Abstract
Background
The Characidium (a Neotropical fish group) have a conserved diploid number (2n = 50), but show remarkable differences among species and populations in relation to sex chromosome systems and location of nucleolus organizer regions (NOR). In this study, we isolated a W-specific probe for the Characidium and characterized six Characidium species/populations using cytogenetic procedures. We analyzed the origin and differentiation of sex and NOR-bearing chromosomes by chromosome painting in populations of Characidium to reveal their evolution, phylogeny, and biogeography.
Results
A W-specific probe for efficient chromosome painting was isolated by microdissection and degenerate oligonucleotide primed-polymerase chain reaction (DOP-PCR) amplification of W chromosomes from C. gomesi. The W probe generated weak signals dispersed on the proto sex chromosomes in C. zebra, dispersed signals in both W and Z chromosomes in C. lauroi and, in C. gomesi populations revealed a proximal site on the long arms of the Z chromosome and the entire W chromosome. All populations showed small terminal W probe sites in some autosomes. The 18S rDNA revealed distinctive patterns for each analyzed species/population with regard to proto sex chromosome, sex chromosome pair, and autosome location.
Conclusions
The results from dual-color fluorescence in situ hybridization (dual-color FISH) using W and 18S rDNA probes allowed us to infer the putative evolutionary pathways for the differentiation of sex chromosomes and NORs, from structural rearrangements in a sex proto-chromosome, followed by gene erosion and heterochromatin amplification, morphological differentiation of the sex chromosomal pair, and NOR transposition, giving rise to the distinctive patterns observed among species/populations of Characidium. Biogeographic isolation and differentiation of sex chromosomes seem to have played a major role in the speciation process in this group of fish.
Background
The Crenuchidae is widespread in freshwater systems of the South and Central Americas [1]. The Characidium is the most representative group in this family, comprising 50 valid species [1]. This is a particularly interesting Neotropical fish group for cytogenetic studies because it presents a diversified model of sex chromosomes [2]. In the Characidium, a diploid number of 50 chromosomes is observed in all studied species and a karyotype of 32 m + 18 sm is most commonly reported [2-10].
Although the diploid number is conserved, the Characidium exhibit remarkable interspecific and interpopulation differences, such as (i) inter- and intraindividual variation of B chromosomes in some species [3-5]; (ii) a sex chromosome system with ZZ/ZW female heterogamety [2-9]; (iii) variation in the location and number of rDNA sites in distinct chromosomal pairs [2-4,8,10]; and (iv) occurrence of natural triploidy in C. gomesi and C. cf. zebra [6,10].
The first cytogenetic report in Characidium was for C. zebra [11] and, so far, this is the only species where heteromorphic sex chromosomes are absent [4,6,10]. Based on morphological analysis, C. zebra exhibits several plesiomorphic traits, being placed as basal in Characidium phylogeny [12]. The chromosomal characters of C. zebra have already been referred to as putatively plesiomorphic for the genus [2]. Therefore, changes in the karyotypes observed in other species of Characidium, when compared with C. zebra, should be the result of secondary chromosomal rearrangements.
The karyotypes of C. lauroi and C. alipioi have been described, and while the latter was characterized by the presence of a ZZ/ZW sex chromosome system, C. lauroi was considered homomorphic because of the lack of differential heterochromatic bands [7]. However, further comparative studies in seven species of this genus (C. zebra, C. lanei, C. pterostictum, C. lauroi, C. oiticicai, Characidium sp. and C. schubarti) showed that only C. zebra lacked differentiated sex chromosomes and the other species presented sex chromosomes in different W-heterochromatin amplification stages and had terminal NORs on both Z and W chromosomes [4]. Characidium gomesi is another species with highly differentiated sex chromosomes, bearing entirely heterochromatic W chromosomes and NORs on autosomes [2,3,6].
A sex proto-chromosome pair for Characidium species was proposed, from which both Z and W chromosomes have evolved by structural rearrangements, such as duplications, deletions, and/or inversions. By contrast, the heterochromatin amplification of W was regarded as a predominant event in the differentiation of the sex chromosomal pair [2]. Later, it was inferred that the NORs on ZW chromosomes in most species of Characidium represented an ancestor state of sex proto-chromosomes, prior to heterochromatin amplification [4]. Heterochromatin amplification in one of the sex chromosomes driven by the accumulation of certain repetitive DNA classes in one homologous is relatively common in fish [13]. Such a process would favor recombination suppression and thereby the independent evolution of chromosomes or chromosomal regions [14-17].
