION
Due to the recent increase in yeast taxonomic studies that focus on Northeast Asia, Thailand has been shown to have a much higher general diversity of yeast species found in natural substrates than previously thought. The number of novel yeast and hyphomycetous yeast-like species described from Thailand has increased drastically in the past few years to more than 70. These include species of Bullera, Candida, Hortaea, Kockovaella, Ogataea, Pseudozyma and Sporobolomyces (Robert et al. 2008, Crous et al. 2009, Nakase et al. 2009, 2010a). This is in contrast to French Guiana, located in the northern part of South America, where very little information is available on the general diversity of novel or even described yeast species found in the natural environment in this country (CBS yeast online database at www.cbs.knaw.nl/yeast/, Robert et al. 2008, Groenewald et al. 2010). Yeast species belonging to Yamadazyma have previously been described as members of the Candida membranifaciens clade (Suh et al. 2005). Isolates of these species are very common and can be isolated from diverse habitats, such as water, plants, animals and guts of insects and termites (Suh et al. 2005, Ganter 2006). Billon-Grand (1989) introduced Yamadazyma to accommodate 16 species of Pichia that formed coenzyme nine (CoQ-9) as their major ubiquinone. Additionally, these isolates also had pseudohyphae, hat-shaped ascospores that were usually liberated from the ascus at maturity and had the ability to ferment sugars but could not grow in the absence of vitamins. Unfortunately the genus was not generally accepted as the polyphyletic nature of Yamadazyma became evident from a D1/D2 sequence analysis a few years later (Kurtzman & Robnett 1998). Two gene regions, namely the D1/D2 domains of the large subunit (LSU) and the nearly complete small subunit (SSU) nrRNA gene, have been used in the past to identify species that belong to the C. membranifaciens (= Yamadazyma) phylogenetic clade (Suh et al. 2005). Recently, Kurtzman & Suzuki (2010) defined the genus Yamadazyma phylogenetically, making use of the D1/D2 and the nearly complete SSU regions, and showed that an additional 11 CoQ-9-forming Candida species are also part of this teleomorphic clade. The genus Yamadazyma was then also placed in the family Debaryomycetaceae, described by Kurtzman & Suzuki (2010).
In this study five strains isolated from plants in French Guiana and Thailand (four and one strains, respectively), were characterised on the basis of morphology, physiology and phylogenetic characters and found to represent three novel species of the genus Yamadazyma. We determined both the ITS and D1/D2 domains of the 26S nrDNA for our isolates, and as the ITS region was only publicly available for a number of the type strains of previously studied Yamadazyma species, we sequenced this region for additional strains during this study to compare the variation among the closely related species between ITS and the D1/D2 domains.
MATERIALS AND METHODS
Strain information
Four strains, CBS 8583T, CBS 8573, CBS 8575T and CBS 8576 were isolated from flowers of an unidentified plant in La Chaumiere, French Guiana, and CBS 8535T was isolated from an unidentified tree in Khao Thalu, Thailand. The strains were collected during an expedition of the British Mycological Society to Khao Yai National Park, Thailand, in August 1997, and to French Guiana in January 1997. The protocols described by Robert et al. (1998) for sample collection, enrichment and isolation were used. Samples were collected in sterile syringes and enriched in dextrose-yeast-peptone (DYP) broth adjusted to pH 3.5. Strains were isolated subsequently on DYP agar augmented with chloramphenicol (200 mg/L). All cultures obtained in this study are maintained in the culture collection of the CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands. Nomenclature descriptions are deposited in MycoBank (www.MycoBank.org; Crous et al. 2004).
Genotypic characterisation
DNA was extracted from cultures grown on GPYA medium (4 % glucose, 0.5 % peptone, 0.5 % yeast autolysate, 2 % agar) for 3 d using the FastDNA kit (BIO101, Carlsbad, CA, USA) with the ‘FastPrep’ Instrument (Q-Biogene). Primers V9G (de Hoog & Gerrits van den Ende 1998) and LR5 (Vilgalys & Hester 1990) were used to amplify the partial nrRNA gene that includes, the 3′ end of the small-subunit nrDNA, the D1/D2 domain of the large-subunit nrDNA, as well as the ITS (internal transcribed spacer) domain (ITS 1, ITS 2 and the intervening 5.8S nrRNA gene) as described by Knutsen et al. (2007). The PCR products were separated by electrophoresis at 80 V for 40 min on a 0.8 % (w/v) agarose gel containing 0.1 μg/mL ethidium bromide in 1 × TAE buffer (0.4 M Tris, 0.05 M NaAc and 0.01 M EDTA, pH 7.85) and examined under UV-light. The amplicons were sequenced in both directions using the primers LR0R (Vilgalys & Hester 1990) and LR5 for the D1/D2 domain, and the primers V9G and ITS4 (White et al. 1990) were used for the ITS domain. The BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems) was used according to the manufacturer’s recommendations and the products were analyzed on an ABI Prism 3730XL DNA Sequencer (Perkin-Elmer). A consensus sequence was computed from the forward and reverse sequences with SeqMan v8 from the Lasergene package (DNASTAR). All sequences were assembled and aligned using the online version of MAFFT (v6, http://mafft.cbrc.jp/alignment/server/index.html; Katoh et al. 2002) and manual adjustments for improvement were made using Sequence Alignment Editor (v2.0a11; Rambaut 2002).
