Abstract
Background
Gene therapy is a promising approach to the treatment of a wide range of diseases. The development of efficient and adequate gene delivery systems could be one of the most important factors. Polyethyleneimine, a cationic polymer, is one of the most successful and widely used vectors for nonviral transfection in vitro and in vivo.
Methods
A novel biodegradable poly(ester amine) copolymer (PEA) was successfully prepared from low molecular weight polyethylenimine (PEI, 2000 Da) and poly(L-lactide) copolymers.
Results
According to the results of agarose gel electrophoresis, particle size and zeta potential measurement, and transfection efficiency, the PEA copolymers showed a good ability to condense plasmid DNA effectively into nanocomplexes with a small particle size (≤150 nm) and moderate zeta potential (≥10 mV) at an appropriate polymeric carrier/DNA weight ratio. Compared with high molecular weight PEI (25kDa), the PEA obtained showed relatively high gene transfection efficiency as well as low cytotoxicity in vitro.
Conclusion
These results indicate that such PEA might have potential application as a gene delivery system.
Keywords: polyethylenimine, poly(L-lactide), gene delivery, cytotoxicity, transfection efficiency
Introduction
In recent decades, gene therapy has attracted attention from researchers in the fields of medicine, pharmaceutical sciences, and biotechnology due to its potential to treat chronic diseases and genetic disorders.1,2 Although viral vectors generally yield higher gene expression, there are concerns over the immunogenicity, cytotoxicity, and recombinogenic potential of viral gene transfer. Due to the improved safety profile and ease of preparation and manipulation, nonviral techniques for gene delivery continue to be explored and optimized. Nonviral vectors have merit because of their low immunogenicity, low cytotoxicity, low production cost, and reproducibility.3 However, the primary obstacle to implementing effective gene therapy using nonviral vectors remains their relatively inefficient gene delivery in vitro and in vivo.
One of the most successful and widely studied cationic polymers for gene delivery reported to date is polyethylenimine (PEI), which contains primary, secondary, and tertiary amines, so has the ability to combine with both DNA and siRNA, a characteristic known as the “proton sponge effect.” Branched 25 kDa PEI with high transfection efficiency has been regarded as the gold standard for nonviral vectors. However, due to its cytotoxicity in many cell lines, its application is limited. Low molecular weight PEI is usually not cytotoxic, but has lower transfection efficiency. In order to enhance the efficiency of gene delivery and minimize the cytotoxicity of PEI, several modified PEI, including degradable hyperbranched poly(ester amine) copolymers (PEA),4 crosslinked low molecular weight PEI,5 PEI-chitosan, and biodegradable PEI,6–8 have been investigated as gene carriers to reduce cytotoxicity and increase transfection efficiency. Arote et al recently synthesized a novel degradable PEA based on low molecular weight of PEI and polycaprolactone. They reported that the PEA obtained had low cytotoxicity and high transfection efficiency due to the hydrophobicity of polycaprolactone.7 However, due to its high degree of crystallinity and considerable hydrophobicity, the degradation rate of polycaprolactone is very slow, so it is not easy to absorb in the body.
Polylactide (PLA) and poly(D, L-lactide-co-glycolide) (PLGA) are the most commonly used biodegradable polymers. These have been approved by the US Food and Drug Administration for some human clinical applications, due to their good biocompatibility and biodegradability.9 PLGA/PEI and PLA/PEI nanoparticles might be useful in delivery of pDNA. Future studies will focus on optimizing PLA/PEI and PLGA/PEI nanoparticles for in vivo use as gene delivery systems.10,11
In this paper, a novel degradable water-soluble branched PEA based on PLA and low molecular weight branched PEI was prepared and characterized. The DNA condensation ability, protective ability, particle sizes, and zeta potentials of PEA were investigated. We observed remarkable transfection efficiency and significantly lower cytotoxicity with these polymers as compared with PEI (25 kDa).
Materials and methods
Reagents
Branched PEI (molecular weight 2000 Da), L-lactide, 1,4- butanediol, isophorone di-isocyanate (IPDI), deoxyribonuclease I (DNase I), Dulbecco’s Modified Eagle’s Medium, and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were obtained from Sigma-Aldrich, St Louis, MO. Dimethyl sulfoxide, dibutyl tin dilaurate, stannous octoate (Sn(Oct)2), 1,1,1-trichloroethane, methanol, petroleum ether, and anhydrous dichloromethane were purchased from Chengdu KeLong Chemicals, Sichuan, People’s Republic of China. All the chemicals used were of analytical grade and used as received.
Cell lines and cell culture
Human embryonic kidney 293 (HEK293) cells and a human hepatocellular carcinoma (HepG-2) cell line were incubated in Dulbecco’s Modified Eagle’s Medium containing 10% fetal bovine serum and 1% antibiotics (penicillin-streptomycin, 100 U/mL) at 37°C in a humidified atmosphere containing 5% CO2.