In the present study, we analyzed the origin, composition, and differentiation of sex and NOR-bearing chromosomes in populations of C. zebra, C. lauroi, and C. gomesi, using chromosomal painting with a W-specific probe from C. gomesi. We also mapped the 18S rDNA to infer the evolution, phylogeny and biogeography of the group.
Results
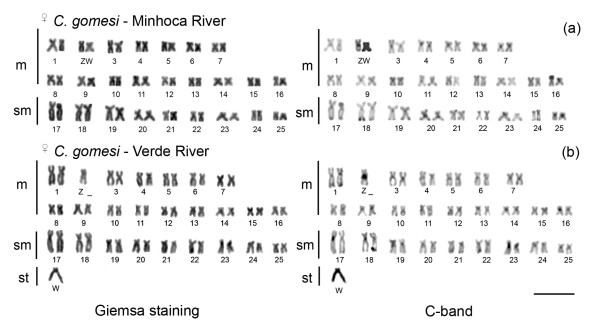
Populations of C. zebra from Passa Cinco River, Upper Paraná basin (SP), C. lauroi from Grande stream, Paraíba do Sul basin (SP), and C. gomesi from Grande River, Upper Paraná basin (SP), Alambari stream, Tietê/Upper Paraná basin (SP), Minhoca stream, São Francisco River basin (MG), and Verde River, Tibagi River/Upper Paraná basin (PR) have a conserved diploid number of 50 chromosomes, most of which are metacentric/submetacentric (m/sm) (table 1). The populations of C. gomesi from Minhoca stream-MG and Verde River-PR are analyzed in the present work for the first time, and showed a differentiated ZZ/ZW system (Figure 1 and table 1). In both populations, the Z chromosome is a medium-sized m chromosome carrying a proximal heterochromatin band on the long arms. The W chromosome is an entirely heterochromatic m, smaller than Z chromosome in specimens from Minhoca stream (Figure 1c), whereas the W chromosome in the population from Verde River is a heterochromatic subtelocentric (st) chromosome, slightly larger than Z (Figure 1d). Therefore, the karyotype formulae differ between both populations because of the morphological differences in the W chromosome (Figure 1a, c and table 1).
Table 1.
Chromosomal formula, sex chromosome system occurrence, W-specific probe sites and 18S rDNA sites on the chromosomes of the Characidium species/populations analysed.
| Species | River | Chromosomal Formula | Sex System | W probe sites | 18S rDNA sites/localization | Figure 3 relation |
|---|---|---|---|---|---|---|
| C. zebra | Passa Cinco | ♂ ♀ 32 m+18 sm | Absent | Dispersed in autosomes | 23 pair/subterminal | (a) |
| C. lauroi | Grande stream | ♂ 32 m+18 sm ♀ 31 m+18 sm+1st |
ZZ/ZW | Autosomal and dispersed in the W chromosome | Z and W/terminal | (b) |
| C. gomesi | Alambari | ♂ ♀ 32 m+18 sm | ZZ/ZW | Z in proximal region of the long arm, entire m W chromosome and some terminal minor sites over autosomes | Z and W/terminal and 19 pair/terminal in just one homologue in males | (c and d) |
| C. gomesi | Grande | ♂ ♀ 32 m+18 sm | ZZ/ZW | Z in proximal region of the long arm, entire m W chromosome and some terminal minor sites over autosomes | 17 pair/terminal | (e and f) |
| C. gomesi | Minhoca stream | ♂ ♀ 32 m+18 sm | ZZ/ZW | Z in proximal region of the long arm, entire m W chromosome and some terminal minor sites over autosomes | 17 pair/terminal | (g) |
| C. gomesi | Verde | ♂ 32 m+18 sm ♀31 m+18 sm+1st |
ZZ/ZW | Z in proximal region of the long arm, entire st W chromosome and some terminal minor sites over autosomes | 17 and 22 pairs/terminal and just one homologue of pairs 1 and 20/terminal | (h) |
Figure 1.
