Abstract
The pancreatic islets of Langerhans play a critical role in maintaining blood glucose homeostasis by secreting insulin and several other important peptide hormones. Impaired insulin secretion due to islet dysfunction is linked to the pathogenesis underlying both Type 1 and Type 2 diabetes. Over the past 5 years, emerging proteomic technologies have been applied to dissect the signaling pathways that regulate islet functions and gain an understanding of the mechanisms of islet dysfunction relevant to diabetes. Herein, we briefly review some of the recent quantitative proteomic studies involving pancreatic islets geared towards gaining a better understanding of islet biology relevant to metabolic diseases.
Keywords: diabetes, insulin resistance, pancreatic β cells, pancreatic islets, quantitative proteomics, signaling pathway
The islets of Langerhans are micro-organs localized within the pancreas that account for nearly 2% of the total pancreatic mass. These islets consist of at least five types of endocrine cells, including glucagon-producing α cells, insulin-producing β cells, somatostatin-producing δ cells, pancreatic polypeptide-producing pancreatic polypeptide cells and ghrelin-producing ε cells [1]. Among these cells, the insulin-producing β cells are the predominant cell type, contributing to approximately 70% of all islet cells [2], and the secreted insulin serves as the primary regulator of blood glucose homeostasis. Elevated blood glucose levels promote insulin synthesis, as well as its secretion from β cell secretory granules. In turn, the secreted insulin promotes glucose uptake in peripheral insulin-sensitive tissues, such as liver, muscle and adipose tissues, thus regulating blood glucose back to normal levels.
Impaired insulin secretion from pancreatic β cells is one of the major factors associated with both Type 1 and Type 2 diabetes mellitus. Type 1 diabetes mellitus (T1DM) is characterized by a loss of insulin-producing β cells due to an autoimmune attack that often leads to complete insulin deficiency. On the other hand, Type 2 diabetes mellitus (T2DM) is characterized by a relative reduction in insulin secretion due to β-cell loss or β-cell dysfunction combined with insulin resistance in the peripheral tissues. A significant loss of β cell mass is characteristic in T1DM and late stages of T2DM [3–5]. There are several lines of evidence pointing to a variety of mechanisms for β cell loss, including autoimmune attack [6], lipotoxicity [7,8], glucose toxicity [9], amyloid deposition [10], glutamate toxicity [11] and altered insulin receptor signaling [12]. Importantly, a more detailed understanding of the molecular pathways associated with β-cell function and the development of diabetes will benefit the development of novel therapeutic strategies for diabetes.
Biological studies geared towards unraveling the molecular mechanisms of islet functions, such as regulation of insulin secretion and β-cell loss, have accelerated as a result of increasingly available genome sequences and continuously improving genomic and proteomic technologies. Benefiting from the recent advances in mass spectrometry (MS) and bioinformatics, proteomics is now capable of identifying and quantifying thousands of proteins from isolated pancreatic islets [13–17], islet-derived primary cell cultures [18], islet-related cell lines [19–21] and subcellular organelles from islet-related cell lines [22–24]. Herein, we review some of the studies involving isolated pancreatic islets, primarily focusing on advances afforded by quantitative proteomic applications in islet biology over the past 5 years. Earlier studies on islet proteomics were covered by a prior review [25]. Before delving into these studies, we will provide a brief overview of the quantitative proteomic technologies.
Quantitative proteomic technologies
Quantitative measurements of differential protein abundances are essential for elucidating molecular pathways associated with a given biological system, such as pancreatic islets. A number of quantitative MS-based strategies have been applied to study pancreatic islets, including gel-based approaches [26,27], as well as label-free [14,16] and stable isotope labeling [15,17] liquid chromatography (LC)-based approaches. In this section we briefly summarize approaches that either have been applied or are applicable to islet studies.
2D gel electrophoresis
Most of the early proteomic studies on islets employed 2DE, a well-established protein separation and visualization technique, for protein profiling and relative quantification [28–31]. The first dimension of 2DE separates proteins on an isoelectric focusing gel based on their isoelectric point, while the second dimension of 2DE separates proteins based on molecular weight [26]. After electrophoresis, the proteins on the gel are visualized and quantified by gel staining, such as Coomassie Brilliant Blue staining and SYPRO Ruby staining, and protein spots of interest are typically excised, digested and then identified using MS. A 2D-DIGE that utilizes fluorescent tags (Cy2, Cy3 and Cy5) to analyze multiple samples simultaneously can further improve protein quantification by 2DE [27,32]. While 2DE is still one of the commonly applied proteomic technologies in diabetes research [33–36], the technique has several inherent limitations, such as the laborious process of identifying proteins from individual gel spots by MS and the low sensitivity for detecting low-abundance proteins, as well as proteins with extreme molecular weight, isoelectric point or hydrophobicity.
Label-free LC–MS or LC–MS/MS quantification
Liquid chromatography coupled with MS or MS/MS affords a highly sensitive analytical tool with a wide dynamic range of detection for identifying and quantifying proteomic changes under different biological conditions [37,38]. Currently there are two general quantification strategies that do not require the use of an isotopic label to discern changes in protein abundance, commonly referred to as spectral counting and LC–MS intensity-based quantification. Both of these approaches have been applied in islets studies [14,16,39]. Spectral counting utilizes the number of MS/MS spectra that identify a given peptide or protein [40–42] as a measure of peptide or protein abundance. While this strategy is simple and straightforward, the quantification of low-abundance proteins is often unreliable because these proteins are typically identified by a small number of spectra. The second strategy utilizes peptide peak intensities or peak areas from LC–MS to quantify relative peptide and protein abundances among different conditions [38,43,44]. An example of the intensity-based LC–MS approach is the accurate mass and time (AMT) tag strategy [45,46], in which peptides are identified by matching the accurate mass and normalized elution time of each detected LC–MS feature to those in a previously established peptide reference database. Following accurate alignment of detected LC–MS features across different analyses, relative quantification of identified peptides is accomplished by comparing integrated MS peak areas among different samples. Similar quantitative approaches that rely on direct LC–MS measurements, feature alignment and peak identifications have also been reported [47–49].
Conceptually, label-free LC–MS quantitative methods have no limit to the number of samples that can be compared and are able to provide good proteome coverage; however, the reliability of label-free approaches is highly dependent on the LC–MS platform reproducibility, which can often be a challenge for analyzing a large-set of samples.
Stable isotope labeling-based quantification
In addition to label-free quantification, stable isotope labeling-based LC–MS quantification has been commonly applied to biological studies [50]. Common stable isotope labeling strategies include metabolic labeling (e.g., stable isotope labeling with amino acids in cell culture [51]), enzymatic labeling (e.g., trypsin-catalyzed 18O labeling [52–54]) and isotopic tagging with chemical reactions (e.g., isobaric tags for relative and absolute quantification [iTRAQ] [55] and tandem mass tags [56]). iTRAQ was recently applied in proteomic studies of islets to identify factors associated with TDM2 diabetes [11,13,50], as well as proteins associated with the islet-like cell differentiation [57].