Synthesis of poly(L-lactide)
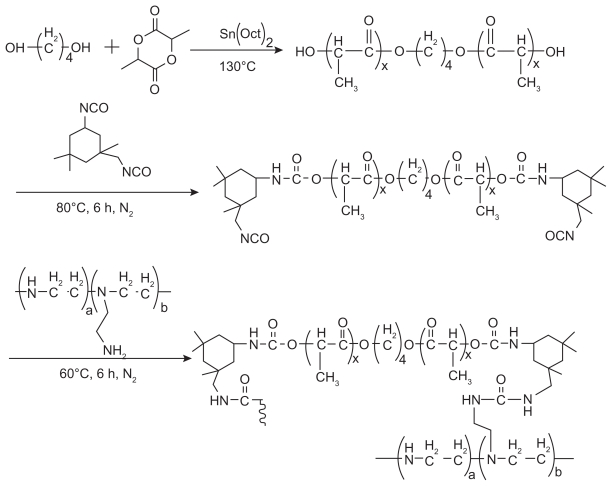
The poly(L-lactide) (PLLA) macromer was prepared by ring-opening copolymerization of L-lactide initiated by 1,4- butanediol, as shown in Figure 1. The calculated L-lactide and 1,4-butanediol were changed to a three-neck round bottle flask in a nitrogen atmosphere. Dry toluene was transferred into the flask, followed by addition of stannous octoate (0.05% w/w in toluene). The system was then kept at 130°C for 14 hours. The resulting copolymer was purified using analytical grade dichloromethane, and then precipitated in excess cold petroleum ether. The mixture was then filtered and dried at room temperature under vacuum for 24 hours. The purified copolymers were kept in a desiccator until further use.
Figure 1.
Synthesis scheme of poly(ester amine) from L-lactide and polyethylenimine.
Synthesis and characterization of NCO-ended PLLA macromer
The PLLA obtained was dissolved in 1,1,1-trichloroethane. A calculated amount of IPDI was added. The solution was then placed in a three-neck glass bottle flask equipped with a reflux condenser, a nitrogen inlet tube, and a magnetic stirrer. The mixture was heated to 80°C for six hours. The reaction system was cooled to room temperature under a nitrogen atmosphere, and the mixture was then precipitated in excess cold petroleum ether, filtered, and dried to a constant weight at 30°C under vacuum. The purified copolymer was kept in a desiccator until further use.
Synthesis and characterization of PLLA-IPDI-PEI
IPDI-PLLA-IPDI and PEI were dissolved in 1,1,1-trichloroethane. The PEI solution was transferred into a three-neck round-bottomed flask equipped with a reflux condenser and stirred at 40°C under nitrogen atmosphere. The IPDI-PLLA-IPDI solution was then added dropwise through a constant pressure infundibulum at 5–10 drops per minute. The system was kept at 80°C for another six hours. The resultant copolymer was cooled to room temperature and then precipitated in excess cold petroleum ether. The mixture was thereafter filtered and dried at room temperature under vacuum for 24 hours. The purified copolymers were kept in a desiccator until further use.
Fourier transform infrared absorption spectra
Fourier transform infrared absorption spectra were recorded on a Nicolet 200SXV infrared spectrophotometer (Nicolet Instruments, Madison, WI). The copolymer samples were prepared after swelling with dichloromethane, mixed with KBr powder, and then pressed into pellets. The spectra were collected over the range of 4000–5000 cm−1.
1H nuclear magnetic resonance analysis
1H nuclear magnetic resonance (NMR) spectra of PLLA and PEA copolymer were recorded on a Varian 400 spectrometer (Varian Medical Systems, Palo Alto, CA) at 400 mHz, using tetramethylsilane as an internal reference and CDCl3 and D2O as the solvent.
Preparation of copolymer/DNA complexes
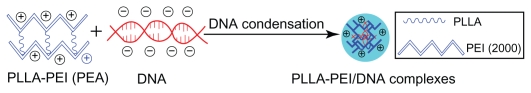
The whole coding sequence of green fluorescent protein was inserted into pcDNA3.1 (Invitrogen, San Diego, CA) which contains a CMV promoter to construct the plasmid green fluorescent protein expressing green fluorescent protein. A plasmid DNA stock solution (2 mg/mL) was prepared in pure water solution. PEA was dissolved in distilled water to form a solution of 2 mg/mL. The PEA/DNA complexes were prepared by adding PEA copolymer solution to the DNA solution at the desired weight ratio and mixed gently. The complexes were incubated at 37°C for 30 minutes before further application. Figure 5 shows possible structure of complexes from plasmid DNA condensation using PLLA-PEI (PEA) copolymers.