Karyotypes of females of C. gomesi. In (a) population from the Minhoca stream, São Francisco River basin (MG). The W chromosome is entirely heterochromatic and easily identifiable by C-banding, used in the chromosomal microdissection and obtaining of W-specific probe. In (b) population from the Verde River, Tibagi River basin (PR). Bar = 10 μm.
DOP-PCR amplification of C-banded W chromosomes from C. gomesi and hybridization of W probe onto the metaphasic chromosomes of C. gomesi from Minhoca stream and Verde River was highly efficient, revealing positive signals at the terminal regions of some autosomes, at the proximal region on the long arms of the Z chromosome, and the entire W chromosome (Figure 2a and 2b).
Figure 2.
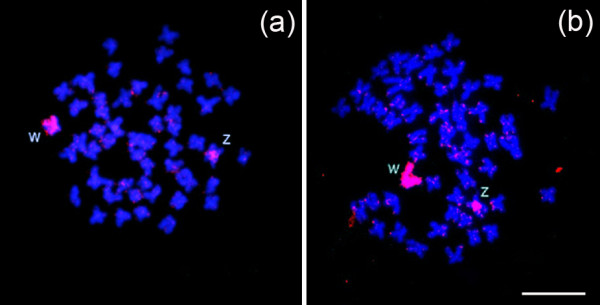
W Characidim probe efficiency. In (a) female metaphases of C. gomesi from the Minhoca stream population and (b) from the population of Verde River (PR) showing the chromosomes hybridised with the W-specific probe. Bar = 10 μm.
Dual-color FISH using 18S rDNA and W probes revealed distinctive patterns for each analyzed population. Thus, C. zebra, presented small sites related to the W probe dispersed through the karyotype and an 18S rDNA site in the sm pair 23 (Figure 3a and table 1). Characidium lauroi presented 18S rDNA signals on the heteromorphic m pair 2, regarded as the sex chromosomes in this species, and hybridization with the W probe revealed weak signals dispersed on both the W and Z chromosomes, in addition to some terminal autosomal regions (Figure 3b and table 1). The population of C. gomesi from the Alambari stream showed 18S rDNA sites in the Z and W chromosomes, besides an additional terminal site in homologous pair 19, exclusively for males (Figure 3c, d and table 1). The W probe hybridized mainly on the proximal region of the Z chromosome and to almost the entire m W chromosome, except for the NORs, besides on some minor terminal sites in the autosomes (Figure 3c, d and table 1). Characidium gomesi from the Grande River and Minhoca stream presented similar 18S rDNA signals in the sm pair 17. The W probe hybridized mainly on the proximal region of the Z chromosome and the entire m W chromosome, besides some small terminal sites on the autosomes (Figure 3e, f, g and table 1). On the other hand, C. gomesi from the Verde River showed six positive 18S rDNA sites. FISH with the W probe revealed small terminal sites in some autosomes, besides a proximal site on the long arms of the Z chromosome and the entire st W chromosome (Figure 3h, table 1).
Figure 3.
Location of signals after hybridization with W-specific (red) and 18S rDNA (green) probes in the karyotypes of species/populations of Characidium stained with DAPI. Bar = 10 μm.
Discussion
The karyotypic analysis
The populations of C. gomesi (Minhoca stream and Verde River-analyzed for the first time in the present work) are highly similar (2n = 50 chromosomes, moistly m/sm) to other previously studied populations. Furthermore, although 32 m + 18 sm represents the most common karyotype in this group, chromosomal formulae variations have already been reported, indicating that some rearrangements, such as pericentric inversions, might have taken place in the chromosomal diversification of this genus [2,4,5]. Nonetheless, in spite of the apparent karyotype conservation, the populations of C. gomesi have diversified mainly in the structure and morphology of their Z and W chromosomes and the number of major rDNA sites [2]. Amongst the several analyzed populations of C. gomesi, only one showed NORs located on sex chromosomes [5]. Therefore, among the Characidium species bearing heteromorphic sex chromosomes, only C. alipioi and C. gomesi (excepting the sample from the Alambari stream) have NORs exclusively on autosomes, suggesting this is a derived feature [4,5].
