Abstract
Viral hepatitis and aflatoxin B1 (AFB1) exposure are common risk factors for hepatocellular carcinoma (HCC). The incidence of HCC in individuals co-exposed to hepatitis C (HCV) or B virus and AFB1 is greater than could be explained by the additive effect, yet the mechanisms are poorly understood due to lack of an animal model. This study investigated the outcomes and mechanisms of combined exposure to HCV and AFB1. We hypothesized that HCV transgenic (HCV-Tg; expressing core, E1, E2, and p7, nucleotides 342–2771) mice will be prone to hepatocarcinogenesis when exposed to AFB1. Neonatal (7 days old) HCV-Tg or C57BL/6J wild-type mice were exposed to AFB1 (6 μg/g bw) or tricaprylin vehicle (15 μl/g bw) and male offspring were followed for up to 12 months. No liver lesions were observed in vehicle-treated wild type or HCV-Tg mice. Tumors (adenomas or carcinomas) and preneoplastic lesions (hyperplasia or foci) were observed in 22.5% (9 of 40) of AFB1-treated wild-type mice. In HCV-Tg, the incidence of tumorous or pre-tumorous lesions was significantly elevated (50%, 18 of 36), with the difference largely due to a 2.5-fold increase in the incidence of adenomas (30.5% vs 12.5%). While oxidative stress and steato-hepatisis were observed in both AFB1-treated groups, molecular changes indicative of the enhanced inflammatory response and altered lipid metabolism were more pronounced in HCV-Tg mice. In summary, HCV proteins core, E1, E2 and p7 are sufficient to reproduce the co-carcinogenic effect of HCV and AFB1 which is a known clinical phenomenon.
Novelty and impact
This work establishes a model for studies of the mechanisms of co-carcinogenesis of HCV and AFB1 and indicates a crucial role for direct viral protein effects in the synergy between these factors. A number of molecular pathways known to play a role in liver carcinogenesis were investigated and the data shows that the HCV structural proteins and AFB1 react synergistically to increase steatosis and tumor incidence whereby HCV and AFB1 may operate to increase liver cancer incidence through different mechanisms. In addition, while tumor occurrence may be also linked to lipid peroxidation consequently to AFB1 exposure, we posit that the conventional explanations (e.g., oxidative stress, lipid accumulation) do not appear to be the cause of the synergy and other more specific, but as yet unidentified effects are likely to be responsible.
Keywords: aflatoxin B1, hepatitis C virus, liver, mouse
Introduction
Hepatocellular carcinoma (HCC) is the most common type of primary malignancy in the liver1. Main risk factors of HCC are well-defined and include hepatitis B virus (HBV); lifestyle, diet and environmental factors (e.g., alcoholic beverages, aflatoxin B1 (AFB1), and tobacco smoking); and metabolic diseases (e.g., obesity, diabetes and non-alcoholic steatohepatitis)2. Importantly, the rise in incidence of HCC in the developed countries has been attributed, at least in part, to an increase in hepatitis C virus (HCV) infections and non-alcoholic steatohepatitis, pathological states whose prevalence is growing in the US and Europe3.
While viral hepatitis is estimated to contribute to as many as 90% of HCC cases worldwide4, several factors have been identified as acting synergistically with both HBV and HCV infection5. Specifically, dietary exposure to aflatoxins, a class of mycotoxins produced by moulds of the genus Aspergillus, is a significant risk factor for HCC in less developed countries with hot and humid climates6. Upon ingestion, AFB1 is metabolized in the liver to a reactive epoxide that covalently binds on the N7 position of guanine7, leading to characteristic mutations in cancer-related genes8. Several recent studies provide emerging epidemiological evidence for the association between AFB1 exposure and advanced liver disease in HCV-infected patients9,10. In mice, AFB1 is only hepatocarcinogenic when injected in neonates11 and the incidence of liver neoplasms varies from ~25 to 90% depending on the mouse strain11–13.