Figure 5.
Possible structure of complexes formed from plasmid DNA condensation using PLLA-polyethylenimine (poly[ester amine]) copolymers.
Abbreviations: PLLA, poly(L-lactide); PEI, polyethylenimine; PEA, poly(ester amine).
Gel retardation assays
The extent and effectiveness of DNA condensation by PEA were evaluated using the gel retardation assay. The PEA and DNA solutions were mixed at various weight ratios and incubated for 30 minutes at room temperature to allow complex formation. The PEA/DNA complexes were dissolved in a 1% (w/v) agarose gel pretreated with 0.5 mg/mL ethidium bromide in Tris-boric acid-ethylenediamine tetra-acetic acid buffer at 80 V.
Determination of complex size and zeta potential
The particle size and zeta potential were measured on a zetasizer 3000 HS (Malvern, Worcestershire, UK) at room temperature. The complexes at various weight ratios were prepared by adding an appropriate volume of DNA solution to appropriate volumes of PEA solution. The complexes were then incubated at room temperature for 30 minutes. After that, the polyplexes were diluted for measurement.
Cytotoxicity determination
Cyototoicity evaluation of the PEA copolymer was measured with an MTT assay using HEK293 and HepG-2 cells. Cells were plated in 96-well plates at an initial density of 5 × 105 cells per well in 200 μL of growth medium for 24 hours. Afterwards, the growth medium was replaced with fresh medium, and different amounts of PEA copolymer were added. After 24 hours, the cell cultures were washed with fresh medium and the MTT assay was conducted. Untreated cells were used as controls with 100% viability. The cytotoxicity of PEA copolymer was defined as the relative viability (%) correlating with the amount of viable cells compared with the control cells.
In vitro transfection
The HEK293 cells were seeded in a 24-well plate at an initial density of 1 × 105 cells per well in 2 mL growth medium and incubated for 20–24 hours to achieve 80% confluency. The following day, wells were washed with l mL of phosphate-buffered saline and the medium was replaced. PEA/DNA complexes formed at various weight ratios were added to the wells for four hours before replacing the medium. After 24 hours, transfection efficiency was estimated by the amount of transfected cells with green fluorescent protein that were observed using a fluorescence inverted microscope (IX71, Olympus), and photographed by Spot Flex. Each experiment was precise quantitatively analyzed by flow cytometry.
Statistical analysis
Quantitative data are expressed as means ± standard deviation. Statistical analysis was performed by single-factor analysis of variance. P values less than 0.05 were considered to be statistically significant.
Results
Synthesis and character of PEA composites
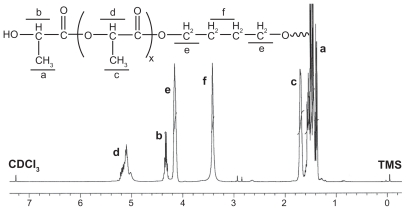
Figure 2 shows the 1H-NMR spectrum of the synthesized PLLA macromer. Peaks C and A were assigned to the methyl protons of –O–CH(CH3)CO– and HO–CH(CH3)–CO–in PLLA. The peaks F and E were attributed to the methylene protons of the two middle –CH2– and the end –CH2– in 1,4-butanediol. The peaks D and B were attributed to the tertiary proton of –O–CH(CH3)CO– and HO–CH(CH3)–CO– in PLLA.
Figure 2.
Representative 1H-nuclear magnetic resonance spectrum (400 mHz) of PLLA copolymer in CDCl3.
Note: In the H-nuclear magnetic resonance spectra, the chemical shifts of the TMS (Si(CH3)4), as relative standard, were defined zero.
Abbreviation: PLLA, poly(L-lactide).
The macromolecular weight (Mn) of the PLLA macromer was calculated from 1H-NMR spectra according to the following equation:
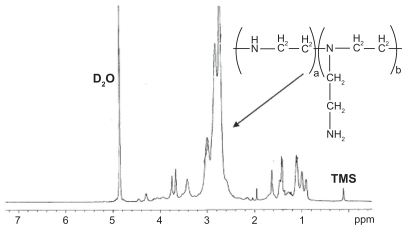
where Id and If were the integral intensities of peaks at 5.193 ppm and 3.608 ppm in the 1H-NMR spectrum of PLLA (Figure 2). The macromolecular weight of the PLLA macromer was calculated to be 490 according to the 1H-NMR spectrum. The 1H-NMR spectrum of the resulting PLLA-PEI based on the reaction of IPDI-PLLA-IPDI and low molecular weight branched PEI (molecular weight 2000 Da) is shown in Figure 3, The broad peaks at 2 ppm and 3 ppm are attributed to the protons of –NHCH2CH2– in the PEI moiety of PLLA-PEI.