The sequence data were analysed using Phylogenetic Analysis Using Parsimony (PAUP) v4.0b10 (Swofford 2003) and the resulting trees were printed as described by Groenewald et al. (2008). Maximum parsimony analyses were performed using the heuristic search option with 1 000 random taxon additions and the robustness of the trees was evaluated by 1 000 bootstrap replicates. Other measures calculated included tree length, consistency index, retention index and rescaled consistency index (TL, CI, RI and RC, respectively). Neighbour-joining analyses using different substitution models (HKY85, Kimura 2-parameter and uncorrected ‘p’) were also performed. All analyses were done where gaps were treated as either missing data or fifth characters (‘new state’). The sequences were deposited in GenBank (Fig. 1) and alignments in TreeBASE (Submission nr 11143, www.treebase.org).
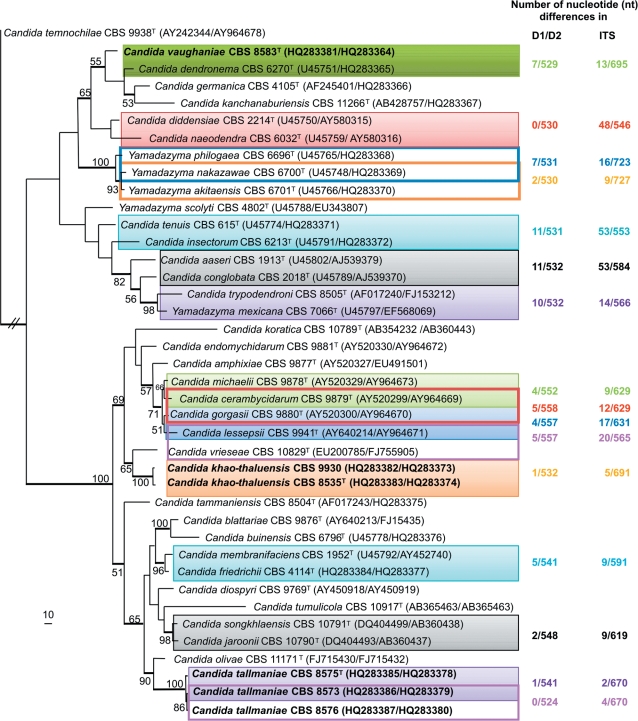
Fig. 1.
The first of five equally most parsimonious trees obtained from a heuristic search with 1 000 random taxon additions of the combined D1/D2 and ITS sequence alignment (TL = 1 521 steps, CI = 0.446, RI = 0.696, RC = 0.311). The scale bar shows 10 changes and bootstrap support values (> 49 %) from 1 000 replicates are shown as percentages at the nodes. Thickened lines indicate the branches present in the strict consensus tree. The three new species, Candida vaughaniae, C. khao-thaluensis and C. tallmaniae, are indicated in bold. The number of nucleotide differences observed between the D1/D2 as well as the ITS regions of the full length sequences present in GenBank of closely related Yamadazyma species are indicated on the right hand side and colour coded with the blocks surrounding the species that the differences refer to. These differences are only indicated for species with 11 nt or less differences in their D1/D2 region. The tree was rooted to Candida temnochilae (GenBank AY242344 and AY964678 for D1/D2 and ITS, respectively).
Morphologic and phenotypic characterisation
The morphology of colonies and cells were determined after growth for 3 d at 25 °C on GPYA for all strains studied. Physiological characteristics were determined using the ID 32C system (bioMe’rieux, Marcy-l’Etoile, France) according to the manufacturer’s instructions and assimilation of nitrogen compounds and fermentation of glucose were tested using the methods described by Yarrow (1998) for all strains listed in Table 1. Data were retrieved after 5 d. Growth at different temperatures ranging from 30–40 °C was determined by incubation on GPYA for 7 d. All tests were replicated.
Table 1.