The mechanisms of the synergism between these etiological factors of HCC are not well understood due to the fact that human HCV is able to cause infection and disease only in non-human primates14, but not other laboratory animal models, such as mice and rats. Studies in transgenic mouse models, in which the HCV proteins are expressed, indicate that HCV is directly pathogenic and oncogenic15, 16. There are several HCV mouse models varying in the viral proteins being constitutively over-expressed17. While hepatic tumors have been observed in some of these models15, 16, 18–20, the incidence varies and tumors develop only in the mice of advanced (>13 months) age. Both HBV12 and H.hepaticus21 have been shown to promote AFB1-induced liver carcinogenesis in mice demonstrating that co-carcinogenesis due to infectious and chemical factors can be reproduced in the animal model, but there have been no studies addressing the purported synergism between AFB1 and HCV.
Liver pathology in HCV can result either as a consequence of the immune response or as a direct effect of viral protein interaction with cellular processes. In this study we hypothesized that, if direct viral protein effects were responsible for the enhanced carcinogenesis, the incidence of HCC due to AFB1 would be elevated in HCV transgenic mice expressing core, E1, E2 and p7 proteins. While expressing viral proteins at levels comparable to human disease, these mice are immune-tolerant to viral antigens and have no viral specific adaptive immune response. We demonstrate that the incidence of liver neoplasia was more than doubled (largely due to an increase in adenomas) in HCV/AFB1 group as compared to AFB1-treated WT mice. Since HCV mice developed no tumors and exhibited no overt liver pathology at 12 month of age, this observation is indicative of a potentiating, but not an additive effect on the molecular pathways to hepatocarcinogenesis. This study establishes a model for studies of the mechanisms of co-carcinogenesis of HCV and AFB1 and indicates a crucial role for direct viral protein effects in the interaction between these factors.
Materials and methods
Animals and treatments
HCV transgenic mice (SL-139 strain, pAlbSVPA-HCV-S, containing the structural genes core, E1, E2, and p7, nucleotides 342–2771 of HCV genotype 1b, strain N, under the control of the murine albumin promoter/enhancer) on C57BL/6J (Jackson Laboratory, Bar Harbor, ME) background were previously reported in Lerat et al16. Transgenic animals were identified after weaning as detailed in Korenaga et al22. C57BL/6J mice were used as a wild type (WT) reference strain in all experiments. Neonatal (7 days old) mice were administered a single dose of AFB1 (6 μg/g bw) or tricaprylin vehicle (15 μl/g bw) by intra-peritoneal injection. Male mice were maintained on the regular animal chow with free access to food and water for up to 12 months. Male mice were selected for these studies since male gender is a risk factor for human HCC4. Three days before sacrifice at 12 month of age, a subset of mice (5 to 8 per group) were given drinking water containing bromodeoxyuridine (0.2 g/L). All animal experiments were approved by the UNC Animal Care and Use Committee.
Chemicals
AFB1, tricaprylin, and other chemicals, unless otherwise noted, were obtained from Sigma (St Louis, MO). AFB1 was dissolved in tricaprylin (400 μg/ml) by heating the solution to 65°C and the solution was then stored at 4°C.
Histopathology
At sacrifice, the livers were examined macroscopically for tumors (carcinomas or adenomas) and then a section of the liver (including gross lesions) and duodenum were fixed in neutral buffered formalin. The remainder of the liver tissue was frozen and stored at −80°C. Histology (from hematoxylin/eosin liver sections) was evaluated by a researcher and an expert veterinary pathologist in a blinded manner. Incidence and multiplicity of the pre-neoplastic lesions (hyperplasia or foci) were evaluated using random sections. Microscopic liver pathology was scored as follows: steatosis (the percentage of hepatocytes containing fat droplets): <25%=1+, <50%=2+, <75%=3+, >75%=4+; inflammation and necrosis: 1 focus per low-power field=1+, 2=2+, ≥3=3+. Formalin-fixed sections were stained with Sirius Red to quantify fibrosis. Oil Red O staining was performed on frozen liver sections (10 μm) to evaluate tissue lipid content.