In general, it is hypothesized that sex chromosomes evolved from a specific pair of autosomes carrying some sex-determining gene(s). Subsequently, the newly formed sex chromosomes stopped recombination in a small region around the sex-determining locus [18]. Sex chromosomes in this early stage of evolution are not cytologically distinguishable (homomorphic). The process of recombination suppression then progressed through almost the entire sex chromosome. The exchange restraint between homologs during meiosis could be caused by structural chromosomal rearrangements [19] or by the accumulation of certain repetitive DNA classes in one homolog in the pair of sex proto-chromosomes [20,21]. The results obtained permit speculation concerning this sex chromosome differentiation.
Sex chromosomes and NOR differentiation in Characidium
Comparative chromosomal painting using the W-specific probe of C. gomesi demonstrated that the repetitive DNAs present on sex chromosomes from different populations of these species are highly conserved, despite the morphological diversification of these chromosomes. In addition, the occurrence of W-specific sites on autosomes, albeit dispersed over terminal regions in all analyzed populations/species, reveals that the repetitive units that compose and differentiate the sex chromosomes in Characidium are widespread in the genus. This corroborates the hypothetical common origin of the ZZ/ZW system [2,5] and its monophyletic status, giving rise to hypothesis of serial transformation of these chromosomes within the genus Characidium.
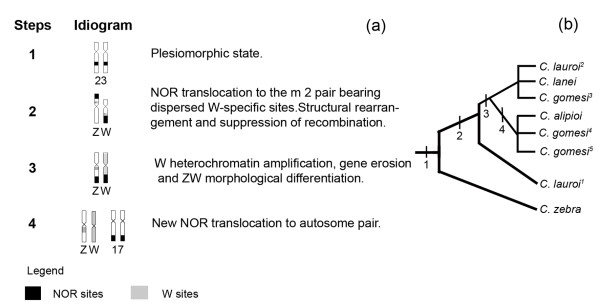
Based on this scenario, it is possible to hypothesize that the sex chromosomes of Characidium had a common origin and diversified in sequential stages (Figure 4). Therefore, the ancestor condition, without morphologically differentiated sex chromosomes, seems to be present in C. zebra, with NORs located at the subterminal region of a small sm pair (23rd) and W-specific regions dispersed at the terminal regions of some chromosomes (Figure 4, step 1).
Figure 4.
Stages in the differentiation of sex and NOR-bearing chromosomes among species and populations of Characidium. In (a) steps and idiogram representation of the NOR and W sequences rearrangements; (b) hypothesised relationship among species analysed. 1C. lauroi of the Ribeirão Grande population; 2C. lauroi of the Ubatuba population; 3C. gomesi of the Alambari population; 4C. gomesi of the Minhoca population and; 5C. gomesi of the Verde population.
The next step probably involved translocation of the NOR to the 2nd m pair, bearing dispersed W-specific sites, along with structural rearrangement of the pair (pericentric inversion), size heteromorphism, and suppression of recombination during meiosis (Figure 4, step 2). This situation is found in the ZW pair of the C. lauroi population analyzed in the present study. Supporting this hypothesis, other populations of C. lauroi also presented NOR-bearing Z and W chromosomes and showed a higher differentiation stage of sex chromosomes because of heterochromatin amplification [4]. Afterwards, gene erosion and heterochromatin amplification of the W chromosome would be more accentuated, leading to an accumulation of W-specific sequences associated with the differentiation of morphological types of Z and W chromosomes reported among species/populations of Characidium. Such a stage can be visualized in the intermediary developmental stages in C. lanei [9,17], or more advanced stages, such as those as in the population of C. gomesi from the Alambari stream (Figure 4, step 3).
Simultaneously with the intense W heterochromatin amplification, such as observed among populations of C. gomesi (Grande River, Minhoca stream, Verde River) and C. alipioi, new translocation events placed the NORs on an autosomal sm pair (17th or 18th) (Figure 4, step 4). The maintenance of NORs on sex chromosomes in the population of C. gomesi (Alambari stream) strongly suggests that NORs have undergone a novel transposition in other populations, moving to an autosomal pair. This condition could be selectively favored by avoiding the W heterochromatin amplification of some populations, again reinforcing the common origin of the ZW system in Characidium.