Physiological characters of the novel strains related to Yamazyma species.
|
C. khao-thaluensis |
C. tallmaniae |
C. vaughaniae |
||||
|---|---|---|---|---|---|---|
| CBS 8535T | CBS 9930 | CBS 8575T | CBS 8573 | CBS 8576 | CBS 8583T | |
| Fermentation | ||||||
| Glucose | + | + | + | + | + | + |
| Assimilation | ||||||
| D-Glucose | + | + | + | + | + | + |
| Sucrose | + | + | + | + | + | + |
| Raffinose | − | − | − | − | − | − |
| Melibiose | − | − | − | − | − | − |
| D-Galactose | + | + | + | + | + | + |
| D-Lactose | − | − | − | − | − | − |
| D-Trehalose | + | + | + | + | + | + |
| Maltose | + | + | + | + | + | + |
| Melezitose | + | + | + | + | + | + |
| Methyl-α-D-Glucoside | + | + | + | + | + | + |
| Cellobiose | + | + | + | + | + | + |
| L-Sorbose | − | − | + | + | + | + |
| L-Rhamnose | + | + | − | − | − | − |
| D-Xylose | + | + | + | + | + | + |
| L-Arabinose | + | + | + | + | + | + |
| D-Ribose | + | w | − | + | + | + |
| Glycerol | + | + | + | + | + | + |
| Erythritol | + | + | + | + | + | + |
| D-Mannitol | + | + | + | + | + | + |
| D-Glucitol | + | + | + | + | + | + |
| Inositol | − | − | − | − | − | − |
| DL-Lactate | − | − | − | − | − | − |
| Potassuim-2-Keto-D-gluconate | − | − | − | − | − | − |
| D-Gluconate | + | + | + | + | + | + |
| D-Glucoronate | − | − | − | − | − | − |
| D-Glucosamine | + | +/w | + | + | + | + |
| Nitrate | − | − | − | − | − | − |
| Ethylamine | + | + | + | + | + | + |
| L-Lysine | + | + | + | + | + | + |
| Cadaverine | + | + | + | + | + | + |
| D-Glucoseamine-HCl | − | − | − | − | − | − |
| D-Tryptophane | − | − | − | − | − | − |
| 0.01 % Cycloheximide | − | − | − | − | − | − |
| Growth at 25 °C | + | + | + | + | + | + |
| Growth at 30 °C | + | + | − | + | + | + |
| Growth at 33 °C | + | + | − | − | + | + |
| Growth at 37 °C | − | − | − | − | − | − |
− = negative; + = positive; w = weak.
Ascospore production of all new strains was examined. Three different media were tested, namely 5 % Difco malt extract, V8 and YM agars (Yarrow 1998). The cultures were incubated at 25 °C and inspected at 3–7 d intervals for 2 mo. All cultures were tested individually. Additionally, ascospore production was also examined in a mixture of CBS 8575T, CBS 8573 and CBS 8576 as well as for CBS 8535T and CBS 9930.
RESULTS
Sequence comparison
All sequences of the studied strains were blasted against sequences in GenBank, EMBL, DDBJ and PDB (http://blast.ncbi.nlm.nih.gov/) as well as against available sequences of strains present in the CBS yeast sequence database (www.cbs.knaw.nl/yeast/; Robert et al. 2008) to make sure these strains indeed represents novel species. The D1/D2 and ITS sequences of the new species are unique and do not match any known yeast species in GenBank or the CBS yeast sequence database except for CBS 8535T whose sequences were similar to an unidentified Candida species (CBS 9930). This strain, that is publicly available from the CBS yeast collection, was isolated from leaves of Schefflera octophylla in Taiwan and deposited in 2004 in the culture collection of the CBS by S.H. Yang.
The combined D1/D2 and ITS sequence alignment (deposited in TreeBASE), containing 40 strains including the outgroup sequence, had a total length of 1 120 characters, of which 704 were constant, 130 were phylogenetically uninformative and 286 were informative. Parsimony analysis resulted in five equally most parsimonious trees, the first of which is shown in Fig. 1. For this tree, alignment gaps were treated as fifth character. The five trees obtained differ with respect to the position of some of the species within the Yamadazyma clade. However, the clustering of the six strains studied were the same in all trees obtained regarding their closely related species (Fig. 1). The trees obtained where gaps were treated as missing also did not differ with respect to the position of the new strains with respect to the neighbouring strains found in Fig. 1 (data not shown). Neighbour-joining analyses, using different substitution models done on this alignment gave similar tree topologies, but differ slightly from the parsimony tree with regard to the position of some of the isolates within the bottom clade (Fig. 1, data not shown). These slight differences in the position of the strains made no significant change in the general topology of the tree with respect to relatedness of species to one another and have therefore no effect on the interpretation of the results.