Both iTRAQ and tandem mass tags, which were developed for multiplexed quantification of four to eight samples in a single LC–MS/MS experiment, are based on specific reactions of isobaric tags with the primary amine groups on peptides, such as, N-termini and lysine side chains [55,56]. Samples are labeled with different versions of isobaric tags and pooled prior to LC–MS/MS. The same peptides labeled with different isobaric tags have exactly the same mass and coelute precisely in LC separations. The quantitative information is obtained from the low-molecular-mass reporter ions with different masses that are generated upon MS/MS fragmentation. For example, in the case of quadruplexed iTRAQ labeling, the masses of reporter ions for different samples are 114, 115, 116 and 117. The intensities of these ions can be used for relative quantification of the peptides across four different conditions.
Compared with label-free LC–MS quantification, stable isotope labeling-based quantification provides more reliable quantitative results; however, the labeling may also lead to less proteome coverage, partially due to the increased sample complexity and complication of peptide fragmentation patterns introduced by the tagging [58].
Proteomic studies on islet biology
Table 1 illustrates the range of islet studies that have applied proteomic techniques to unravel islet biology. In this section, we review these studies with regard to islet proteome profiling, glucose-stimulated islet proteome response, mechanisms of β-cell failure, T2DM and insulin resistance, islet development and regeneration, and diabetic drug response. One of the concerns for MS-based islet proteome profiling is the potential contamination of exocrine acinar tissue in islet preparations as reported by Ahmed et al., and it is important to take such potential contamination into consideration for data interpretation [59].
Table 1.
List of selected literature reports on proteomics studies involving pancreatic islets.
| Study (year) | Species | Condition | Biological replicates |
Technique | Altered proteins | Ref. |
|---|---|---|---|---|---|---|
| Islet proteome profiling | ||||||
| Petyuk et al. (2008) | Mouse | NA | 8 pooled in 1 | 2DLC–MS/MS | [14] | |
| Metz et al. (2006) | Human | NA | 5 pooled in 1 | 2DLC–MS/MS | [13] | |
| Ahmed et al. (2005) | Human | NA | 5 | 2DE | [60] | |
| Glucose-stimulated islet proteome response | ||||||
| Martens et al. (2010) | Rat | Glucose | 3 | LC–MS/MS | 93 | [39] |
| Waanders et al. (2009) | Mouse | Glucose | 1 | LC–MS/MS | 142 | [16] |
| Ahmed et al. (2005) | Mouse | Glucose | 6 | 2DE | 77 | [59] |
| β -cell failure | ||||||
| Karlsen et al. (2006) | Human, rat | IL-1β | 3 | 2DE | 1 | [72] |
| Xie et al. (2008) | Mouse | Streptozotocin | 3 | 2DE | 7 | [35] |
| Johnson et al. (2006) | Human | Insulin | 4 | 2D-DIGE | 36 | [36] |
| T2DM & insulin resistance | ||||||
| Lu et al. (2010) | Mouse | MKR | 3 pools | iTRAQ, 2D LC–MS/MS | 36 | [76] |
| Lu et al. (2008) | Mouse | MKR | 3 pools | iTRAQ, 2D LC–MS/MS | 159 | [17] |
| Han et al. (2010) | Rat | Zucker rat | 3 | iTRAQ, 2D LC–MS/MS | 112 | [15] |
| Qiu et al. (2005) | Mouse | High fat | 3 | 2DE | 4 | [75] |
| Nyblom et al. (2009) | Human | T2DM | 5 | LC–MS/MS, SELDI | 31 from SELDI, 20 from LC–MS |
[78] |
| Islet development & regeneration | ||||||
| Jin et al. (2009) | Human | Development | 2 | iTRAQ, 2D LC–MS/MS | 97/91 | [57] |
| Hong et al. (2005) | Pig | Development | 4 | 2DE | 13 | [18] |
| Diabetic drug response | ||||||
| Kim et al. (2008) | Rat | EPS | 6 | 2DE | 34 | [33] |
| Jagerbrink et al. (2007) | Rat | Imidazoline | 6 | 2DE | 22 | [34] |
EPS: Extracellular polysaccharide; iTRAQ: Isobaric tags for relative and absolute quantification; LC: Liquid chromatography; MKR: Muscle IGF-1 receptor–lysine–arginine; MS: Mass spectrometry; NA: Not applicable; T2DM: Type 2 diabetes.
Islet proteome profiling
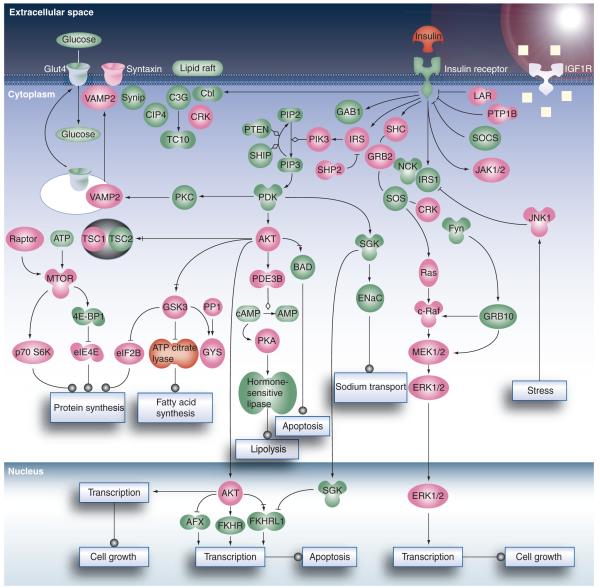
Although the application of 2DE in early proteomic profiling of islets revealed thousands of islet protein spots, only small sets of proteins of interest were excised for identification by MS [35,59–61]. Later studies attained significantly improved islet proteome coverage through the use of 2D LC (i.e., strong cation exchange chromatography coupled with reversed phase LC)–MS/MS. For example, initial LC–MS/MS-based profiling of the human islet proteome by Metz et al. resulted in approximately 3400 confident protein identifications [13]. Similarly, application of 2D LC–MS/MS to profile the mouse islet proteome resulted in 2612 proteins identified with at least two unique peptides [14]. Figure 1 shows the coverage of insulin receptor signaling pathways obtained from this dataset, which illustrates the ability of LC–MS/MS-based proteomics to detect a good percentage of signaling proteins (~45%) in a major signaling pathway. Moreover, a comparison of the islet proteome with the proteomes of eight other mouse tissues (i.e., brain, liver, heart, muscle, kidney, lung, placenta and adipocyte) revealed 133 proteins either specifically or predominantly expressed in pancreatic islets; 68 of which have never been identified in islets previously. In addition to known peptide hormones (insulin, glucagon, peptide YY, pancreatic polypeptide and urocortin-3), these islet-specific proteins included many proteins known to be involved in the regulation of hormone secretion (chromogranins, secretogranins, syntaxins and synaptotagmins), providing confidence in the approach. These observations were consistent with the islet specific transcriptome [62]. Importantly, these islet-specific proteins are potentially interesting candidates for further detailed biological studies in order to investigate their roles in islet biology and their relevance to metabolic diseases.