Figure 3.
Representative 1H-nuclear magnetic resonance spectrum (400 mHz) of poly(ester amine) copolymer in D2O.
Note: In the H-nuclear magnetic resonance spectra, the chemical shifts of the TMS (Si(CH3)4), as relative standard, were defined zero.
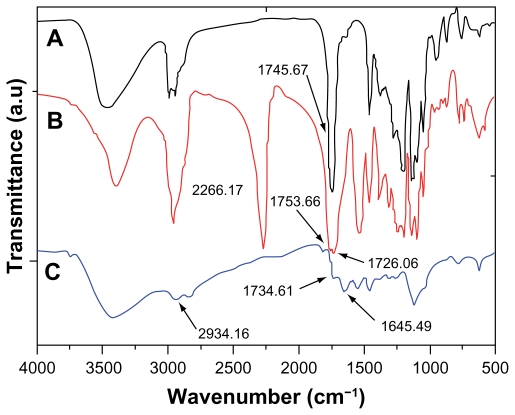
Figure 4 shows the typical Fourier transform infrared spectra of the PLLA, IPDI-PLLA-IPDI, and PEA composite. In Figure 4A, the absorption peaks at 2991.00 cm−1 and 2994.56 cm−1 represent the bending vibration of –CH3– and –CH2–. The peaks at 1745.61 cm−1 belong to the ester carbonyl (C=O). The wide absorption band at 3476.28 cm−1 is assigned to the terminal hydroxyl groups (–OH) in the copolymer. In Figure 4B, there is a strong absorption in 2266.17 cm−1 which is attributed to the –NCO stretching vibrations, and the absorption peak at 1726.06 cm−1 represents the bending vibration of urethane carbonyl (C=O). It is clear that PLLA only reacted with one –NCO in IPDI, which is more active than the other. In Figure 4C, there is no absorption at 2250–2270 cm−1, which indicates that the –NCO groups of IPDI disappears completely due to the coupling reaction of –NCO with –NH2. A weak peak at 1734 cm−1 belongs to ester carbonyl (C=O) which could be seen and enhanced obviously in composite. The absorption band at 1645.69 cm−1 is attributed to the stretching vibration of urea carbonyl (C=O) in the PEA macromer.
Figure 4.
Fourier transform infrared absorption spectra of (A) PLLA, (B) isophorone di-isocyanate-PLLA-isophorone di-isocyanate, and (C) poly(ester amine) copolymer.
Abbreviation: PLLA, poly(L-lactide).
Gel retardation assay
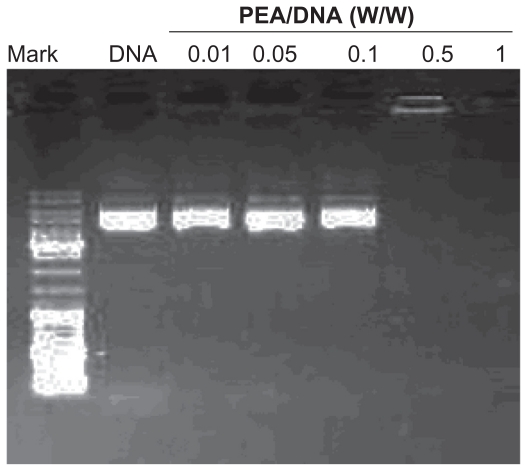
In this study, to determine the optimal concentration for complete inhibition of DNA, the PEA and DNA solutions were mixed at various w/w PEA/DNA ratios keeping the amount of DNA (2 μg) constant, and incubated for 30 minutes at room temperature to allow formation of the complexes. Figure 6 shows the results of agarose gel electrophoresis of the PEA/DNA complexes at various weight ratios (from 0.01 to 1). Naked DNA was used as the control group. It is obvious that increasing amounts of PEA in the complexes led to decreased electrophoretic mobility. When the weight ratio (carrier to gene) was 0.5, the PEA/DNA complexes showed complete retardation and the band completely disappeared.
Figure 6.
Agarose gel electrophoresis of poly(ester amine) (PeA)/DNA complexes at various weight ratios.
Particle size and zeta potential
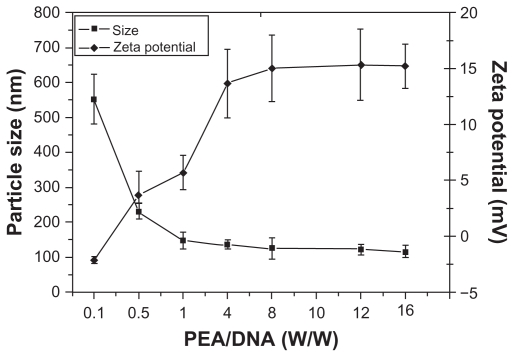
The particle size of the PEA/DNA complexes at various weight ratios (carrier to DNA) ranging from 0.1 to 16 were measured by adding different volumes of PEA solution to DNA solutions at physiological ionic strength (in 0.9% NaCl solution). As shown in Figure 7, the particle size gradually decreases with increasing weight ratio due to the net positive electrostatic repulsion between complexes. At a PEA/DNA weight ratio of 0.1, the particle size is 551 nm, indicating poor complexation between DNA and PEA. At a PEA/DNA weight ratio of 1, the size of the PEA/DNA nanoparticles reduced to 140 nm. At a PEA/DNA weight ratio of 1 and 16, the particle sizes of PEA/DNA complexes are less than 150 nm.