For all analyses done, the close relatives of CBS 8583T are C. dendronema (CBS 6270T), C. germanica (CBS 4105T), C. kanchanaburiensis (CBS 11266T), C. diddensiae (CBS 2214T) and C. naeodendra (CBS 6032T) (Fig. 1) and differs in the 529 nucleotide (nt) D1/D2 region with seven substitutions, 12 substitutions and one gap, 12 substitutions and four gaps, three substitutions and three substitutions from these species respectively. In the ITS region eight substitutions and five gaps in a 695 nt region, 12 substitutions and seven gaps in a 709 nt region, 20 substitutions and 20 gaps in a 706 nt region, 16 substitutions and 10 gaps in a 548 nt region and 19 substitutions and 27 gaps in a 548 nt region were found respectively.
Isolates CBS 8535T, CBS 8573, CBS 8575T and CBS 8576 cluster in all analyses within the C. membranifaciens clade with 100 % bootstrap support (Fig. 1). Isolates CBS 8573, CBS 8575T and CBS 8576 were all obtained from the same location but from different plants. CBS 8575T differs from CBS 8573 and CBS 8576 with one and two substitutions in the D1/D2 region respectively, and with two and four nucleotide differences in the ITS region respectively. These three strains are therefore seen as conspecific. Their closest relative is C. olivae (CBS 11171T) from which CBS 8575T differs with five substitutions and nine gaps in the 548 nt D1/D2 region and 23 substitutions and 18 gaps in the 588 nt ITS region.
Isolate CBS 8535T differs with one substitution in the D1/D2 region and one substitution and four gaps in the ITS region from CBS 9930, indicating that these two strains are also conspecific. CBS 8535T differs with eight substitutions in the 551 nt D1/D2 region and 21 substitutions and 19 gaps in the 567 nt ITS region from CBS 10829T, the type strain of C. vrieseae.
Comparing the sequence variation within the D1/D2 and ITS regions between two closely related Yamadazyma species as well as among strains of two of the novel species, shows that the ITS region is more variable than that of the D1/D2 domain. None to one nucleotide difference in the D1/D2 domain was found among strains of the same species tested during this study, whereas up to 5 nt differences could be found in the ITS region of CBS 9930 and CBS 8535T. The interspecies variation tested for these two loci indicates little variation within the D1/D2 domain as in some cases only two nucleotide differences between two species are found, where nine or more nucleotide differences can be found between the ITS regions (Fig. 1).
Mating studies and physiology
No asci with ascospores were found for CBS 8583T tested alone and no conjugation of cells or asci with ascospores were found in any combination among CBS 8573, CBS 8575T and CBS 8576 or in a mixture between CBS 8535T and CBS 9930.
The physiological characters of all the strains tested in this study are presented in Table 1. These strains were found to be similar in their abilities to ferment glucose and assimilate different carbon and nitrogen sources, apart for the assimilation of D-ribose, L-rhamnose and L-sorbose where CBS 8575 was the only strain negative for D-ribose and CBS 8535T and CBS 9930 the only strains positive for L-rhamnose and negative for L-sorbose. The maximum temperatures where growth was detected for the three conspecific strains CBS 8573 (30 °C), CBS 8575 (25 °C) and CBS 8576 (33 °C) also differ. The additional conspecific strains (CBS 8535T and CBS 9930) were found to be similar in all the physiological and temperature analyses done.
Taxonomy
Based on their morphology, physiology and the two molecular markers, ITS and D1/D2, used in this study, the strains above are well supported to represent novel species within Yamadazyma. The three species, Candida khao-thaluensis, C. tallmaniae and C. vaughaniae are described formally below.
Candida khao-thaluensis M. Groenew., M.T. Sm. & V. Robert, sp. nov. — MycoBank MB517412; Fig 2
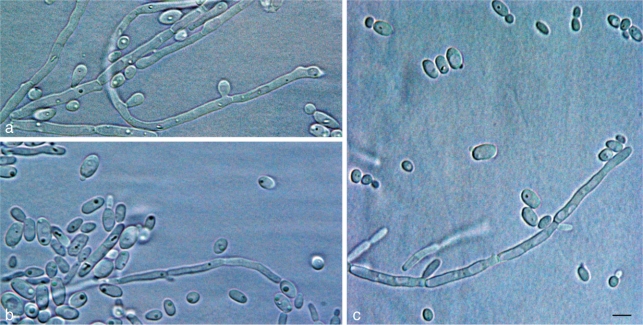
Fig. 2.
Vegetative cells and pseudohyphae. a. Candida vaughaniae (CBS 8583T); b. Candida khao-thaluensis (CBS 8535T); c. Candida tallmaniae (CBS 8575T). — Scale bar = 5 μm.