Even though NORs proved to be highly mobile in the genomes of Characidium, most species present a single NOR-bearing pair. Likewise, C. zebra, placed as basal in Characidium phylogeny [12] shows a single NOR pair. Only two populations of C. gomesi from Southern Brazil were divergent, one from the Quebra Perna stream (PR) bearing 10 ribosomal sites [2], and another from the Verde River (PR) bearing six autosomal NORs (present study), clearly representing derived and rare conditions in this genus.
Biogeoghrapical features and interspecific divergence
The evolutionary history and the chromosomal diversification of Characidium are closely related to the isolation of populations in headwaters of distinct hydrographic systems [2,4,5,7]. The main rivers in South and Southeaster Brazil are currently separated by barriers to the dispersal of species and populations, favoring isolation and gene flow restraints [22]. Thus, the populations were adapted to a particular environmental subset, and the occurrence of small-sized species of limited spatial dispersal favor the fixation of interpopulation genetic divergences [1]. Therefore, the karyotype structure and differences in both morphology and composition of sex chromosomes in Characidium could have been fixed from the common ancestor of different species/populations (demonstrated in Figure 4), promoting inter- and intraspecific variation and contributing to the speciation process.
Conclusions
Comparative chromosomal painting using W-specific and 18S rDNA probes in species/populations of Characidium, allowed us to infer the differentiation of sex and NOR-bearing chromosomes in this group, gaining a better understanding of the chromosomal and biogeographic features acting on the population differentiation, reproductive isolation, and speciation of this group.
Methods
Analyzed species and chromosomal preparations
Chromosomal preparations of C. zebra, C. lauroi, and C. gomesi were analyzed (Table 2). Voucher specimens were deposited at the fish collection of the Museu Nacional (MNRJ), Rio de Janeiro, under catalog numbers (MNRJ 22212, 29185) and Laboratório de Biologia e Genética de Peixes at UNESP in Botucatu, São Paulo, Brazil, under the numbers (LBP 6733 and 8709).
Table 2.
Species/populations of the Characidium analysed, sampled localities/state, hydrographic basin, number of females (F) and males (M).
| Species | Localities (State) | Hydrographic basin | F | M | Total |
|---|---|---|---|---|---|
| C. zebra | Passa Cinco River (SP) | Alto Paraná | 4 | 8 | 12 |
| C. lauroi | Grande Stream (SP) | Paraíba do Sul | 2 | 3 | 5 |
| C. gomesi | Alambari Stream (SP) | Alto Paraná | 22 | 8 | 30 |
| C. gomesi | Grande River (SP) | Alto Paraná | 7 | 10 | 17 |
| C. gomesi | Minhoca Stream (MG) | São Francisco | 5 | 2 | 7 |
| C. gomesi | Verde River (PR) | Alto Paraná | 36 | 8 | 44 |
The chromosomal preparations were obtained from anterior kidney cells by an air-drying technique [23]. The metaphase chromosomes were conventionally stained and C-banded [24] to determine diploid number, chromosome formula, and heteromorphism of the sex chromosomes prior to further analyses using fluorescence in situ hybridization (FISH). The procedures were performed in compliance with the Ethics Committee on Animal Experimentation (process number: 04741/08) of the Universidade Estadual de Ponta Grossa (Brazil).
Chromosomal probes
Two probes were used to map regions/chromosomes in Characidium species: (1) An 18S rDNA probe isolated from the total DNA of Prochilodus argenteus [25] and; (2) a W-specific probe of C. gomesi (population from Minhoca stream, São Francisco River basin) isolated in this study by microdissection of C-banded chromosomes [17], here named the W probe.
