Abstract
Alternative lysine and methionine residues at position 44 in the D1 domain determine the specificities of human lineage III killer cell immunoglobulin-like receptors (KIR) for the C1 and C2 epitopes of HLA-C. KIR having glutamate 44 are also present in orangutans (Popy2DLB) and chimpanzees (Pt-2DL9) but notably absent from humans. Popy2DLB exhibits broad specificity for both the C1 and C2 epitopes, whereas Pt-2DL9 has narrow specificity for C2. Mutation of phenylalanine 45 in Popy2DLB to the cysteine residue present in Pt-2DL9 was sufficient to narrow the Popy2DLB specificity to be like that of Pt-2DL9. In contrast, replacement of cysteine 45 in Pt-2DL9 by phenylalanine had no effect on its C2 specificity, but reduced the avidity. In a similar manner, replacement of phenylalanine 45 with cysteine in Popy2DLA, which has lysine 44 and recognizes C1, maintained this specificity while reducing avidity. Position 45 is exceptionally variable, exhibiting twelve residues that distinguish KIR of different lineages and species. Our study demonstrates the potential for variation at position 45 to modulate KIR avidity and specificity for HLA-C. The various effects of position 45 mutation are consistent with a model in which a Popy2DLB-like receptor, having glutamate 44 and broad specificity for C1 and C2, facilitated the evolution of the C2 epitope from the C1 epitope and C2-specific KIR from C1-specific KIR. With the acquisition of C2 and C2-specific receptors, the selection against this broadly specific receptor led to its loss from the human line and narrowing of its specificity on the chimpanzee line.
Keywords: KIR receptors, MHC, Non-human primates, Evolution
Killer cell immunoglobulin-like receptors (KIR) are expressed by natural killer (NK) cells and recognize polymorphic epitopes of HLA-A, B and C (Moretta et al. 2006). These cognate interactions educate NK cells to be tolerant of self and responsive to infected and malignant cells displaying abnormal expression of HLA class I (Karre 2008). KIR can evolve rapidly and in species-specific fashion, but their diversification appears restricted to simian primates and cattle (Guethlein et al. 2007). The study of great apes is particularly informative for the human system, because they are the only species having strict orthologs of HLA-A, B and C. Of these three isoforms, HLA-C is most recently evolved and has become dominant in human populations as the source of the C1 and C2 epitopes recognized by lineage III KIR (Abi-Rached et al. 2010b). Every HLA-C allotype carries either the C1 or C2 epitope, defined by asparagine and lysine at position 80, respectively. In complementary manner, dimorphism at position 44 in the D1 domain of KIR2D determines the specificity for HLA-C: with lysine and methionine imparting specificity for C1 and C2, respectively (Winter and Long 1997). Lineage III is one of four human KIR lineages defined on the basis of phylogenetic relationship (Rajalingam et al. 2004). HLA-G is recognized by lineage I KIR; HLA-A and HLA-B epitopes are recognized by lineage II KIR, and for lineage V KIR, a ligand has yet to be defined.
Allotypes of Patr-C, the chimpanzee ortholog of HLA-C, similarly carry the C1 and C2 epitopes and are recognized by lineage III KIR having lysine and methionine at position 44, respectively. In addition, Pt-2DL9 has glutamate 44, a residue never observed in human KIR, associated with C2 specificity (Moesta et al. 2009). All allotypes of Popy-C, the orangutan ortholog of HLA-C, carry the C1 epitope, and correspondingly, orangutans have C1-specific lineage III KIR with lysine 44, but none with C2 specificity and methionine 44 (Adams et al. 1999; Guethlein et al. 2002; Older Aguilar et al. 2010). The orangutan also has lineage III KIR with glutamate 44, such as Popy2DLB, which exhibit a novel, broad reactivity for HLA-C that does not discriminate between C1 and C2 (Older Aguilar et al. 2010). Paradoxically, glutamate 44 is seen to confer broad HLA-C specificity on orangutan KIR but narrow C2 specificity on chimpanzee KIR. Here, we report on the investigation of the molecular basis for this intriguing difference.