Post 3 dies 25 °C in agaro cum dextroso, peptono et extracto levedinis, cultura alba, nitens, butyrosa, infirmo-convexa. Cellulae vegetativae globosae vel ovoidae, 1.5–4 × 2–6 μm, singulae aut binae. Pseudohyphae formantur. Ascosporae non formantur. Glucosum fermentatur. Glucosum, galactosum, sucrosum, maltosum, cellobiosum, trehalosum, melezitosum, D-ribosum, D-xylosum (lente), L-arabinosum, L-rhamnosum, glycerolum, erythritolum, α-methyl-D-glucosidum, D-glucosaminum (lente), D-mannitolum, D-glucitolum, et D-gluconas, assimilantur at non L-sorbosum, lactosum, melibiosum, raffinosum, inositolum, potassium-2-keto-D-gluconas et D-glucuronas, DL-acidum-lacticum. Ethylaminum, lysinum et cadaverinum assimilantur at non potasii nitras, D-glucosaminum et tryptophanum. Non crescit in medio 0.01 % cycloheximido addito. Augmentum ad 33 °C at non ad 37 °C.
Etymology. Epithet is chosen after the collection site Khao Thalu in Thailand.
After 3 d at 25 °C on glucose-yeast extract-peptone agar, the culture is white, smooth, glossy, butyrous, and low-convex. The cells are spherical to ovoid, 1.5–4 × 2–6 μm, single or in pairs. Pseudohyphae are formed. Asci with ascospores are not produced. Fermentation and growth on various carbon and nitrogen compounds are presented in Table 1.
Specimens examined. Taiwan, Pingtung, on leaves of Schefflera octophylla, 29 July 2001, S.H. Yang, culture CBS 9930. – Thailand, Khao Thalu, on an unidentified tree, Jan. 1997, V. Robert, holotype CBS H-20540, culture ex-type CBS 8535.
Candida tallmaniae M. Groenew., M.T. Sm. & V. Robert, sp. nov. — MycoBank MB517413; Fig 2
Post 3 dies 25 °C in agaro cum dextroso, peptono et extracto levedinis, cultura alba, nitens, butyrosa, and infirm-convexa. Cellulae vegetativae globosae vel ovoidae, 1–4 × 3–6 μm, vel cylindricae, 1–2 × 5–11 μm, singulae vel binae. Pseudohyphae formantur. Ascosporae non formantur. Glucosum fermentatur. Glucosum, galactosum, L-sorbosum, sucrosum, maltosum, cellobiosum, trehalosum, melezitosum, D-ribosum, D-xylosum, L-arabinosum, glycerolum, erythritolum, α-methyl-D-glucosidum, D-glucosaminum, D-mannitolum, D-glucitolum, et D-gluconas, assimilantur at non lactosum, melibiosum, raffinosum, L-rhamnosum, inositolum, potassium-2-keto-D-gluconas et D-glucuronas, DL-acidum-lacticum. Ethylaminum, lysinum et cadaverinum assimilantur at non potasii nitras, D-glucosaminum et tryptophanum. Non crescit in medio 0.01 % cycloheximido addito. Augmentum negativum ad 37 °C at variabile ad 30 °C et 33 °C.
Etymology. Named in honour of the valuable contribution of A. Statzell-Tallman in yeast taxonomy.
After 3 d at 25 °C on glucose-yeast extract-peptone agar, the culture is white, smooth, glossy, butyrous, and low-convex. The cells are spherical to ovoid, 1–4 × 3–6 μm, to cylindrical, 1–2 × 5–11 μm, single or in pairs. Pseudohyphae are formed. Ascospores are not produced. Fermentation and growth on various carbon and nitrogen compounds are presented in Table 1.
Specimens examined. French Guiana, La Chaumiere, on flower of an unidentified plant, Jan. 1997, V. Robert, holotype CBS H-20541, culture type CBS 8575; on flower of an unidentified plant, Jan. 1997, V. Robert, culture CBS 8573; on flower of an unidentified plant, Jan. 1997, V. Robert, culture CBS 8576.
Candida vaughaniae M. Groenew., M.T. Sm. & V. Robert, sp. nov. — MycoBank MB517411; Fig 2
Post 3 dies 25 °C in agaro cum dextroso, peptono et extracto levedinis, cultura alba, stolida, butyrosa, convexa. Cellulae vegetativae globosae, ovoidae, vel ellipsoidae, 2–4 × 2–7 μm, vel cylindricae, 1.5–2 × 7–13 μm, singulae aut binae. Pseudohyphae formantur. Ascosporae non fiunt. Glucosum fermentatur. Glucosum, galactosum, L-sorbosum, sucrosum, maltosum, cellobiosum, trehalosum, melezitosum, D-ribosum, D-xylosum, L-arabinosum, glycerolum, erythritolum, α-methyl-D-glucosidum, D-glucosaminum, D-mannitolum, D-glucitolum, et D-gluconas, assimilantur at non lactosum, melibiosum, raffinosum, L-rhamnosum, inositolum, potassium-2-keto-D-gluconas et D-glucuronas, DL-acidum-lacticum. Ethylaminum, lysinum et cadaverinum assimilantur at non potasii nitras, D-glucosaminum et tryptophanum. Non crescit in medio 0.01 % cycloheximido addito. Augmentum ad 33 °C at non ad 37 °C.