Immunohistochemistry
Formalin-fixed, paraffin-embedded liver sections (5μm) were deparaffinized, rehydrated, treated for antigen retrieval and incubated in Peroxidase Blocking Reagent (Dako, 10 min). Dako EnVision System HRP kit (all antibodies were diluted in saline containing 1% bovine serum albumin) was used for the detection of bromodeoxyuridine (monoclonal anti-bromodeoxyuridine antibody, clone Bu20a, Dako, 1:200 dilution, 10 min), 4-hydroxynonenal (primary rabbit anti-4-HNE antibody, Alpha Diagnostics, San Antonio, TX, 1:500 dilution, 1 h), or F4/80 (primary rat anti-mouse F4/80 antibody, Serotec, Raleigh, NC, 1:200 dilution, 30 min). Quantitative analysis, where applicable, was performed using Image-Pro® Plus software (Media Cybernetics, Silver Spring, MD) on images at 200×.
Quantification of glutathione
Reduced (GSH) and oxidized (GSSG) glutathione were assessed in liver mitochondria and whole liver as previously described23 using high performance liquid chromatography. Liver mitochondria were isolated from ~100 mg of fresh tissue, using Mitochondria Isolation Kit for Tissue (Pierce, Rockford, IL) following the manufacturer’s instructions (Option A, protocol 1). The mitochondria were stored −80°C in 250 μl of Mitochondria Isolation Reagent C containing 10 μg/ml of protease inhibitors cocktail (Pierce).
Determination of hepatic triglycerides content
Hepatic triglycerides were extracted by homogenizing 20 mg of frozen liver tissue in 500 μl of isopropyl alcohol and 4 μl of the extract was used in subsequent analysis. The level of triglycerides was determined by using L-Type TG-M Assay Kit (Wako Diagnostic, Richmond, VA) according to the manufacturer’s instructions.
Quantitative real time reverse polymerase chain reaction
Total RNA was isolated from frozen liver samples using the RNeasy (Qiagen, Valencia, CA) kit according to the manufacturer’s instructions. RNA concentration and quality were assessed using ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), respectively. Ten micrograms of total RNA were reverse transcribed in a final volume of 100 μl using the High Capacity Archive kit and random hexamers (Applied Biosystems, Foster City, CA), diluted 4-fold in water and stored at −80°C. For each sample, 2 μl of cDNA, corresponding to ~25 ng of reverse transcribed RNA, was analyzed in duplicate, using the LightCycler®480 Instrument (Roche Applied Science, Indianapolis IN). Primers were obtained from Applied Biosystems and are listed as Supplemental Table 1. The relative amount of target gene mRNAs was normalized to internal control Tbp.
Microarray experiments
RNA amplifications and labeling were performed using Low RNA Input Linear Amplification kits (Agilent Technologies). For hybridization, 750 ng of total RNA from each mouse liver was amplified and labeled with a fluorescent dye Cy5. In parallel, 750 ng of a common reference RNA (Icoria Inc., RTP, NC) was labeled with a fluorescent dye Cy3. Labeled cRNA was then processed, hybridized to Agilent Mouse Toxicology Arrays (catalog# 4121A, 22,575 features), washed and scanned according to the manufacturer’s protocol. The resulting images were analyzed by determining the Cy5 fluorescence intensity of all gene spots (features) on each array using the Agilent Feature Extraction Software (Version 9.5). Raw data is available from Gene Expression Omnibus (GSE26838). The median fluorescence intensity of all pixels within one feature was taken as the intensity value for that feature. The raw intensity values were then normalized using 75th percentile channel scaling normalization setting of ArrayTrack. The list of differentially expressed genes was generated using a t-test at p-value <0.01 and a fold change at >1.5. The Ingenuity Pathway Analysis Software (Ingenuity Systems, Redwood City, CA) was used for pathway analyses. Ingenuity Pathways Analysis (IPA v. 7.1; Ingenuity Systems, Redwood City, CA) was used to determine canonical pathways that are enriched by the significant transcripts identified by the ANCOVA model for each factor.
Mitochondrial DNA damage
Long-polymerase chain reaction method was used to assess integrity of mitochondrial DNA 24. Primers L15322 (5′-CTCTGGTCTTGTAAACCTG-3′) and H3266 (5′-CAGGCTGGCAGAAGTAATCAT-3′) yield the product of 4277 bp24, while primers mt316 (forward: 5′-CGACAGCTAAGACCCAAACTGGG-3′, nucleotides 470–492; and reverse 5′-CCCATTTCTTCCCATTTCATTGGC-3′, nucleotides 785–762) amplify a 316 bp fragment25. The products were separated on 1% agarose gels and visualized with ethidium bromide.