Figure 7.
Particle sizes and zeta potentials of poly(ester amine) (PEA)/DNA complexes at various weight ratios.
Figure 7 indicates that the majority of PEA condensed DNA into nanoparticles with significant surface charges. At a carrier to DNA weight ratio of 0.1, the zeta potential was −2.1 mV, indicating poor complexation between DNA and PEA. As expected, the zeta potential rapidly increased, with an increasing carrier to DNA weight ratio from 0.5 to 16. Average zeta potentials of the PEA/DNA complexes ranged from 3.7 to 15.2 mV.
In vitro cytotoxicity
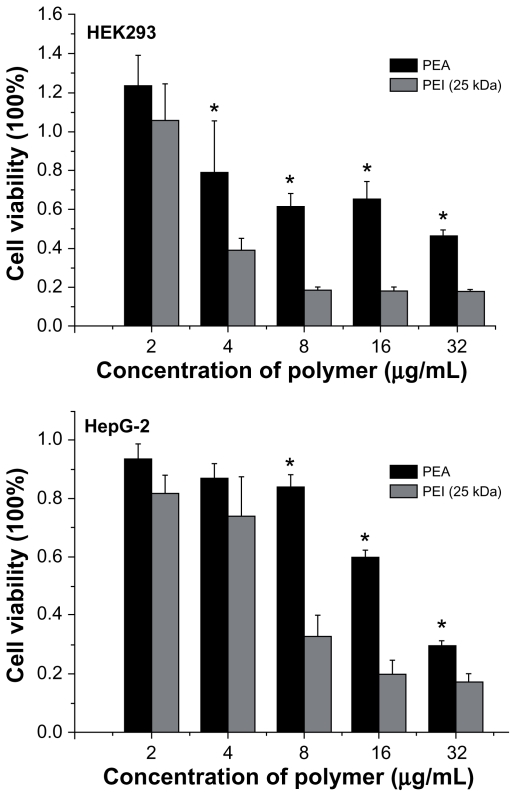
Figure 8 shows the results for the in vitro cytotoxicity of PEA and PEI (25 kDa) analyzed by the MTT method in the HEK293 and HepG-2 cell lines. Cell viability was calculated according to the following equation:
Figure 8.
Cytotoxicity of poly(ester amine) (PEA) and polyethylenimine (PEI) (25 kDa) at various weight ratios in HEK293 and HepG-2 cell lines.
Notes: Cells were seeded in 96-well plates at an initial density of 5 × 105 cells/well in 0.2 mL of growth media; cell viability were detected by MTT assay (mean ± standard deviation, n = 6); significance were assessed compared with the polyethylenimine (25 kDa) control for each time point assayed; *P < 0.05.
where OD570(control) was obtained in the absence of polymers and OD570(sample) was obtained in the presence of polymers. As shown in Figure 8, PEA and PEI (25 kDa) showed a strong dose-dependent effect on cytotoxicity. No significant cytotoxicity was found among the various polymers at the low concentration (≤2 ug/mL). However, with increasing concentration, PEA was found to have much lower cytotoxicity than PEI (25 kD) in the HEK293 and HepG-2 cell lines (P < 0.05).