Etymology. Named in honour of the valuable contribution of A. Vaughan-Martini in ascomycetous yeast taxonomy.
After 3 d at 25 °C on glucose-yeast extract-peptone agar, the culture is white, smooth, butyrous, dull, and convex. The cells are spherical, ovoid, to ellipsoidal, 2–4 × 2–7 μm, to cylindrical, 1.5–2 × 7–13 μm, single or in pairs. Pseudohyphae are formed. Asci with ascospores are not produced. Fermentation and growth on various carbon and nitrogen compounds are presented in Table 1.
Specimen examined. French Guiana, La Chaumiere, on flower of unidentified plant, Jan. 1997, V. Robert, holotype CBS H-20539, culture ex-type CBS 8583.
DISCUSSION
The drastic increase of novel yeast species described and general diversity of yeast species found in Thailand during the past few years are possibly due to the recent increase in yeast taxonomic studies focusing on isolates obtained specifically from this region. The species described during this study from French Guiana represent two of only six species described to date from this country. The four species already described in an earlier study from French Guiana by Groenewald et al. (2010) are Candida eppingiae, Candida pseudoflosculorum, Candida robnettiae and Wickerhamomyces chaumierensis, and were obtained from the same locality from which the strains were isolated for this study. The explanation for the low number of species obtained from this country thus far can be attributed to the fact that intensive collecting and isolating of yeast strains have not been done or published so far from this area. The fact that six novel species have been isolated from plants in the same region gives the impression that this country could have a high diversity of yeast species on natural substrates and that novel species are just waiting to be isolated and identified.
Within the Yamadazyma clade, both homothallic (Y. philogaea) and heterothallic (Y. mexicana and Y. scolyti) species are present (Kurtzman & Fell 1998). As no ascospores were found on ascospores-inducing medium for the C. vaughaniae strain, the three C. tallmaniae strains as well as the two C. khao-thaluensis strains, the reasons for the unsuccessful mating can be that the conditions used to induce mating were not optimal, or that the strains could have lost their ability to produce ascospores, or that they represent only one mating type. Although mating could not be achieved and there were no significant differences found within the physiological characters tested for these strains except for the variation in the maximum growth temperature for CBS 8573, CBS 8575T and CBS 8576, these novel species and their phylogenetic positions within Yamadazyma are well supported based on the molecular markers used.
Significant variation in the nucleotide differences among the novel strains studied and their closest relatives was observed, with three to 16 nucleotide differences for the D1/D2 region and 13 to 46 differences for the ITS region. The three nucleotide differences found in the D1/D2 region in contrast to the 46 differences in the ITS region between CBS 8583T and C. naeodendra can be seen as a good example why the D1/D2 region, a region that is still commonly used as an exclusive marker in studies for the identification and delimitation of species belonging to commonly known yeast genera (Nakase et al. 2008, 2010a, b, Ganter et al. 2010, de Garcia et al. 2010, Mestre et al. 2010), alone is often not sufficient for species delimitation in yeasts; especially within the Yamadazyma clade. Several other yeast taxonomic studies have also proven this to be true in species from other genera such as Blastobotrys (Kurtzman 2007), Debaryomyces (Groenewald et al. 2008), Saturnispora (Canelhas et al. 2010) and Rhodotorula (Libkind et al. 2010), and shown that sequences of additional regions are necessary in order to separate the closely related species from one another.