In the context of previous studies relating KIR structure to function, we compared the sequences of chimpanzee (Pt-2DL9) and orangutan KIR (Popy2DLB and 2DSB) having glutamate 44 (Biassoni et al. 1997; Boyington et al. 2000; Graef et al. 2009). The extracellular, ligand-binding domains of Pt-2DL9 differ from orangutan 2DLB and 2DSB at 29 positions, 20 in the D1 domain and 9 in the D2 domain (Fig. 1). Of these, mutation at positions 45, 70, 71 and 72 in D1 have been shown previously to influence the interaction of KIR2D with HLA-C (Biassoni et al. 1997; Graef et al. 2009; Saulquin et al. 2003; Winter et al. 1998). Accordingly, at these four positions, we made point mutants of Popy2DLB having the corresponding residue from Pt-2DL9. Fc-fusion proteins were constructed from mutant and wild-type Popy2LB, as well as Pt-2DL9, and tested for binding to a panel of beads, each coated with one of 95 HLA class I allotypes (Fig. 2).
Fig. 1.
Alignment of positions of difference within the D1 and D2 domains of 13 human, chimpanzee and orangutan lineage III KIR. Popy2DLA is used as the consensus sequence, and identity with it is marked by a dot. Position 44 is marked in dark grey, and the residues selected for mutational analysis, positions 45, 70, 71, and 72, are marked in light grey
Fig. 2.
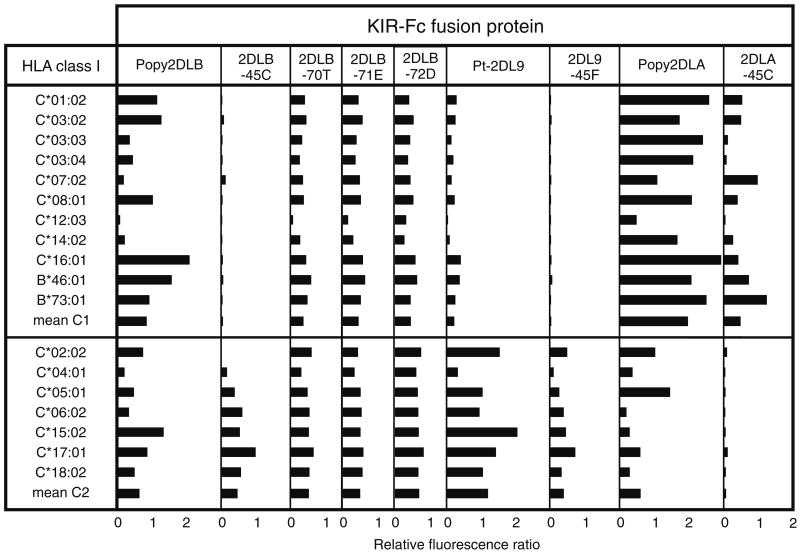
Mutation at position 45 changes the specificity of Popy2DLB, while affecting only the avidity of Popy2DLA and Pt-2DL9. Shown are results of assays to measure the binding of nine KIR-Fc fusion proteins to beads coated with one of 18 HLA class I allotypes: eleven C1 and seven C2. Binding is shown for individual allotypes and for mean C1 and C2. Experiments were performed as described previously (Moesta et al. 2009; Older Aguilar et al. 2010). A minimum of three independent assays were performed for each fusion protein. Shown are the averaged data for all experiments
The results demonstrated that substitution of cysteine for phenylalanine at position 45 is sufficient to change the specificity of Popy2DLB from its broad reactivity with C1+ and C2+ HLA-C to a narrow C2 reactivity, resembling that of Pt-2DL9 (Fig. 2). Interaction with C1+ HLA-C was abrogated, while binding to C2+ HLA-C was preserved, with the exception of HLA-C*02:02. Neither were the latter reactions as strong as those of Pt-2DL9, a difference we attribute to one or more of the 34 other substitutions in the D1 and D2 domains that distinguish Popy2DLB from Pt-2DL9 (Fig. 1). In this context, the functional effect of the mutation at position 45 is particularly striking. By contrast, mutation at positions 70, 71 and 72 had a similar effect in reducing Popy2DLB avidity for HLA-C, while preserving its broad reactivity for HLA-C (Fig. 2). These results are consistent with the observation that natural variation at these positions, which are in or near the binding site for HLA class I, principally affects the avidity, but not the specificity, of the interaction (L. Vago, personal communication).