Quantification of genomic 8-oxo-deoxyguanine
Capillary liquid chromatography tandem mass spectrometry method was used for detection of 8-oxo-deoxyguanine adducts in liver DNA isolated and processed as detailed elsewhere26.
Statistical analysis
The GraphPad Prism software v.5 (GraphPad Software Inc., San Diego, CA) was used for analyses. Incidence of tumors, steatosis and inflammatory foci between groups were compared by Fisher’s exact test. In other experiments, unpaired t-test or Welch’s t-test (when variances were unequal) were used. Quantitative values are expressed as mean ± standard deviation.
Results
HCV transgene doubled tumor incidence in mice treated with AFB1
All animals treated with AFB1 or vehicle neonatally survived to weaning. A total of 161 male mice were maintained for 6 (n=20), 9 (n=20), or 12 (n=121) months. No neoplastic or pre-neoplastic lesions were observed in WT and HCV-Tg mice treated with the vehicle alone at any of the three time points (Table 1). Liver neoplasias were found only in 12-month old animals treated with AFB1, with an incidence of 22.5% in WT, and 50% (p<0.05) in HCV-Tg group, indicating a potentiating effect. In HCV-Tg/AFB1 mice, increased incidence of liver neoplasms was mainly due to a significantly greater (p<0.05) incidence of liver adenomas. While transgene expression varied between individual mice in HCV-Tg groups, tumor incidence did not correlate with the viral protein levels (data not shown). In both wild-type and HCV-Tg mice, all types of neoplasms (e.g., hyperplasia, foci, adenoma and carcinoma) were observed (Supplemental Figure 1, Table 1) and there was no difference in tumor multiplicity. Interestingly, three different subtypes of liver adenomas (vacuolated, well-differentiated and basophilic focus-like) were observed in both groups (Supplemental Figure 1).
Table 1.
Liver injury and tumors in WT and HCV-Tg mice treated with AFB1.
| Vehicle | Aflatoxin B1 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild-type | HCV-Tg | Wild-type | HCV-Tg | |||||||||
| 6 mo n=5 |
9 mo n=5 |
12 mo n=24 |
6 mo n=5 |
9 mo n=5 |
12 mo n=21 |
6 mo n=5 |
9 mo n=5 |
12 mo n=40 |
6 mo n=5 |
9 mo n=5 |
12 mo n=36 |
|
| Inflammatory foci [number of animals for each grade (% of total)] | ||||||||||||
| 1+ | 1 (20) | 1 (20) | 5 (21) | 1 (20) | 1 (20) | 9 (43) | 3 (8) | 1 (20) | 6 (17) | |||
| 2+ | 3 (13) | 2 (9) | 4 (10) | 6 (17) | ||||||||
| 3+ | 3 (8) | |||||||||||
| Total | 1 (20) | 1 (20) | 8 (33) | 1 (20) | 1 (20) | 11 (52) | 7 (18) | 1 (20) | 15 (42)* | |||
|
| ||||||||||||
| Steatosis [number of animals for each grade (% of total)] | ||||||||||||
| 1+ | 1 (20) | 1 (4) | 1 (5) | 4 (10) | 2 (40) | 10 (28) | ||||||
| 2+ | 1 (4) | 1 (5) | 4 (10) | 3 (8) | ||||||||
| 3+ | 2 (5) | |||||||||||
| Total | 1 (20) | 2 (8) | 2 (10) | 10 (25) | 2 (40) | 13 (36)† | ||||||
|
| ||||||||||||
| Tumor incidence# [number of animals for each type (% of total)] | ||||||||||||
| Hyperplasia | 2 (5) | 2 (6) | ||||||||||
| Focus | 4 (13) | 8 (22) | ||||||||||
| Adenoma | 4 (13) | 11 (31)‡ | ||||||||||
| Carcinoma | 2 (5) | 1 (3) | ||||||||||
| Total# | 9 (23) | 18 (50)§ | ||||||||||
| Multiplicity | 1.6 | 1.4 | ||||||||||
p-value = 0.02 (Fisher’s exact test between WT/AFB1 mice and HCV-Tg/AFB1 mice)
p-value = 0.03 (Fisher’s exact test between HCV-Tg mice and HCV-Tg/AFB1 mice)
Some mice displayed several types of neoplastic lesions; thus, the “total” reflects incidence of any lesion in each mouse to avoid double counting
p-value = 0.04 (Fisher’s exact test between WT/AFB1 mice and HCV-Tg/AFB1 mice)
p-value = 0.02 (Fisher’s exact test between WT/AFB1 mice and HCV-Tg/AFB1 mice)
Effects of HCV-Tg and AFB1 on liver inflammation and steatosis
Hepatic steatosis and inflammation are commonly observed in patients with HCV27. HCV-Tg mice also develop steatosis which is thought to be mediated by HCV core protein28, 29. There are reports that AFB1 exposure leads to steatohepatitis in humans30, rats31, and mice32. Thus, we evaluated liver histopathology, cell proliferation, as well as markers of oxidative stress and steatosis.