The results obtained from the ITS region show that this gene region is a useful marker for species delimitation within the Yamadazyma clade. For closely related species within Yamadazyma (Fig. 1) none to only two substitutions can be found in the D1/D2 domains of the ex-type strains of the closest related species, whereas the ITS regions show more variation between two closely related well-defined species. This is true for C. diddensiae and C. naeodendra, Y. akitaensis and Y. nakazawae as well as C. jaroonii and C. songkhlaensis, where in contrast to the low nucleotide differences in the D1/D2 domain, nine to 48 differences can be found between the ITS regions of these species. The four and five nucleotide differences found between the strains of C. tallmaniae and C. khuo-thaluensis respectively indicate that the nine and more differences found between different Yamadazyma species are most likely not population differences but sufficient in separating the strains at species level. An approximately 1 750 nt region of the SSU region, have been used in the past as an alternative region in combination with the D1/D2 domain to identify species that belong to the Yamadazyma phylogenetic clade (Suh et al. 2005, Kurtzman & Suzuki 2010). Only four nucleotide differences were found within the SSU regions between Y. nakazawae (AB054287) and Y. akitaensis (AB054279) as well as between C. membranifaciens (AB013551) and C. friedrichii (AB013531) with the nine differences found between each of these species in the much smaller ITS regions. This is an additional indication that the ITS fragment, which can be easily amplified as part of a fragment containing also the D1/D2 region, is as sufficient to separate closely related species from one another. It is therefore a good region, in combination with the commonly used D1/D2 region, for species delimitation within Yamadazyma and most likely for other yeast genera as well (see studies by Kurtzman 2007, Canelhas et al. 2010, Statzell-Tallman et al. 2010, Wang et al. 2010). Although D1/D2 is still an appropriate region to use for higher level taxon delimitations, it is clear from this study that the D1/D2 region alone is not sufficient for species delimitation in the Yamadazyma clade also showing that the ITS region is a good alternative marker to obtain a better understanding of relatedness of the different Yamadazyma species, including its anamorphs.
Acknowledgments
Wendy Epping is thanked for technical assistance, Ewald (J.Z.) Groenewald for valuable discussions. S.H. Yang is thanked for depositing the strain designated as CBS 9930 in the CBS culture collection and making it available for public use.
REFERENCES
- Billon-Grand G.1989. A new ascosporogenous yeast genus: Yamadazyma gen. nov. Mycotaxon 35: 201 – 204 [Google Scholar]
- Canelhas MR, Barbosa AC, Medeiros AO, Lee C-F, Huang L-Y, Lachance M-A, Rosa CA.2010. Saturnispora serradocipensis sp. nov. and Saturnispora gosingensis sp. nov., two ascomycetous yeasts from ephemeral habitats. Antonie van Leeuwenhoek doi: 10.1007/s10482-010-9482-9 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G.2004. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19 – 22 [Google Scholar]
- Crous PW, Schoch CL, Hyde KD, Wood AR, Gueidan C, Hoog GS de, Groenewald JZ.2009. Phylogenetic lineages in the Capnodiales. Studies in Mycology 64: 17 – 47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganter PF.2006. Yeast and invertebrate associations. In: Rosa CA, Peter G. (eds), The yeast handbook, Biodiversity and ecophysiology of yeasts, 1st edn.: 303 – 370 Heidelberg, Springer: [Google Scholar]
- Ganter PF, Cardinali G, Boundy-Mills K.2010. Pichia insulana sp. nov., a novel cactophilic yeast from the Caribbean. International Journal of Systematic and Evolutionary Microbiology 60: 1001 – 1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V de, Brizzio S, Libkind D, Rosa CA, Broock M van.2010. Wickerhamomyces patogonicus sp. nov., an ascomycetous yeast species from Patagonia, Argentina. International Journal of Systematic and Evolutionary Microbiology 60: 1693 – 1696 [DOI] [PubMed] [Google Scholar]
- Groenewald M, Daniel H-M, Robert V, Poot GA, Smith MTh.2008. Polyphasic re-examination of Debaryomyces hansenii strains and re-instatement of D. hansenii, D. fabryi and D. subglobosus. Persoonia 21: 17 – 27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenewald M, Robert V, Smith MTh.2010. Novel species related to Wickerhamomyces and Metschnikowia isolated in Thailand and Guiana. International Journal of Systematic and Evolutionary Microbiology doi: 10.1099/ijs.0.026062-0 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Hoog GS de, Gerrits van den Ende AHG.1998. Molecular diagnostics of clinical strains of filamentous basidiomycetes. Mycoses 41: 183 – 189 [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T.2002. MAFFT: a novel method for rapid multiple sequence alignment based on Fast Fourier Transform (describes the FFT-NS-1, FFT-NS-2 and FFT-NS-i strategies). Nucleic Acids Research 30: 3059 – 3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutsen AK, Robert V, Poot GA, Epping W, Figge M, Holst-Jensen A, Skaar I, Smith MTh.2007. Polyphasic re-examination of Yarrowia lipolytica strains and the description of three novel Candida species: Candida osloensis sp. nov., Candida alimentaria sp. nov. and Candida hollandica sp. nov. International Journal of Systematic and Evolutionary Microbiology 57: 2426 – 2435 [DOI] [PubMed] [Google Scholar]