Figure 1. Coverage of the insulin receptor signaling pathways.
Proteins identified are highlighted as pink. Undetected proteins and low-molecular-weight chemical compounds are shown in green.
Adapted with permission from [14]. © 2008 American Chemical Society.
More recently, LC–MS/MS using an LTQ-Orbitrap mass spectrometer demonstrated sufficient sensitivity for profiling single pancreatic islets [16]; nearly 2000 proteins were identified in single islets. By analyzing pooled islets in the same study, 6873 proteins were confidently identified, representing the most comprehensive islet proteome coverage to date [16]. In general, this extensive islet proteome coverage provides a valuable resource with respect to the proteins and potential signaling pathways that regulate islet function. This information contributes towards the understanding of normal islet function and the pathogenesis of diabetes.
Glucose-stimulated islet proteome response
Since the major function of islets is the secretion of hormones in response to alterations in the concentrations of metabolites (e.g., glucose and free fatty acids), the identification of protein factors or pathways associated with glucose-stimulated insulin secretion is crucial for understanding the underlying mechanisms of diabetes. For this reason, significant efforts have been centered on studying glucose-responsive proteins in islets and islet-related cell lines. In the early 1990s, 2DE studies by Collins et al. revealed numerous islet proteins altered after glucose stimulation; however, these glucose-responsive proteins were not identified at that time [63,64]. More than a decade later, Ahmed et al. applied 2DE coupled with MALDI-TOF-MS to characterize changes in the global mouse islet proteome following stimulation with 11 mM glucose [59]. Among the approximately 1000 protein spots observed, 379 of them were differentially expressed. A total of 77 proteins corresponding to 124 protein spots were identified using MS, including α enolase, endoplasmin, glucose-regulated proteins, heat shock proteins, peroxiredoxins, prohormone convertase 2, protein disulfide isomerase and superoxide dismutase. These results indicated that after glucose stimulation, the activity of insulin synthesis, granular mobilization and maturation and stress response are enhanced in mouse islets.
Hu et al. used a 2D-DIGE approach to identify the islet proteins related to insulin secretion activated by glucose stimulation [65]. Based on a previous observation that α2A adrenergic receptor (α2AAR) attenuates glucose-stimulated insulin release from islet β cells [66], this study compared proteomic differences between islets from α2AAR knockout mice (α2AAR KO) and wild-type mice. The authors were able to identify a small set of proteins significantly upregulated in α2AAR KO mice, including bile salt activated lipase (Bsdl), pancreatic lipase related protein 1 (Plrp1), D-3-phosphogycerate dehydrogenase (3-Pgdh), pancreatic triglyceride lipase (Ptl), carboxypeptidase B1 (Cpb1) and A1 (Cpa1) and peroxiredoxin-4 (Prdx4). Many of the identified upregulated proteins were involved in biosynthesis, enzyme secretion and other pancreatic functions; however, several of these proteins, such as Bsdl, Ptl, Cpb1 and Cpa1, are known as digestive enzymes, potentially from pancreatic acinar tissue [59].
As most of these studies were based on 2DE–MS, the number of glucose-responsive proteins identified was limited by the laborious protein identification process from individual gel spots. The application of LC–MS/MS-based methodologies has the potential to reveal additional glucose-responsive proteins [16].
Mechanisms of β-cell failure
Type 1 diabetes results from irreversible selective destruction of the insulin-producing β cells within the islets. The gradual development of β-cell dysfunction can lead to β-cell failure, with approximately 70–80% loss of β cell mass at the time of diagnosis [4]. Currently, the most accepted hypothesis for β-cell failure in T1DM is that β cells are destroyed by an autoimmune attack. In this hypothesis, the immune system invades the islet space, releases toxic cytokines such as IL-1β, IFN-γ and TNF-α, consequently triggering the apoptosis of β cells [67]. However, the exact molecular mechanism behind this hypothesis is still unclear. Application of quantitative proteomics has the potential to significantly improve our understanding of the mechanisms of β-cell failure and provide potential candidates for therapeutic interventions.
Most of the T1DM-related quantitative proteomic studies on islets to date have been performed using islets isolated from animal models [68] and β cell derived cell lines [69], and involved the use of 2DE-based approaches. For example, Andersen et al. applied 2DE to compare the rat islet proteome change after exposure to IL-1β and identified 33 proteins as being differentially regulated [70]. In a subsequent study that utilized 2DE to profile 35S-methionine-labeled proteins from IL-1β-treated rat islets, abundance alterations for 89 proteins were reported [71]. In 2006, Karlsen et al. also used 2DE to compare rat islet proteome changes after IL-1β treatment and found that gal-3 was the most highly upregulated protein [72]. Further investigation revealed that over expression of gal-3 protected β cells against IL-1β toxicity by completely blocking JNK phosphorylation, which is essential for IL-1-mediated apoptosis. A haplotype comprising three coding single nucleotide polymorphisms showed significantly increased transmission to unaffected offspring in 257 T1DM families and this result was replicated in an independent set of 170 T1DM families [72]. These data suggest gal-3 as a candidate gene/protein that promotes T1DM susceptibility; however, the effect of gal-3 may not be limited to T1DM, since it was also reported to be associated with amyotrophic lateral sclerosis [73].
In a more recent study, Xie et al. generated a mouse model by multiple injections of low-dose streptozotocin to mimic the β-cell failure as observed in T1DM [35,74]. They subsequently analyzed the proteomic changes in the islets using 2DE and MALDI-TOF-MS/MS. Seven proteins were observed to be significantly altered in diabetic mice – that is, ATP synthase subunit β (Atpb), calreticulin (Crtc), lithostathine 1 (Lit1) and 2 (Lit2), Prdx4, ubiquinol-cytochrome-c reductase complex core protein I (Uqcr1) and voltage-dependent anion-selective channel protein 1 (Vdac1).
Besides those proteomic studies on drug-induced β-cell failure, Johnson et al. found that physiological concentrations of insulin can protect islet cells from apoptosis through the antiapoptotic transcription factor Pdx1 [36]. Further 2D-DIGE study on human islets identified 36 protein spots significantly changed by insulin stimulation. More importantly, biological studies on these candidates revealed that Bridge-1 was a positive regulator of Pdx1 after low-concentration insulin stimulation.
Collectively, these data suggest that the balance between β cell proliferation and cell death is important for the maintenance of islet mass and function, and that oxidative stress plays an important role in diabetes [35].