Microdissection of the W chromosome in Characidium gomesi and DOP-PCR
The W chromosomes of C. gomesi from Minhoca stream, São Francisco River basin, (MG) were microdissected from C-banded metaphase chromosomes to allow precise identification of sex chromosomes. The chromosomal microdissections were carried out using an inverted microscope IX51 (Olympus®) equipped with a mechanical micromanipulator (Narishige®). Glass capillaries of about 0.7 μm in diameter were prepared using a micropipette puller (Narishige®) and were used in the microdissection. Sixteen C-banded W chromosomes of C. gomesi were microdissected. The chromosomes were then transferred to a microtube and used in (DOP-PCR). The DOP-PCR amplification [26] and the changes procedures to C-banded chromosomes [17] were used to obtain the W-probe. The DOP-PCR of C. gomesi W chromosome consists of 1 × ThermoSequenase reaction buffer, 40 μM dNTPs and 2 μM DOP primer (5' ccg act cga gnn nnn nat gtg g 3'). The microtube was heated to 95°C for 10 min, followed by the addition of 10 U of ThermoSequenase enzyme. The first amplification is carried out through RAMP-PCR: 5 min 94°C; 12 cycles of low stringency (94°C 1 min and 30 s, 32°C 2 min, increasing 0.2°C/s until reaching 72°C and 72°C 2 min); followed by 35 cycle of high stringency (94°C 1 min and 30 s, 52°C 1 min and 30 s and 72°C 1 min and 30 s).
Fluorescence in situ hybridization (FISH)
FISH experiments were carried out in Characidium representatives using 18S rDNA and W-chromosome probes. The 18S rDNA probe was labeled by PCR using 16-dUTP-biotin (Roche®). In this probe, the PCR mix comprised 20 ng template, 1× Taq polymerase buffer (2 mM MgCl2), 40 μM of dATP, dGTP and dCTP, 28 μM of dTTP, 12 μM of 16-dUTP biotin, 2 μM of each M13 primer and 0,05 U/μL of Taq polymerase. The W probe was labeled by DOP-PCR using 11-dUTP digoxigenin (Roche®). The PCR mix comprised 1x Taq polymerase buffer (2 mM MgCl2), 40 μM of dATP, dGTP and dCTP, 28 μM of dTTP, 12 μM of 11-dUTP digoxigenin, 2 μM of DOP primer, and 0,05 U/μL of Taq polymerase under the following conditions: (1×) 94°C 5 min; (35×) 90°C 1 min and 30 s, 52°C 1 min and 30 s, 72°C 1 min and 30 s, and (1×) 72°C 5 min. The FISH procedure [27] was performed under high stringency conditions (2.5 ng/μL of each probe, 50% formamide, 2 × SSC, 10% dextran sulfate). Signal detection was accomplished using AlexaFluor-Streptavidin 488 (Molecular Probes®) and antidigoxigenin-Rhodamine (Roche®) antibodies. The chromosomes were counterstained with 4',6'-diamino-2-phenylindole (DAPI-0.2 μg/mL) in Vectashield mounting medium (Vector®), and analyzed using an Olympus BX41 epifluorescence microscope equipped with the DP 71 image capture system (Olympus®). The chromosomes were organized into metacentric (m), submetacentric (sm), and subtelocentric (st), according to arm ratio, and arranged in decreasing order in the karyotype [28].
List of abbreviations
bp: base pairs; DAPI: 4',6'-diamino-2-phenylindole; DOP-PCR: Degenerate Oligonucleotide primed-Polymerase Chain Reaction; FISH: fluorescence in situ hybridization; m: metacentric; MG: Minas Gerais State; NOR: nucleolus organizer region; PR: Paraná State; sm: submetacentric; SP: São Paulo State; st: subtelocentric
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
TCM, JCP and MBPA collected the samples, collaborated on all cytogenetic procedures, undertook the bibliographic review, and coordinated the writing of this paper. VN and MCA participated in developing the laboratory techniques, cytogenetic analyses and writing. CO, FF, LACB, OMF and RFA participated in the design of the study, in the inter institution integration study and drafted the manuscript. MRV coordinated the study, helped develop the laboratory techniques, performed specific W-probe of Characidium, and reviewed the manuscript. All authors read and approved the final manuscript.
Contributor Information
Tatiana C Machado, Email: tati_crm@yahoo.com.br.
José C Pansonato-Alves, Email: pansonato@ibb.unesp.br.
Marcela B Pucci, Email: marcelabaer@hotmail.com.
Viviane Nogaroto, Email: vivianenogaroto@hotmail.com.
Mara C Almeida, Email: almeidamara@uol.com.br.
Claudio Oliveira, Email: claudio@ibb.unesp.br.
Fausto Foresti, Email: fforesti@ibb.unesp.br.