In vitro transfection
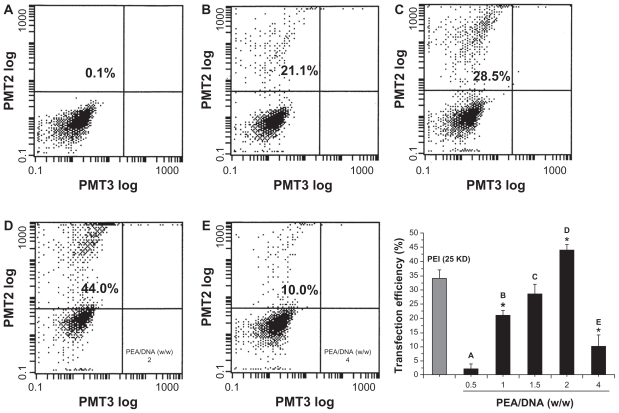
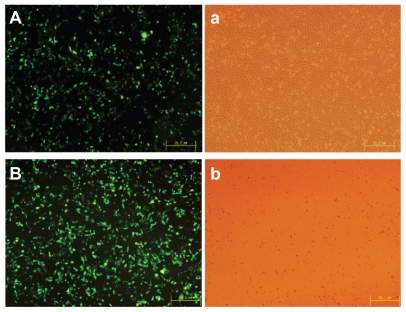
The gene delivery efficiency of PEA based on PLA and low molecular weight PEI was evaluated by in vitro transfection experiments in HEK293 cells using plasmid DNA. For rapid examination, PEA/DNA polyplexes formed at various carrier to DNA weight ratios were added to HEK293 cell cultures followed by incubation. After transient transfection and additional incubation for 24 hours, transfection efficiency was analyzed quantitatively using flow cytometry, and transfection images were observed by fluorescence inverted microscopy and photographed by Spot Flex. Surprisingly, the most efficient gene expression of PEA was observed at the carrier to DNA weight ratio of 2:1. The highest transfection efficiency of PEA was 44% ± 3%, as shown in Figure 9, while the optimized transfection efficiency of the control PEI (25 kDa) was 34% ± 4% at the carrier to DNA weight ratio of 1.5:1.0, which equals an N/P ratio of 10:1. Figure 10 shows the high transfection efficiency images for PEI (25 kDa) and PEA in the HEK293 cell lines.
Figure 9.
Flow cytometry graphs typical of the transfection efficiency in HEK293 cells were incubated with poly(ester amine)/DNA complexes at different weight ratios for 24 hours, polyethylenimine (25 kDa) and an optimized carrier to gene weight ratio of 1.5 as the control.
Note: P < 0.05.
Abbreviations: PEI, polyethylenimine; PEA, poly(ester amine); PMT, photo multiplier tube.
Figure 10.
High transfection efficiency images of (A) polyethylenimine (25 kDa) and (B) poly(ester amine)/DNA were shown in cell lines HEK293.
Notes: Cells were incubated with polyethylenimine (25 kDa)/DNA and poly(ester amine)/DNA complexes at carrier to gene weight ratios of 2 and 1.5 for 24 hours; green fluorescent protein expression was observed under fluorescent microscopy.
Discussion
The aim of this research was to develop and optimize delivery systems for plasmid DNA. In this paper, we successfully synthesized a novel biodegradable PEA copolymer based on PLA and low molecular weight PEI, which showed degradability, a good ability to form complexes with DNA, and suitable physicochemical properties for a gene delivery system. These polymers had less cytotoxicity and exhibited much higher gene transfer efficiency than PEI (25 kDa) itself. Therefore, it is expected that this PEA will have the potential to be a safe gene carrier owing to its degradability, low cytotoxicity, and high transfection efficiency.
The condensation of anionic DNA into small particles is an important prerequisite for gene carriers employing polycations.12 From Figure 6, when the carrier to DNA weight ratio is greater than 0.5, the PEA polymer can concentrate DNA, and can protect against DNase I. Meanwhile, the small and compact complex particle is critically important for polycations used as gene vectors,13 and particle size would apparently affect the transfection efficiency of gene vectors.14 Particle size depends on many parameters, including DNA concentration, sequence of addition of polycations or DNA during complex preparation, and the ionic strength of the solvent. 15 Large particles in the micrometer range in the vascular periphery are poorly internalized by the cells, so the desirable in vivo vectors should be small (below 150 nm) and compact for good systemic circulation and effective endocytosis.16–18 From Figure 7, showing a weight ratio between 1 and 16, the particle sizes of PEA/DNA complexes are smaller than the 150 nm limit for effective endocytosis. The particle size decreased with increasing charge ratio. This was due to the strong electrostatic interactions between the polymer and DNA. The positively charged complexes would bind to the negatively charged cellular membrane, facilitating the entry of the complexes into cells by cellular uptake.19–21 From Figure 7, when the carrier to DNA weight ratio is greater than 0.5, the average zeta potential of the PEA/DNA complexes ranged from 3.67 mV to 15.2 mV. Therefore, PEA is expected to be a suitable candidate for effective transfection owing to its ideal particle size and significant surface charge.
The cytotoxicity of a polymeric gene vector may be an important factor affecting transfection efficiency. It is generally thought that the polycation cytotoxicity is caused by polymer aggregation on the cell surface due to strong electrostatic interactions in the plasma membrane, which impairs cell membrane function.22,23 Bieber and Elsasser suggested that there is a correlation between cytotoxicity and molecular weight.12 It has already been reported that high molecular weight PEI is significantly more cytotoxic than low molecular weight PEI.20 In this research, the cytotoxicity of PEA and PEI (25 kDa) was also analyzed using the MTT assay. From Figure 8, with increasing concentrations, PEA complexes were found to be much less cytotoxic than PEI (25 kDa) in the HEK293 and HepG-2 cells, which indicates that a wider dose range of polymer could be selected for gene transfer. PEI (25 kDa), which displays cytotoxicity, has no degradation or excretion pathway in vivo. Therefore, it may accumulate in vivo, resulting in increased cytotoxicity.24 Owing to the backbone of the PLA and low molecular weight PEI (2000 Da), the synthesized PEA can be expected to have better biocompatibility.