T2DM & insulin resistance
Type 2 diabetes is characterized by two major defects: β-cell dysfunction and insulin resistance in peripheral tissues. The exact alterations in molecular pathways associated with β-cell dysfunction in insulin-resistant and diabetic states are not clear. Proteomic studies of human islets are generally challenged by the availability of islets from living donors, and potentially large interindividual variation. As a result, most of the islet proteomic studies involving T2DM and insulin resistance tend to utilize islets from insulin-resistant or diabetic animal models, such as Zucker fatty (ZF) and Zucker diabetic fatty rats (ZDF) [15], high-fat-fed mice [75], muscle IGF-1 receptor–lysine–arginine (MKR) mice [17,76] and lep/lep mice [77]. A common feature of these animal models is that all models have manifested insulin resistance, and often exhibit islet dysfunction as it occurs in the early stages of diabetes.
Lu et al. used iTRAQ and an mRNA microarray approach to identify islet protein factors associated with the development of T2DM by using a 10-week-old MKR mouse model [17]. Among the 590 identified islet proteins, 159 were differentially expressed in MKR compared with control islets. Some previously reported proteins associated with insulin-secretion pathways or T2DM such as glucose transporter 2, Dnajc3, vesicle-associated membrane protein 2 (Vamp2), ras-related protein (Rab3a) and prohormone convertases (Pc1, Pc3) were significantly altered in the MKR islets. Many proteins associated with protein folding, endoplasmic reticulum (ER)-associated protein degradation and mitochondrial energy metabolism were also identified as differentially expressed. The comparison between proteomic data and mRNA microarray data suggested that approximately 54% of differentially expressed proteins in MKR islets showed changes in the proteome but not the transcriptome, suggesting that post-transcriptional regulation played an important role in disease development. More recently, the same group applied iTRAQ and LC–MS/MS to identify mitochondrial proteomic changes in MKR mouse islets [76]. A total of 36 mitochondrial proteins, including inner membrane proteins of the electron transport chain, were differentially expressed in 10-week-old MKR mice, indicating that mitochondrial dysfunction played a key role in the development of T2DM.
In another recent study, Han et al. compared islet proteomes from Zucker Lean (ZL), ZF and ZDF rats, representing control, obese and obese/diabetes conditions, respectively [15]. A total of 54 and 58 proteins were observed as differentially expressed in ZDF versus ZL rats and in ZF versus ZL rats, respectively. Impaired insulin secretion, mitochondrial dysfunction, dysregulation of triglyceride/free fatty acid cycling and lipotoxicity and microvascular dysfunction were proposed as potential factors mediating the progress from insulin resistance to T2DM [15].
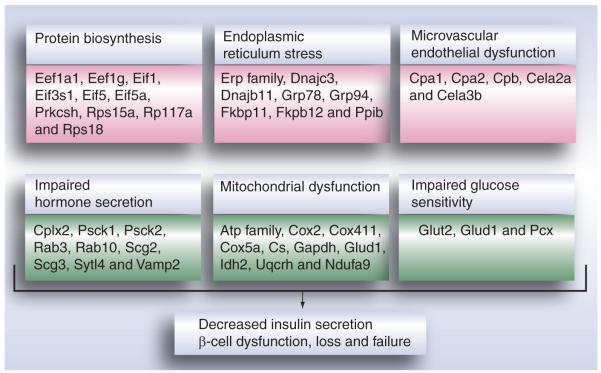
Figure 2 illustrates the significant alterations in protein abundances in protein biosynthesis, ER stress, microvascular endothelial dysfunction, mitochondrial dysfunction, impaired hormone secretion and impaired glucose sensitivity, all of which are potential contributing factors to islet dysfunction and T2DM. Similar results were observed from our own study of pancreatic islets [Zhou et al., Unpublished Data] from insulin-resistant mice, including high-fat diet mice.
Figure 2. Proteomic alterations observed from studies using insulin-resistant and diabetic animal models.
Data were summarized from proteomic studies in diabetic animal models [15,17,76]. A maximum of ten representative proteins were presented for each category.
Pink background: upregulated proteins; green background: downregulated proteins.
In a study performed with human islets, Nyblom et al. used LC–MS and SELDI methodologies to compare the differences in the proteome between islets isolated from patients with T2DM and age- and weight-matched controls [78]. In this study, approximately 20 proteins were observed as significantly changed in T2DM and several pathways, including cell arrest and apoptosis, immune-response and cell proliferation and regeneration were suggested to be activated in the islets of T2DM patients.
Altogether these studies have provided important insights into the pathways and factors induced by insulin resistance, as well as the factors linked to the transition from insulin resistance to β cell dysfunction in diabetes.
Islet development & regeneration
Islet transplantation has long been recognized as a potentially useful therapeutic strategy for both T1DM and advanced T2DM; however, successful islet transplantation still faces many, challenges despite significant recent advances [79]. A better understanding of the molecular mechanisms of islet development and regeneration may lead to new strategies for islet cell generation and transplantation. To this end, Hong et al. applied 2DE–MS to discover protein factors associated with the EGF-stimulated growth of neonatal porcine pancreatic cell clusters that are potential alternative sources of cells for islet transplantation [18]. They found that EGF-stimulated proliferation was mediated by the activation of the MAPK and PI3K pathways.
In another study, Jin et al. used iTRAQ and mRNA microarrays to identify proteins involved in islet-like differentiation of a human β cell line [57]. A number of proteins involved in cell cycle, cell structure and developmental processes were significantly downregulated, while proteins involved in lipid, fatty acid, steroid and nucleotide metabolism were upregulated, suggesting these molecular functions are likely to be associated with β-cell differentiation. To identify bioactive human peptides that might trigger islet neogenesis, Levetan et al. evaluated the peptide sequences within a variety of mammalian pancreas-specific regenerating genes from GenBank and all available proteomic databases to develop large-scale protein-to-protein interaction maps, which led to the discovery of human pro-islet peptide as a potential factor for islet neogenesis [80].
In general, these studies provide important information for developing new strategies to manipulate the process of islet development or regeneration.
Diabetic drug response
Another objective of proteomic studies on islets centers on the development of novel therapeutic strategies to treat diabetes. For example, Sanchez et al. used 2DE to investigate the effect of rosiglitazone on islet protein differential expression and found that the modulation of actin-binding protein levels by rosiglitazone may be involved in the protection of islet cell structure and function [77,81]. Jagerbrink et al. used 2DE to investigate changes in the islet proteome in response to an imidazoline derivative BL1128 and discovered 22 proteins with abundance changes, including four upregulated calcium-binding proteins – that is, calreticulin, calgizzarin, calcyclin and annexin I [34]. The upregulation of calcium-binding proteins is potentially linked to BL1128-stimulated insulin release at high glucose concentrations. While these studies provide initial insights into the islet response to potential therapeutic candidates, additional biological information remains to be acquired by using proteomic methodologies other than 2DE, which limits the number of identified proteins.