Luiz AC Bertollo, Email: bertollo@power.ufscar.br.
Orlando Moreira-Filho, Email: omfilho@power.ufscar.br.
Roberto F Artoni, Email: rfartoni@uepg.br.
Marcelo R Vicari, Email: vicarimr@pq.cnpq.br.
Acknowledgements and Funding
The authors are grateful to the Ministério do Meio Ambiente/Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis (MMA/IBAMA-License number: 10538-1) for authorization to collect the biological material. This study was financed by Fundação Araucária (Fundação Araucária de Apoio ao Desenvolvimento Científico e Tecnológico do Estado do Paraná), CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) and the FAPESP (Fundacão de Amparo à Pesquisa do Estado de São Paulo).
References
- Buckup PA. In: Checklist of the freshwater fish of South and Central America. Reis RE, Kullander SO, Ferraris CJ Jr, editor. Porto Alegre: Edipucrs; 2003. Family Crenuchidae (South American Darters) pp. 87–95. [Google Scholar]
- Vicari MR, Artoni RF, Moreira-Filho O, Bertollo LAC. Diversification of a ZZ/ZW sex chromosome system in Characidium fish (Crenuchidae, Characiformes) Genetica. 2008;134:311–317. doi: 10.1007/s10709-007-9238-2. [DOI] [PubMed] [Google Scholar]
- Maistro EL, Mata EP, Oliveira C, Foresti F. Unusual occurrence of a ZZ/ZW sex-chromosome system and supernumerary chromosomes in Characidium cf. fasciatum (Pisces, Characiformes, Characidiinae) Genetica. 1998;104:1–7. doi: 10.1023/A:1003242020259. [DOI] [PubMed] [Google Scholar]
- Pansonato-Alves JCP, Paiva LRS, Oliveira C, Foresti F. Interspecific chromosomal divergences in the genus Characidium (Teleostei: Characiformes: Crenuchidae) Neotrop Ichthyol. 2010;8:77–86. doi: 10.1590/S1679-62252010000100010. [DOI] [Google Scholar]
- Pansonato-Alves JCP, Vicari MR, Oliveira C, Foresti F. Chromosomal diversification in populations of Characidium cf. gomesi (Teleostei, Crenuchidae) J Fish Biol. 2011;78:183–194. doi: 10.1111/j.1095-8649.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Centofante L, Bertollo LAC, Moreira-Filho O. Comparative cytogenetics among sympatric species of Characidium (Pisces, Characiformes): Diversity analysis with the description of a ZW sex chromosome system and natural triploidy. Caryologia. 2001;5:253–260. [Google Scholar]
- Centofante L, Bertollo LAC, Buckup PA, Moreira Filho O. Chromosomal divergence and maintenance of sympatric Characidium fish species (Crenuchidae, Characidiinae) Hereditas. 2003;138:213–218. doi: 10.1034/j.1601-5223.2003.01714.x. [DOI] [PubMed] [Google Scholar]
- Maistro EL, Jesus CM, Oliveira C, Moreira Filho O, Foresti F. Cytogenetic analysis of A, -B chromosomes and ZZ/ZW sex chromosomes of Characidium gomesi (Teleostei, Characiformes, Crenuchidae) Cytologia. 2004;69:181–186. doi: 10.1508/cytologia.69.181. [DOI] [Google Scholar]
- Noleto RB, Amorin AP, Vicari MR, Artoni RF, Cestari MM. An unusual ZZ/ZW sex chromosome system in Characidium fishes (Crenuchidae, Characiformes) with the presence of rDNA sites. J Fish Biol. 2009;75:448–453. doi: 10.1111/j.1095-8649.2009.02342.x. [DOI] [PubMed] [Google Scholar]
- Pansonato-Alves JCP, Oliveira C, Foresti F. Karyotypic conservatism in samples of Characidium cf. zebra (Teleostei, Characiformes, Crenuchidae): Physical mapping of ribosomal genes and natural triploidy. Genet Mol Biol. 2011;34:208–213. doi: 10.1590/S1415-47572011005000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa CS, Galetti PM Jr. First cytogenetical studies in Characidium species (Pisces: Characiformes, Characidiinae) Cytologia. 1994;59:73–79. [Google Scholar]