PEIs with a molecular weight of 25 kDa or greater have shown good transfection efficiency both in vitro and in vivo.25,26 However, these higher molecular weight cationic polymers induce rapid necrotic-like changes resulting from perturbation of the plasma membrane and the activation of mitochondrial-mediated apoptosis.27 Low molecular weight PEI (2000 Da) displayed much less cytotoxicity but almost no transfection activity.28 Thus, PEI (≤2000 Da) was used in the current study not just because of its endosomolytic function, but also because of its lower cytotoxicity and lower background transfection efficiency. Relatively, compared with PEI (25 kDa), we attempted to achieve low cytotoxicity and to increase the transfection efficiency of the nonviral gene carriers. From Figure 9, it is clear that the transfection efficiency of PEA is higher than that of the gold standard, PEI (25 kDa). Our results demonstrate that degradable, disulfide-crosslinked, low molecular weight PEI has higher gene transfection efficiency and lower cytotoxicity than nondegradable high molecular weight PEI (25 kDa). Therefore, these PEAs can be used as efficient vectors, providing a versatile platform for further investigation of their structure-property relationship, along with fair stability, significantly low cytotoxicity, and high transfection efficiency.
Acknowledgment
This research was funded by the New Century Excellent Talents in University and Chinese Key Basic Research Program.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Verma M, Somia N. Gene therapy – promises, problem and prospects. Nature. 1997;389:239–242. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 2.Edelstein ML, Abedi MR, Wixon J. Gene therapy clinical trials worldwide to 2007: an update. J Gene Med. 2007;9:833–842. doi: 10.1002/jgm.1100. [DOI] [PubMed] [Google Scholar]
- 3.Lee M, Kim SW. Polyethylene glycol-conjugated copolymers for plasmid DNA delivery. Pharm Res. 2005;22:1–10. doi: 10.1007/s11095-004-9003-5. [DOI] [PubMed] [Google Scholar]
- 4.Fischer D, Bieber T, Li Y, Elsasser HP, Kissel T. A novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: effect of molecular weight on transfection and cytotoxicity. Pharm Res. 1999;16:1273–1279. doi: 10.1023/a:1014861900478. [DOI] [PubMed] [Google Scholar]
- 5.Gosselin MA, Guo W, Lee RJ. Efficient gene transfer using reversibly cross-linked low molecular weight polyethylenimine. Bioconjug Chem. 2001;12:989–994. doi: 10.1021/bc0100455. [DOI] [PubMed] [Google Scholar]
- 6.Wong K, Sun G, Zhang X, et al. PEI-g-chitosan, a novel gene delivery system with transfection efficiency comparable to polyethylenimine in vitro and after liver administration in vivo. Bioconjug Chem. 2006;17:152–158. doi: 10.1021/bc0501597. [DOI] [PubMed] [Google Scholar]
- 7.Arote R, Kim TH, Kim YK, et al. A biodegradable poly(ester amine) based on polycaprolactone and polyethylenimine as a gene carrier. Biomaterials. 2007;28:735–744. doi: 10.1016/j.biomaterials.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 8.Peterson H, Merdan T, Kunath F, et al. Poly(ethylenimine-co- L-lactamide- co-succinamide): a biodegradable poly-ethylenimine derivative with an advantageous pH-dependent hydrolytic degradation for gene delivery. Bioconjug Chem. 2002;13:812–821. doi: 10.1021/bc0255135. [DOI] [PubMed] [Google Scholar]
- 9.Sahoo SK, Panyam J, Prabha S, Labhasetwar V. Residual polyvinyl alcohol associated with poly(D, L-lactide-co-glycolide) nanoparticles affects their physical properties and cellular uptake. J Control Release. 2002;82:105–114. doi: 10.1016/s0168-3659(02)00127-x. [DOI] [PubMed] [Google Scholar]
- 10.Bivas-Benita M, Romeijn S, Junginger HE, Borchard G. PLGA-PEI nanoparticles for gene delivery to pulmonary epithelium. Eur J Pharm Biopharm. 2004;58:1–6. doi: 10.1016/j.ejpb.2004.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim IS, Lee SK, Park YM, et al. Physicochemical characterization of poly(l-lactic acid) and poly(d,l-lactide-co-glycolide) nanoparticles with polyethylenimine as gene delivery carrier. Int J Pharm. 2005;298:255–262. doi: 10.1016/j.ijpharm.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 12.Bieber T, Elsasser HP. Preparation of a low molecular weight polyethylenimine for efficient cell transfection. Biotechniques. 2001;30:74–81. doi: 10.2144/01301st03. [DOI] [PubMed] [Google Scholar]