Conclusion
Recent proteomic studies of pancreatic islets have provided insights into the complexity of the islet proteome and the molecular pathways that regulate islet function, including a significant number of proteins that exhibit changes in expression levels in response to different biological or disease conditions. From these studies, it is becoming clear that mitochondrial dysfunction and impaired hormone secretion are two important processes that are linked to islet dysfunction. Further proteomic investigations of the mechanisms leading to islet dysfunction, as well as the functional roles of specific key proteins, should help pave the way for the development of novel therapeutic strategies for treating diabetes.
Expert commentary
Given the essential role of pancreatic islets in the regulation of glucose homeostasis, there is a major interest in gaining an understanding of the molecular mechanisms that modulate insulin secretion, islet cell proliferation, survival and apoptosis. The rapidly evolving science of proteomics has enabled investigation of some of these mechanisms at the proteome level, and several studies performed over the past decade have afforded new insights into the molecular pathways that regulate islet functions. Nevertheless, many of these studies were limited by the proteome coverage that could be achieved by the existing proteomic technology. This is especially true considering that a majority of the published proteomic studies on islets are based on 2DE, where often only a small number of proteins were identified. Although recent LC–MS/MS-based applications have significantly expanded coverage of the proteome, many important signaling proteins or transcription factors have yet to be identified. Moreover, it is well recognized that alterations in posttranslational protein modifications such as phosphorylation play a potentially even more important role in regulating cellular functions compared with alterations in protein abundances. In spite of its importance, proteomic analysis of protein phosphorylation in pancreatic islets has not yet been reported, most likely due to limitations associated with islet sample size and instrument sensitivity. Lastly, while proteomics is advantageous for measuring a large number of proteins simultaneously, this technology is often regarded as a discovery or hypothesis-generating tool. To make full use of this approach, selecting essential protein factors for further detailed functional studies following initial proteomic discovery of a large number of differentially expressed proteins remains a significant challenge. Further advances in bioinformatics and a larger knowledgebase will likely facilitate a more effective discovery of the key protein factors relevant to disease functions.
Five-year view
Within the next 5 years, we anticipate a significantly broader interest in studying the molecular mechanisms of islet hormone secretion, islet cell regeneration and islet cell death as a result of more advanced quantitative proteomic strategies, such as multidimensional LC–MS/MS [13,14], coupled with either label-free or stable isotope labeling-based quantification. Further improvements in the sensitivity of proteomic technologies will allow the application of subcellular fractionation approaches, such as mitochondrial enrichment [76] and secretory granule separation [22,24], to identify low-abundance signaling proteins. Moreover, studies will start to explore post-translational protein modifications, such as phosphorylation [82], oxidation [83], ubiquitination [84,85] and sumoylation [86] in pancreatic islets to reveal the respective roles of such modifications in regulating islet biology. We also foresee targeted quantitative proteomic technologies, such as selected reaction monitoring-MS [87] becoming an important tool for validating therapeutic candidates discovered in large-scale proteomic efforts. With these integrated efforts, one might expect the discovery of novel factors or therapeutic targets for treating both T1DM and T2DM.
Key issues.
Pancreatic islets play an essential role in maintaining blood glucose homeostasis; islet dysfunction is linked to both Type 1 and Type 2 diabetes.
Proteomic studies of islets have revealed alterations in many proteins under conditions of insulin resistance, diabetes, glucose stimulation or drug treatment.
Early islet studies that employed 2DE to separate proteins only provided a small number of protein identifications. Studies using liquid chromotography–MS/MS have illustrated the potential for increased proteome coverage.
Future islet studies are expected to include protein phosphorylation and other posttranslational modifications involved in cell signaling.
Key protein factors may be identified as therapeutic targets for treating diabetes.
Acknowledgments
Portions of this research were supported by National Institutes of Health grants R01DK074795 and RR018522. Experimental work was performed in the Environmental Molecular Sciences Laboratory, a US Department of Energy (DOE) Office of Biological and Environmental Research national scientific user facility on the Pacific Northwest National Laboratory (PNNL) campus. PNNL is a multiprogram national laboratory operated by Battelle for the DOE under Contract No. DE-AC05–76RLO1830.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Elayat AA, el-Naggar MM, Tahir M. An immunocytochemical and morphometric study of the rat pancreatic islets. J. Anat. 1995;186(Pt 3):629–637. [PMC free article] [PubMed] [Google Scholar]
- 2.Cabrera O, Berman DM, Kenyon NS, Ricordi C, Berggren PO, Caicedo A. The unique cytoarchitecture of human pancreatic islets has implications for islet cell function. Proc. Natl Acad. Sci. USA. 2006;103(7):2334–2339. doi: 10.1073/pnas.0510790103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC. β cell deficit and increased β-cell apoptosis in humans with Type 2 diabetes. Diabetes. 2003;5(1):102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 4.Gillespie KM. Type 1 diabetes: pathogenesis and prevention. CMAJ. 2006;175(2):165–170. doi: 10.1503/cmaj.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meisterfeld R, Ehehalt F, Saeger HD, Solimena M. Pancreatic disorders and diabetes mellitus. Exp. Clin. Endocrinol. Diabetes. 2008;116(Suppl. 1):S7–S12. doi: 10.1055/s-2008-1080918. [DOI] [PubMed] [Google Scholar]
- 6.Wellen KE, Hotamisligil GS. Inflammation, stress and diabetes. J Clin. Invest. 2005;115(5):1111–1119. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diakogiannaki E, Dhayal S, Childs CE, Calder PC, Welters HJ, Morgan NG. Mechanisms involved in the cytotoxic and cytoprotective actions of saturated versus monounsaturated long-chain fatty acids in pancreatic β-cells. J. Endocrinol. 2007;194(2):283–291. doi: 10.1677/JOE-07-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Assaad W, Buteau J, Peyot ML, et al. Saturated fatty acids synergize with elevated glucose to cause pancreatic β-cell death. Endocrinology. 2003;144(9):4154–4163. doi: 10.1210/en.2003-0410. [DOI] [PubMed] [Google Scholar]
- 9.Federici M, Hribal M, Perego L, et al. High glucose causes apoptosis in cultured human pancreatic islets of Langerhans: a potential role for regulation of specific Bcl family genes toward an apoptotic cell death program. Diabetes. 2001;50(6):1290–1301. doi: 10.2337/diabetes.50.6.1290. [DOI] [PubMed] [Google Scholar]
- 10.Guardado-Mendoza R, Davalli AM, Chavez AO, et al. Pancreatic islet amyloidosis, β-cell apoptosis and α-cell proliferation are determinants of islet remodeling in Type-2 diabetic baboons. Proc. Natl Acad. Sci. USA. 2009;106(33):13992–13997. doi: 10.1073/pnas.0906471106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Cairano ES, Davalli AM, Perego L, et al. The glial glutamate transporter 1 (GLT1) is expressed by pancreatic β-cells and prevents glutamate-induced β-cell death. J Biol. Chem. 2011;286(16):14007–14018. doi: 10.1074/jbc.M110.183517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hribal ML, Tornei F, Pujol A, et al. Transgenic mice overexpressing human G97 R IRS-1 show impaired insulin action and insulin secretion. J Cell Mol. Med. 2008;12(5B):2096–2106. doi: 10.1111/j.1582-4934.2008.00246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metz TO, Jacobs JM, Gritsenko MA, et al. Characterization of the human pancreatic islet proteome by 2D LC/MS/MS. J Proteome Res. 2006;5(12):3345–3354. doi: 10.1021/pr060322n. • The most complete profiling of the human islet proteome.