- Buckup PA. The monophyly of the Characidiinae, a Neotropical group of characiform fishes (Teleostei: Ostariophysi) Zool J Linn Soc. 1993;108:224–245. [Google Scholar]
- Oliveira C, Foresti F, Almeida-Toledo LF. In: Fish Cytogenetics. Pisano E, Ozouf-Costaz C, Foresti F, Kapoor BG, editor. Enfield: Science Publisher; 2007. Karyotypic evolution in Neotropical fishes; pp. 111–164. [Google Scholar]
- Steinemann S, Steinemann M. Biased distribution of repetitive elements: a landmark for neo-Y chromosome evolution in Drosophila miranda. Cytogenet Cell Genet. 2001;93:228–233. doi: 10.1159/000056988. [DOI] [PubMed] [Google Scholar]
- Steinemann S, Steinemann M. Retroelements: tools for sex chromosome Evolution. Cytogenet Genome Res. 2005;110:134–143. doi: 10.1159/000084945. [DOI] [PubMed] [Google Scholar]
- Silva DS, Milhomem SSR, Pieczarka JC, Nagamachi CY. Cytogenetic studies in Eigenmannia virescens (Sternopygidae, Gymnotiformes) and new inferences on the origin of sex chromosomes in the Eigenmannia genus. BMC Genetics. 2009;10:74. doi: 10.1186/1471-2156-10-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicari MR, Nogaroto V, Noleto RB, Cestari MM, Cioffi MB, Almeida MC, Moreira-Filho O, Bertollo LAC, Artoni RF. Satellite DNA and chromosomes in Neotropical fishes: Methods, applications and perspectives. J Fish Biol. 2010;76:1094–1116. doi: 10.1111/j.1095-8649.2010.02564.x. [DOI] [PubMed] [Google Scholar]
- Reed KM, Phillips RB. Polymorphism of the nucleolus organizer region (NOR) on the putative sex chromosomes of Arctic char (Salvelinus alpinus) is not sex related. Chromosoma. 1997;5:221–227. doi: 10.1023/A:1018411417816. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- Beçak W, Beçak ML. Cytotaxonomy and chromosomal evolution in Serpents. Cytogenetics. 1969;8:247–262. doi: 10.1159/000130037. [DOI] [PubMed] [Google Scholar]
- Singh L, Purdom IF, Jones KW. Sex chromosome associated satellite DNA: evolution and conservation. Chromosome Res. 1980;79:137–157. doi: 10.1007/BF01175181. [DOI] [PubMed] [Google Scholar]
- Weitzman SH, Menezes NA, Weitzman MJ. In: Proceedings of a Workshop on Neotropical Distribution Patterns. Vanzolini PE, Heyer WR, editor. Rio de Janeiro: Academia Brasileira de Ciências; 1988. Phylogenetic biogeography of the Glandulocaudini (Teleostei, Characiformes, Characidae) with comments on the distributions of other freshwater fishes in Eastern and Southeastern Brazil; pp. 379–427. [Google Scholar]
- Bertollo LAC, Takahash CS, Moreira-Filho O. Cytotaxonomic considerations on Hoplias lacerdae (Pisces, Erythrinidae) Braz J Genet. 1978;1:103–120. [Google Scholar]
- Sumner AT. A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res. 1972;75:304–306. doi: 10.1016/0014-4827(72)90558-7. [DOI] [PubMed] [Google Scholar]
- Hatanaka T, Galetti PM Jr. Mapping of the 18S and 5S ribosomal RNA genes in the fish Prochilodus argenteus Agassiz, 1829 (Characiformes, Prochilodontidae) Genetica. 2004;122:239–244. doi: 10.1007/s10709-004-2039-y. [DOI] [PubMed] [Google Scholar]
- Telenius H, Carter NP, Bebb CE, Nordenskjold M, Ponder BA, Tunnacliffe A. Degenerate oligonucleotide primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-K. [DOI] [PubMed] [Google Scholar]
- Pinkel D, Straume T, Gray JW. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA. 1986;83:2934–2938. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levan A, Fredga K, Sandberg AA. Nomenclature for centromeric position on chromosomes. Hereditas. 1964;52:201–220. [Google Scholar]