We examined the effects of substitution at position 45 further by mutating phenylalanine 45 to cysteine in Popy2DLA, the orangutan C1-specific receptor, and mutating cysteine 45 to phenylalanine in chimpanzee Pt-2DL9. Neither of these mutations gave the results anticipated from the effect of the phenylalanine 45 to cysteine substitution on the specificity of Popy2DLB. Mutant Popy2DLA-45C exhibited reduced avidity for C1+ HLA-C and loss of the cross-reactivity for C2+ HLA-C that distinguishes Popy-2DLA (Fig. 2). This result was, however, consistent with previous mutational analysis showing that substitution at position 44 exclusively altered Popy2DLA specificity, whereas substitution at 12 other sites affected only avidity (Older Aguilar et al. 2010). Changing cysteine 45 in Pt-2DL9 to phenylalanine produced a similar effect to that of changing phenylalanine 45 in Popy2DLA to cysteine. The avidity for C2+ HLA-C was reduced, and the weak cross-reactions with C1+ HLA-C were eliminated. Thus, one or more of the 33 substitutions (apart from position 45) that distinguish Pt-2DL9 and 2DLB must be responsible for Pt-2DL9 having C2-specificity and Popy2DLB recognizing both C1 and C2 (Fig. 1). In summary, for both 2DLA and 2DL9, the effect of mutation at position 45 was to reduce the strength of the receptors, but not to alter their specificity.
Interaction of C1-specific lineage III KIR with the C1 epitope evolved under natural selection for several million years before emergence of the C2 epitope and C2-specific KIR. The latter advance required mutations at both position 80 in MHC-C and position 44 in KIR. Moreover, the intermediate form, in which only one of the mutations was in place, must have had useful biological function to be selected. We have previously hypothesized that a Popy2DLB-like receptor, having glutamate 44 and specificity for C1 and C2, was such an intermediate, which allowed C2 to function when it first emerged (Older Aguilar et al. 2010). In turn, the presence of C2 would then enable C2-specific receptors with methionine 44 to function, when they first emerged. The acquisition of specialized C1-specific and C2-specific receptors would then have allowed the functional interaction between C2 and C2-specific KIR to evolve and diversify differently from the interaction between C1 and C1-specific KIR. Such evolution has expanded the functional NK cell repertoire (Andersson et al. 2009; Yawata et al. 2008). Because Popy2DLB-like receptors do not discriminate C1 and C2, their continuing presence would have hindered such divergence. Consequently, Popy2DLB-like receptors became subject to negative selection, leading to the elimination of KIR with glutamate 44 on the human line and their conversion from C1+C2-specific to C2-specific receptors on the chimpanzee line through mutation at position 45. This hypothetical scheme is entirely consistent with the effects observed here of mutation at position 45, which impart new function to Popy2DLB and loss of function to Popy2DLA and Pt-2DL9.
Orangutans have inhibitory (Popy2DLA and Popy2DLB) and activating (Popy2DSA and Popy2DSB) receptors with similar specificity and avidity for MHC-C1 and C2 (Older Aguilar et al. 2010). Likewise, chimpanzees have potent inhibitory and activating KIR specific for C1 and C2 (Moesta et al. 2010). In striking contrast, the human species has inhibitory receptors for C1 (KIR2L2/3) and C2 (KIR2DL1), but only C2 is recognized by an activating receptor (KIR2DS1). KIR2DS2, which has similar D1 and D2 domains to KIR2DL2/3, acquired and fixed a fatal mutation: tyrosine at position 45. Wild-type KIR2DS2 has no detectable affinity for C1 or any HLA class I, but when position 45 was mutated to phenylalanine, the residue present in other lineage III human KIR, KIR2DS2 demonstrated good avidity for C1, comparable to that of KIR2DL2/3 (Winter et al. 1998). This provocative observation indicates there was selection to eliminate C1 recognition by KIR2DS2, exemplifying a more general trend on the human line for activating KIR to lose affinity for HLA class I (Abi-Rached et al. 2010b; Moesta et al. 2009, 2010). This trend, associated with the human-specific evolution of A and B haplotypes, is believed to be a consequence of distinctive selection pressures caused by the functions KIR serve in immunity and reproduction (Abi-Rached and Parham 2005; Parham 2005).