- 14.Petyuk VA, Qian WJ, Hinault C, et al. Characterization of the mouse pancreatic islet proteome and comparative analysis with other mouse tissues. J. Proteome Res. 2008;7(8):3114–3126. doi: 10.1021/pr800205b. •• The first extensive profiling of the mouse islet proteome by 2D-liquid chromotography (LC)–MS/MS.
- 15.Han D, Moon S, Kim H, et al. Detection of differential proteomes associated with the development of Type 2 diabetes in the Zucker rat model using the iTRAQ technique. J Proteome Res. 2010;10(2):564–577. doi: 10.1021/pr100759a. •• A LC–MS/MS based quantitative proteomic study on islets comparing control and diabetic rat models to identify factors associated with the development of diabetes.
- 16.Waanders LF, Chwalek K, Monetti M, Kumar C, Lammert E, Mann M. Quantitative proteomic analysis of single pancreatic islets. Proc. Natl Acad. Sci. USA. 2009;106(45):18902–18907. doi: 10.1073/pnas.0908351106. • High-sensitivity LC–MS/MS proteomics at the single islet level.
- 17.Lu H, Yang Y, Allister EM, Wijesekara N, Wheeler MB. The identification of potential factors associated with the development of Type 2 diabetes: a quantitative proteomics approach. Mol. Cell. Proteomics. 2008;7(8):1434–1451. doi: 10.1074/mcp.M700478-MCP200. •• An extensive quantitative proteomic study on islets from a diabetic model.
- 18.Hong OK, Suh SH, Kwon HS, et al. Proteomic analysis of differential protein expression in response to epidermal growth factor in neonatal porcine pancreatic cell monolayers. J. Cell Biochem. 2005;95(4):769–781. doi: 10.1002/jcb.20482. • EGF-stimulated intracellular signaling relevant to islet development revealed by differential proteomics.
- 19.Dowling P, O’Driscoll L, O’Sullivan F, et al. Proteomic screening of glucose-responsive and glucose nonresponsive MIN-6 β cells reveals differential expression of proteins involved in protein folding, secretion and oxidative stress. Proteomics. 2006;6(24):6578–6587. doi: 10.1002/pmic.200600298. • Differential protein expressions relevant to glucose-stimulated response in β-cell line.
- 20.Maziarz M, Chung C, Drucker DJ, Emili A. Integrating global proteomic and genomic expression profiles generated from islet α cells: opportunities and challenges to deriving reliable biological inferences. Mol. Cell. Proteomics. 2005;4(4):458–474. doi: 10.1074/mcp.R500011-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.D’Hertog W, Overbergh L, Lage K, et al. Proteomics analysis of cytokine-induced dysfunction and death in insulin-producing INS-1E cells: new insights into the pathways involved. Mol. Cell. Proteomics. 2007;6(12):2180–2199. doi: 10.1074/mcp.M700085-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Brunner Y, Coute Y, Iezzi M, et al. Proteomics analysis of insulin secretory granules. Mol. Cell. Proteomics. 2007;6(6):1007–1017. doi: 10.1074/mcp.M600443-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Nyblom HK, Thorn K, Ahmed M, Bergsten P. Mitochondrial protein patterns correlating with impaired insulin secretion from INS-1E cells exposed to elevated glucose concentrations. Proteomics. 2006;6(19):5193–5198. doi: 10.1002/pmic.200600137. [DOI] [PubMed] [Google Scholar]
- 24.Hickey AJ, Bradley JW, Skea GL, et al. Proteins associated with immunopurified granules from a model pancreatic islet β-cell system: proteomic snapshot of an endocrine secretory granule. J. Proteome Res. 2009;8(1):178–186. doi: 10.1021/pr800675k. • Proteome profiling of purified secretory granules from the INS-1E β-cell model.
- 25.Ortsater H, Bergsten P. Protein profiling of pancreatic islets. Expert Rev. Proteomics. 2006;3(6):665–675. doi: 10.1586/14789450.3.6.665. [DOI] [PubMed] [Google Scholar]
- 26.Lopez JL. 2D electrophoresis in proteome expression analysis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2007;849(1–2):190–202. doi: 10.1016/j.jchromb.2006.11.049. [DOI] [PubMed] [Google Scholar]
- 27.Unlu M, Morgan ME, Minden JS. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis. 1997;18(11):2071–2077. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- 28.Ahmed M. Proteomics and islet research. Adv. Exp. Med. Biol. 2010;654:363–390. doi: 10.1007/978-90-481-3271-3_16. [DOI] [PubMed] [Google Scholar]
- 29.Sundsten T, Ortsater H. Proteomics in diabetes research. Mol. Cell Endocrinol. 2009;297(1–2):93–103. doi: 10.1016/j.mce.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 30.Maris M, Overbergh L, Mathieu C. Type 2 diabetes: gaining insight into the disease process using proteomics. Proteomics Clin. Appl. 2008;2(3):312–326. doi: 10.1002/prca.200780093. [DOI] [PubMed] [Google Scholar]
- 31.Sparre T, Larsen MR, Heding PE, Karlsen AE, Jensen ON, Pociot F. Unraveling the pathogenesis of Type 1 diabetes with proteomics: present and future directions. Mol. Cell. Proteomics. 2005;4(4):441–457. doi: 10.1074/mcp.R500002-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Minden JS, Dowd SR, Meyer HE, Stuhler K. Difference gel electrophoresis. Electrophoresis. 2009;30(Suppl. 1):S156–S161. doi: 10.1002/elps.200900098. [DOI] [PubMed] [Google Scholar]
- 33.Kim SW, Hwang HJ, Baek YM, Lee SH, Hwang HS, Yun JW. Proteomic and transcriptomic analysis for streptozotocin-induced diabetic rat pancreas in response to fungal polysaccharide treatments. Proteomics. 2008;8(11):2344–2361. doi: 10.1002/pmic.200700779. [DOI] [PubMed] [Google Scholar]
- 34.Jagerbrink T, Lexander H, Palmberg C, et al. Differential protein expression in pancreatic islets after treatment with an imidazoline compound. Cell. Mol. Life Sci. 2007;64(10):1310–1316. doi: 10.1007/s00018-007-7136-5. • Proteome profiling of pancreatic islet proteome alterations in response to a diabetic drug.