- Kurtzman CP.2007. Blastobotrys americana sp. nov., Blastobotrys illinoisensis sp. nov., Blastobotrys malayseinsis sp. nov., Blastobotrys muscicola sp. nov., Blastobotrys peoriensis sp. nov. and Blastobotrys raffinosifermentans sp. nov., novel anamorphic yeast species. International Journal of Systematic and Evolutionary Microbiology 57: 1154 – 1162 [DOI] [PubMed] [Google Scholar]
- Kurtzman CP, Fell JW. (eds). 1998. The yeasts: a taxonomic study. 4th ed Elsevier, Amsterdam: [Google Scholar]
- Kurtzman CP, Robnett CJ.1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek 73: 331 – 371 [DOI] [PubMed] [Google Scholar]
- Kurtzman CP, Suzuki M.2010. Phylogenetic analysis of ascomycete yeasts that form coenzyme Q-9 and the proposal of the new genera Babjeviella, Meyerozyma, Millerozyma, Priceomyces, and Scheffersomyces. Mycoscience 51: 2 – 14 [Google Scholar]
- Libkind D, Sampaio JP, Broock M van.2010. Cystobasidiomyces yeasts from Patagonia (Argentina): description of Rhodotorula meli sp. nov. from glacial meltwater. International Journal of Systematic and Evolutionary Microbiology 60: 2251 – 2256 [DOI] [PubMed] [Google Scholar]
- Mestre MC, Ulloa JR, Rosa CA, Lachance MA, Fontenla S.2010. Lachancea nothofagi sp. nov., a yeast associated with Nothofagus species in Patagonia, Argentina. International Journal of Systematic and Evolutionary Microbiology 60: 2247 – 2250 [DOI] [PubMed] [Google Scholar]
- Nakase T, Jindamorakot S, Am-In S, Ninomiya S, Kawasaki H.2010a. Candida tanticharoeniae sp. nov., a novel anamorphic yeast species found in Thailand. Journal of General and Applied Microbiology 56: 89 – 92 [DOI] [PubMed] [Google Scholar]
- Nakase T, Jindamorakot S, Ninomiya S, Imanishi Y, Kawasaki H.2009. Candida wancherniae sp. nov. and Candida marokotiae sp. nov., two novel ascomycetous anamorphic yeast species found in Thailand. Journal of General and Applied Microbiology 55: 93 – 100 [DOI] [PubMed] [Google Scholar]
- Nakase T, Jindamorakot S, Ninomiya S, Imanishi Y, Kawasaki H, Potacharoen W.2008. Candida kanchanaburiensis sp. nov., a new ascomycetous yeast species related to Pichia nakazawae isolated in Thailand. Journal of General and Applied Microbiology 54: 259 – 265 [DOI] [PubMed] [Google Scholar]
- Nakase T, Jindamorakot S, Tanaka K, Ninomiya S, Imanishi Y, Kawasaki H, Limtong S, Lee C-F.2010b. Vanderwaltozyma tropicalis sp. nov., a novel ascomycetous yeast species found in Thailand. Journal of General and Applied Microbiology 56: 31 – 36 [DOI] [PubMed] [Google Scholar]
- Rambaut A.2002. SE-AL Sequence Alignment Editor v2.0a11. University of Oxford, Oxford, UK: [Google Scholar]
- Robert V, Bonjean B, Untereiner W.1998. Problems and prospects linked to studies of yeast biodiversity. Proceedings of the Asia-Pacific Mycology Conference: 56 – 67 [Google Scholar]
- Robert V, Groenewald M, Epping W, Boekhout T, Smith MTh, Stalpers J.2008. CBS yeasts database. Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands: [Google Scholar]
- Statzell-Tallman A, Scorzetti G, Fell JW.2010. Three species of yeasts: Candida spencermartinsiae, Candida taylori and Pseudozyma abaconensis spp. nov. from mangrove and coral reef ecosystems. International Journal of Systematic and Evolutionary Microbiology 60: 1978 – 1984 [DOI] [PubMed] [Google Scholar]
- Suh S-O, Nguyen NH, Blackwell M.2005. Nine new Candida species near C. membranifaciens isolated from insects. Mycological Research 109: 1045 – 1056 [DOI] [PubMed] [Google Scholar]
- Swofford DL.2003. PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, Massachusetts, USA: [Google Scholar]
- Vilgalys R, Hester M.1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238 – 4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S-A, Li F-L, Bai F-Y.2010. Candida laoshanensis sp. nov. and Candida qingdaonensis sp. nov., novel anamorphic, ascomycetous yeast species isolated from decayed wood. International Journal of Systematic and Evolutionary Microbiology 60: 1697 – 1701 [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J.1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenics. In: Innis N, Gelfand D, Sninsky J, White T. (eds), PCR protocols: a guide to methods and applications: 315 – 322 Academic Press, New York, USA: [Google Scholar]
- Yarrow D.1998. Methods for the isolation, maintenance and identification of yeasts. In: Kurtzman CP, Fell JW. (eds), The yeasts: a taxonomic study, 4th ed.: 77 – 100 Elsevier, Amsterdam: [Google Scholar]