Within the lineage III KIR, there is little variation at position 45, apart from that in KIR2DS2, Pt-2DL9 and gorilla Gg-2DLe (Table 1). Consequently, this position emits no detectable signal of positive selection (Abi-Rached et al. 2010a). Position 45 does, however, stand out for its variability between KIR lineages and species of simian primate, with no less than 12 of the 20 natural amino acids being represented (Table 1). The lineage II, III and V hominoid KIR are all defined by the residues at position 45, which can be used with confidence to assign KIR lineage. This unique property suggests that position 45 has played a formative role in the emergence and differentiation of KIR lineages, while in the process becoming ‘fixed’.
Table 1. Natural variation at position 45 in KIR.
| KIR lineage | KIR | Pos. 45 | ||
|---|---|---|---|---|
| KIR3DL | Hominoids | LIII | Common; all species | F |
| LIII | 2DS2 | Y | ||
| LIII | Pt-2DL9 | C | ||
| LIII | Gg-2DLe | V | ||
| LII/IIIa | Pt-3DL1/2b, Pp-3DLa, Gg-3DLa | V | ||
| LII | Human, chimpanzeeb, bonobob, orangutan, gibbons | S | ||
| LV | Human, chimpanzee, gorilla, orangutan, gibbons | T | ||
|
| ||||
| Old World monkeys | LII, LIII | Macaque, Sabeus | F | |
| LII | Cs-3DL3, Cs-3DS4c | L | ||
| LIII | Cs-1Dd | L | ||
| LV | T | |||
|
| ||||
| New World monkeys | S, T, A, D, N | |||
|
| ||||
| Horse | M | |||
| Mouse, rat | I, L | |||
|
| ||||
| KIR3DX | Human, chimpanzee | I | ||
| Macaque, cattle | T | |||
lineage II and III recombinant sequences
except recombinant sequences
originally named Cs-3DH4
some alleles have F
The first observed functional effect of variation at position 45 in lineage III KIR was that tyrosine 45 in KIR2DS2, compared to phenylalanine in C1-specific KIR2DL2, prevented recognition of C1 (Winter et al. 1998). This important functional difference, which distinguishes humans from great apes, was achieved with very small perturbations to the KIR2D structure (Saulquin et al. 2003). Here, we show that variation at position 45 can change the specificity of lineage III KIR for HLA-C, as well as modulating KIR avidity in a subtler manner than seen for KIR2DS2. The extraordinary variety of residues found at position 45 in the D1 domain and the extent of their species specificity point to the importance of substitution at position 45 being a critical factor in the rapid co-evolution of KIR with MHC class I.
Acknowledgments
We thank the Yerkes Regional Primate Center at Emory University for the chimpanzee and orangutan blood samples. This work was supported by NIH grants AI31168 and AI24258 to P.P.