- 13.Lin C, Zhong ZY, Lok MC, et al. Linear poly(amido amine)s with secondary and tertiary amino groups and variable amounts of disulfide linkages: synthesis and in vitro gene transfer properties. J Control Release. 2006;116:130–137. doi: 10.1016/j.jconrel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 14.Xu D, Yao S, Liu Y, et al. Size-dependent properties of M-PEIs nanogels for gene delivery in cancer cells. Int J Pharm. 2007;338:291–296. doi: 10.1016/j.ijpharm.2007.01.050. [DOI] [PubMed] [Google Scholar]
- 15.Kircheis R, Wightman L, Wagner E, et al. Design and gene delivery activity of modified polyethylenimines. Adv Drug Deliv Rev. 2001;53:341–358. doi: 10.1016/s0169-409x(01)00202-2. [DOI] [PubMed] [Google Scholar]
- 16.Park MR, Han KO, Han IK, et al. Degradable polyethylenimine-alt- poly(ethylene glycol) copolymers as novel gene carriers. J Control Release. 2005;105:367–380. doi: 10.1016/j.jconrel.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 17.Nakai T, Kanamori T, Sando S, Aoyama Y. Remarkably size-regulated cell invasion by artificial viruses saccharide-dependent self-aggregation of glycoviruses and its consequences in glycoviral gene delivery. J Am Chem Soc. 2003;125:8465–8475. doi: 10.1021/ja035636f. [DOI] [PubMed] [Google Scholar]
- 18.Vinogradov SV, Bronich TK, Kabanov AV. Self-assembly of polyamine-poly(ethylene glycol) copolymers with phosphorothioate oligonucleotides. Bioconjug Chem. 1998;9:805–812. doi: 10.1021/bc980048q. [DOI] [PubMed] [Google Scholar]
- 19.Kim T, Baek J, Yoon JK, et al. Synthesis and characterization of a novel arginine-grafted dendritic block copolymer for gene delivery and study of cellular uptake pathway leading to transfection. Bioconjug Chem. 2007;18:309–317. doi: 10.1021/bc0601525. [DOI] [PubMed] [Google Scholar]
- 20.Kunath K, Harpe AV, Fischer D, et al. Low-molecular-weight polyethylenimine as a non-viral vector for DNA delivery: comparison of physicochemical properties, transfection efficiency and in vivo distribution with high-molecular-weight polyethylenimine. J Control Release. 2003;89:113–125. doi: 10.1016/s0168-3659(03)00076-2. [DOI] [PubMed] [Google Scholar]
- 21.Tian H, Xiong W, Wei JZ, et al. Gene transfection of hyperbranched PEI grafted by hydrophobic amino acid segment PBLG. Biomaterials. 2007;28:2899–2907. doi: 10.1016/j.biomaterials.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 22.Fischer D, Bieber T, Li Y, Elsasser H, Kissel T. A novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: effect of molecular weight on transfection efficiency and cytotoxicity. Pharm Res. 1999;16:1273–1279. doi: 10.1023/a:1014861900478. [DOI] [PubMed] [Google Scholar]
- 23.Ryser HJ. A membrane effect of basic polymers dependent on molecular size. Nature. 1967;215:934–936. doi: 10.1038/215934a0. [DOI] [PubMed] [Google Scholar]
- 24.Huang HL, Yu H, Li D, et al. A novel copolymer based on hydroxypropyl a-cyclodextrin conjugated to low molecular weight polyethylenimine as an in vitro gene delivery vector. Int J Mol Sci. 2008;9:2278–2289. doi: 10.3390/ijms9112278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boussif O, Lezoualch F, Zanta MA, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc Natl Acad Sci U S A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neu M, Fischer D, Kissel T. Recent advances in rational gene transfer vector design based on poly(ethylenimine) and its derivatives. J Gene Med. 2005;7:992–1009. doi: 10.1002/jgm.773. [DOI] [PubMed] [Google Scholar]
- 27.Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. A two-stage poly(ethylenimine)-mediated cytotoxicity: implications for gene transfer/therapy. Mol Ther. 2005;11:990–995. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Fischer D, Bieber T, Li Y, Elsasser HP, Kissel T. A novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: effect of molecular weight on transfection efficiency and cytotoxicity. Pharm Res. 1999;16:1273–1279. doi: 10.1023/a:1014861900478. [DOI] [PubMed] [Google Scholar]