- 35.Xie X, Li S, Liu S, Lu Y, Shen P, Ji J. Proteomic analysis of mouse islets after multiple low-dose streptozotocin injection. Biochim. Biophys. Acta. 2008;1784(2):276–284. doi: 10.1016/j.bbapap.2007.11.008. • Proteomic study of pancreatic islets revealed proteins relevant to β-cell failure.
- 36.Johnson JD, Bernal-Mizrachi E, Alejandro EU, et al. Insulin protects islets from apoptosis via Pdx1 and specific changes in the human islet proteome. Natl Acad. Sci. USA. 2006;103(51):19575–19580. doi: 10.1073/pnas.0604208103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 38.Qian WJ, Jacobs JM, Liu T, Camp DG, 2nd, Smith RD. Advances and challenges in liquid chromatography-mass spectrometry-based proteomics profiling for clinical applications. Mol. Cell. Proteomics. 2006;5(10):1727–1744. doi: 10.1074/mcp.M600162-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martens GA, Jiang L, Verhaeghen K, et al. Protein markers for insulin-producing β cells with higher glucose sensitivity. PLoS One. 2010;5(12):e14214. doi: 10.1371/journal.pone.0014214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004;76(14):4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 41.Zhou JY, Schepmoes AA, Zhang X, et al. Improved LC–MS/MS spectral counting statistics by recovering low-scoring spectra matched to confidently identified peptide sequences. J. Proteome Res. 2010;9(11):5698–5704. doi: 10.1021/pr100508p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qian WJ, Jacobs JM, Camp DG, 2nd, et al. Comparative proteome analyses of human plasma following in vivo lipopolysaccharide administration using multidimensional separations coupled with tandem mass spectrometry. Proteomics. 2005;5(2):572–584. doi: 10.1002/pmic.200400942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang W, Shaler TA, Norton SM, Hill LR, Becker CH. Direct quantitative profiling of proteins in complex biological systems by nano electrospray mass spectrometry without tagging or isotopic standards. Presented at: 50th ASMS Conference on Mass Spectrometry and Allied Topics; Orlando, FL, USA. 2–6 June 2002. [Google Scholar]
- 44.Metz TO, Qian WJ, Jacobs JM, et al. Application of proteomics in the discovery of candidate protein biomarkers in a Diabetes Autoantibody Standardization Program (DASP) sample subset. J. Proteome Res. 2008;7(2):698–707. doi: 10.1021/pr700606w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith RD, anderson GA, Lipton MS, et al. An accurate mass tag strategy for quantitative and high-throughput proteome measurements. Proteomics. 2002;(5):513–523. doi: 10.1002/1615-9861(200205)2:5<513::AID-PROT513>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 46.Monroe ME, Tolic N, Jaitly N, Shaw JL, Adkins JN, Smith RD. VIPER: an advanced software package to support high-throughput LC–MS peptide identification. Bioinformatics. 2007;3(15):2021–2023. doi: 10.1093/bioinformatics/btm281. [DOI] [PubMed] [Google Scholar]
- 47.Li XJ, Yi EC, Kemp CJ, Zhang H, Aebersold R. A software suite for the generation and comparison of peptide arrays from sets of data collected by liquid chromatography-mass spectrometry. Mol. Cell. Proteomics. 2005;4(9):1328–1340. doi: 10.1074/mcp.M500141-MCP200. [DOI] [PubMed] [Google Scholar]
- 48.Jaffe JD, Mani DR, Leptos KC, Church GM, Gillette MA, Carr SA. PEPPeR, a platform for experimental proteomic pattern recognition. Mol. Cell. Proteomics. 2006;5(10):1927–1941. doi: 10.1074/mcp.M600222-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;6(12):1367–1137. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 50.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 2005;1(5):252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 51.Ong SE, Blagoev B, Kratchmarova I, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 2002;1(5):376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 52.Qian WJ, Monroe ME, Liu T, et al. Quantitative proteome analysis of human plasma following in vivo lipopolysaccharide administration using 16O/18O labeling and the accurate mass and time tag approach. Mol. Cell. Proteomics. 2005;4(5):700–709. doi: 10.1074/mcp.M500045-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petritis BO, Qian WJ, Camp DG, 2nd, Smith RD. A simple procedure for effective quenching of trypsin activity and prevention of 18O-labeling back-exchange. J. Proteome Res. 2009;8(5):2157–2163. doi: 10.1021/pr800971w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qian WJ, Liu T, Petyuk VA, et al. Large-scale multiplexed quantitative discovery proteomics enabled by the use of an (18)O-labeled ‘universal’ reference sample. J. Proteome Res. 2009;8(1):2290–2299. doi: 10.1021/pr800467r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ross PL, Huang YN, Marchese JN, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics. 2004;3(12):1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 56.Thompson A, Schafer J, Kuhn K, et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 2003;75(8):1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 57.Jin J, Park J, Kim K, et al. Detection of differential proteomes of human β-cells during islet-like differentiation using iTRAQ labeling. J. Proteome Res. 2009;8(3):1393–1403. doi: 10.1021/pr800765t. [DOI] [PubMed] [Google Scholar]
- 58.Pichler P, Kocher T, Holzmann J, et al. Peptide labeling with isobaric tags yields higher identification rates using iTRAQ 4-plex compared with TMT 6-plex and iTRAQ 8-plex on LTQ Orbitrap. Anal. Chem. 2010;8(15):6549–6558. doi: 10.1021/ac100890k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ahmed M, Bergsten P. Glucose-induced changes of multiple mouse islet proteins analysed by 2D gel electrophoresis and mass spectrometry. Diabetologia. 2005;48(3):477–485. doi: 10.1007/s00125-004-1661-7. [DOI] [PubMed] [Google Scholar]
- 60.Ahmed M, Forsberg J, Bergsten P. Protein profiling of human pancreatic islets by 2D gel electrophoresis and mass spectrometry. J. Proteome Res. 2005;4(3):931–940. doi: 10.1021/pr050024a. [DOI] [PubMed] [Google Scholar]