References
- Abi-Rached L, Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J Exp Med. 2005;201:1319–1332. doi: 10.1084/jem.20042558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abi-Rached L, Kuhl H, Roos C, ten Hallers B, Zhu B, Carbone L, de Jong PJ, Mootnick AR, Knaust F, Reinhardt R, et al. A small, variable, and irregular killer cell Ig-like receptor locus accompanies the absence of MHC-C and MHC-G in gibbons. J Immunol. 2010a;184:1379–1391. doi: 10.4049/jimmunol.0903016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abi-Rached L, Moesta AK, Rajalingam R, Guethlein LA, Parham P. Human-specific evolution and adaptation led to major qualitative differences in the variable receptors of human and chimpanzee natural killer cells. PLoS Genet. 2010b;6:e1001192. doi: 10.1371/journal.pgen.1001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams EJ, Thomson G, Parham P. Evidence for an HLA-C-like locus in the orangutan Pongo pygmaeus. Immunogenetics. 1999;49:865–871. doi: 10.1007/s002510050566. [DOI] [PubMed] [Google Scholar]
- Andersson S, Fauriat C, Malmberg JA, Ljunggren HG, Malmberg KJ. KIR acquisition probabilities are independent of self-HLA class I ligands and increase with cellular KIR expression. Blood. 2009;114:95–104. doi: 10.1182/blood-2008-10-184549. [DOI] [PubMed] [Google Scholar]
- Biassoni R, Pessino A, Malaspina A, Cantoni C, Bottino C, Sivori S, Moretta L, Moretta A. Role of amino acid position 70 in the binding affinity of p50.1 and p58.1 receptors for HLA-Cw4 molecules. Eur J Immunol. 1997;27:3095–3099. doi: 10.1002/eji.1830271203. [DOI] [PubMed] [Google Scholar]
- Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405:537–543. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- Graef T, Moesta AK, Norman PJ, Abi-Rached L, Vago L, Older Aguilar AM, Gleimer M, Hammond JA, Guethlein LA, Bushnell DA, et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J Exp Med. 2009;206:2557–2572. doi: 10.1084/jem.20091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guethlein LA, Flodin LR, Adams EJ, Parham P. NK cell receptors of the orangutan (Pongo pygmaeus): a pivotal species for tracking the coevolution of killer cell Ig-like receptors with MHC-C. J Immunol. 2002;169:220–229. doi: 10.4049/jimmunol.169.1.220. [DOI] [PubMed] [Google Scholar]
- Guethlein LA, Abi-Rached L, Hammond JA, Parham P. The expanded cattle KIR genes are orthologous to the conserved single-copy KIR3DX1 gene of primates. Immunogenetics. 2007;59:517–522. doi: 10.1007/s00251-007-0214-x. [DOI] [PubMed] [Google Scholar]
- Karre K. Natural killer cell recognition of missing self. Nat Immunol. 2008;9:477–480. doi: 10.1038/ni0508-477. [DOI] [PubMed] [Google Scholar]
- Moesta AK, Abi-Rached L, Norman PJ, Parham P. Chimpanzees use more varied receptors and ligands than humans for inhibitory killer cell Ig-like receptor recognition of the MHC-C1 and MHC-C2 epitopes. J Immunol. 2009;182:3628–3637. doi: 10.4049/jimmunol.0803401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moesta AK, Graef T, Abi-Rached L, Older Aguilar AM, Guethlein LA, Parham P. Humans differ from other hominids in lacking an activating NK cell receptor that recognizes the C1 epitope of MHC class I. J Immunol. 2010;185:4233–4237. doi: 10.4049/jimmunol.1001951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretta L, Bottino C, Pende D, Castriconi R, Mingari MC, Moretta A. Surface NK receptors and their ligands on tumor cells. Semin Immunol. 2006;18:151–158. doi: 10.1016/j.smim.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Older Aguilar AM, Guethlein LA, Adams EJ, Abi-Rached L, Moesta AK, Parham P. Coevolution of killer cell Ig-like receptors with HLA-C to become the major variable regulators of human NK cells. J Immunol. 2010;185:4238–4251. doi: 10.4049/jimmunol.1001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- Rajalingam R, Parham P, Abi-Rached L. Domain shuffling has been the main mechanism forming new hominoid killer cell Ig-like receptors. J Immunol. 2004;172:356–369. doi: 10.4049/jimmunol.172.1.356. [DOI] [PubMed] [Google Scholar]
- Saulquin X, Gastinel LN, Vivier E. Crystal structure of the human natural killer cell activating receptor KIR2DS2 (CD158j) J Exp Med. 2003;197:933–938. doi: 10.1084/jem.20021624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter CC, Long EO. A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA-C allotypes. J Immunol. 1997;158:4026–4028. [PubMed] [Google Scholar]
- Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol. 1998;161:571–577. [PubMed] [Google Scholar]
- Yawata M, Yawata N, Draghi M, Partheniou F, Little AM, Parham P. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 2008;112:2369–2380. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]