- 61.Sanchez JC, Chiappe D, Converset V, et al. The mouse SWISS- D PAGE database: a tool for proteomics study of diabetes and obesity. Proteomics. 2001;1(1):136–163. doi: 10.1002/1615-9861(200101)1:1<136::AID-PROT136>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 62.Kutlu B, Burdick D, Baxter D, et al. Detailed transcriptome atlas of the pancreatic β cell. BMC Med. Genomics. 2009;3 doi: 10.1186/1755-8794-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Collins H, Najafi H, Buettger C, Rombeau J, Settle RG, Matschinsky FM. Identification of glucose response proteins in two biological models of β-cell adaptation to chronic high glucose exposure. J. Biol. Chem. 1992;67(2):1357–1366. [PubMed] [Google Scholar]
- 64.Collins HW, Buettger C, Matschinsky F. High-resolution 2D polyacrylamide gel electrophoresis reveals a glucose-response protein of 65 kDa in pancreatic islet cells. Proc. Natl Acad. Sci. USA. 1990;87(14):5494–5498. doi: 10.1073/pnas.87.14.5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu X, Friedman D, Hill S, et al. Proteomic exploration of pancreatic islets in mice null for the α2A adrenergic receptor. J. Mol. Endocrinol. 2005;35(1):73–88. doi: 10.1677/jme.1.01764. [DOI] [PubMed] [Google Scholar]
- 66.Yamada M, Ohata H, Momose K, Richelson E. Pharmacological characterization of SR 48692 sensitive neurotensin receptor in human pancreatic cancer cells, MIA PaCa-2. Res. Commun. Mol. Pathol. Pharmacol. 1995;90(1):37–47. [PubMed] [Google Scholar]
- 67.Eizirik DL, Mandrup-Poulsen T. A choice of death – the signal-transduction of immune-mediated β-cell apoptosis. Diabetologia. 2001;44(12):2115–2133. doi: 10.1007/s001250100021. [DOI] [PubMed] [Google Scholar]
- 68.Van Belle TL, Taylor P, von Herrath MG. Mouse models for Type 1 diabetes. Drug Discov. Today Dis. Models. 2009;6(2):41–45. doi: 10.1016/j.ddmod.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hohmeier HE, Newgard CB. Cell lines derived from pancreatic islets. Mol. Cell Endocrinol. 2004;228(1–2):121–128. doi: 10.1016/j.mce.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 70.Andersen HU, Larsen PM, Fey SJ, Karlsen AE, Mandrup-Poulsen T, Nerup J. 2D gel electrophoresis of rat islet proteins. Interleukin 1 β-induced changes in protein expression are reduced by l-arginine depletion and nicotinamide. Diabetes. 1995;44(4):400–407. doi: 10.2337/diab.44.4.400. [DOI] [PubMed] [Google Scholar]
- 71.Andersen HU, Fey SJ, Larsen PM, et al. Interleukin-1β induced changes in the protein expression of rat islets: a computerized database. Electrophoresis. 1997;18(11):2091–2103. doi: 10.1002/elps.1150181136. [DOI] [PubMed] [Google Scholar]
- 72.Karlsen AE, Storling ZM, Sparre T, et al. Immune-mediated β-cell destruction in vitro and in vivo-A pivotal role for galectin-3. Biochem. Biophys. Res. Commun. 2006;344(1):406–415. doi: 10.1016/j.bbrc.2006.03.105. [DOI] [PubMed] [Google Scholar]
- 73.Zhou JY, Afjehi-Sadat L, Asress S, et al. Galectin-3 is a candidate biomarker for amyotrophic lateral sclerosis: discovery by a proteomics approach. J. Proteome Res. 2010;9(10):5133–5141. doi: 10.1021/pr100409r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Karabatas LM, Pastorale C, de Bruno LF, et al. Early manifestations in multiple-low-dose streptozotocin-induced diabetes in mice. Pancreas. 2005;30(4):318–324. doi: 10.1097/01.mpa.0000161888.02244.7a. [DOI] [PubMed] [Google Scholar]
- 75.Qiu L, List EO, Kopchick JJ. Differentially expressed proteins in the pancreas of diet-induced diabetic mice. Mol. Cell. Proteomics. 2005;4(9):1311–1318. doi: 10.1074/mcp.M500016-MCP200. [DOI] [PubMed] [Google Scholar]
- 76.Lu H, Koshkin V, Allister EM, Gyulkhandanyan AV, Wheeler MB. Molecular and metabolic evidence for mitochondrial defects associated with β-cell dysfunction in a mouse model of Type 2 diabetes. Diabetes. 2010;59(2):448–459. doi: 10.2337/db09-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sanchez JC, Converset V, Nolan A, et al. Effect of rosiglitazone on the differential expression of diabetes-associated proteins in pancreatic islets of C57Bl/6 lep/lep mice. Mol. Cell. Proteomics. 2002;1(7):509–516. doi: 10.1074/mcp.m200033-mcp200. [DOI] [PubMed] [Google Scholar]
- 78.Nyblom HK, Bugliani M, Fung E, et al. Apoptotic, regenerative and immune-related signaling in human islets from Type 2 diabetes individuals. J. Proteome Res. 2009;8(12):5650–5656. doi: 10.1021/pr9006816. •• Proteomic change in human islets from Type 2 diabetes patients.
- 79.Noguchi H. Pancreatic islet transplantation. World J Gastrointest. Surg. 2009;1(1):16–20. doi: 10.4240/wjgs.v1.i1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Levetan CS, Upham LV, Deng S, et al. Discovery of a human peptide sequence signaling islet neogenesis. Endocr. Pract. 2008;14(9):1075–1083. doi: 10.4158/EP.14.9.1075. [DOI] [PubMed] [Google Scholar]
- 81.Verges B. Effects of glitazones in the treatment of diabetes and/or hyperlipidaemia: glycaemic control and plasma lipid levels. Fundam. Clin. Pharmacol. 2007;21(Suppl. 2):15–18. doi: 10.1111/j.1472-8206.2007.00532.x. [DOI] [PubMed] [Google Scholar]
- 82.Yang F, Wu S, Stenoien DL, et al. Combined pulsed-Q dissociation and electron transfer dissociation for identification and quantification of iTRAQ-labeled phosphopeptides. Anal. Chem. 2009;81(10):4137–4143. doi: 10.1021/ac802605m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Butterfield DA, Sultana R. Redox proteomics: understanding oxidative stress in the progression of age-related neurodegenerative disorders. Expert Rev. Proteomics. 2008;5(2):157–160. doi: 10.1586/14789450.5.2.157. [DOI] [PubMed] [Google Scholar]
- 84.Zhou J, Livak MF, Bernier M, et al. Ubiquitination is involved in glucose-mediated downregulation of GIP receptors in islets. Am. J Physiol. Endocrinol. Metab. 2007;293(2):E538–E547. doi: 10.1152/ajpendo.00070.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jeram SM, Srikumar T, Pedrioli PG, Raught B. Using mass spectrometry to identify ubiquitin and ubiquitin-like protein conjugation sites. Proteomics. 2009;9(4):922–934. doi: 10.1002/pmic.200800666. [DOI] [PubMed] [Google Scholar]
- 86.Ehninger A, Mziaut H, Solimena M. Emerging role of SUMO in pancreatic β-cells. Horm. Metab. Res. 2007;39(9):658–664. doi: 10.1055/s-2007-985372. [DOI] [PubMed] [Google Scholar]
- 87.Hossain M, Kaleta DT, Robinson EW, et al. Enhanced sensitivity for selected reaction monitoring-mass spectrometry-based targeted proteomics using a dual-stage electrodynamic ion funnel interface. Mol. Cell. Proteomics. 2010;10(2):M000062–MCP201. doi: 10.1074/mcp.M000062-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]