In vehicle treated animals, mild inflammatory and steatotic changes were observed in some, but not all animals, with most of the changes manifesting in 12-month old mice (Table 1). Interestingly, HCV-Tg mice showed no difference from WT in either incidence or severity of the liver histopathological changes. In AFB1-treated mice, no adverse liver pathology was observed in 6- or 9-months groups; however, at 12 months, HCV-Tg/AFB1 mice exhibited greater inflammatory change than WT/AFB1 mice. It should be noted, however, that these lesions were comprised of small lobular inflammatory foci which are different from the portal-based inflammatory lesions observed in human HCV patients.
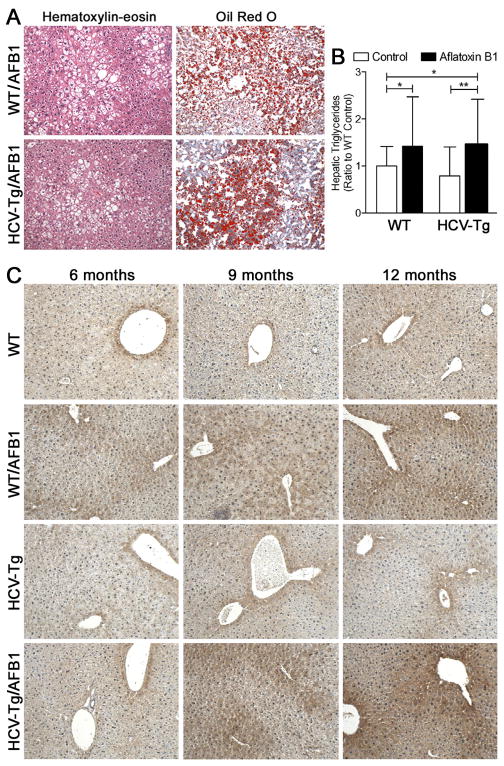
Furthermore, liver steatosis, as evidenced from the histopathological evaluation (Table 1, Figure 1A), Oil Red O staining (Figure 1A) and hepatic triglyceride content (Figure 1B), was observed in both AFB1 groups at 12 months. Concomitantly, the extent of hepatic lipid peroxidation, as evidenced by presence of 4-hydroxynonenal-adducted proteins, was also elevated at 12 months (Figure 1C).
Figure 1. Liver steatosis in WT and HCV-Tg mice treated with AFB1.
(A) Representative hematoxylin-eosin and Oil Red O stained sections (100x magnification) from 12 months old animals of WT/AFB1 and HCV-Tg/AFB1 groups showing mixed macrovesicular and microvesicular characteristics of steatosis. (B) Hepatic triglyceride content in 12 months old animals. Asterisks (* and **) denote statistical significance (p<0.05 and p<0.01, respectively; n=18–37 per group) between the groups as indicated. (C) Representative photomicrographs (100x) of 4-hydroxy-nonenal stained liver sections.
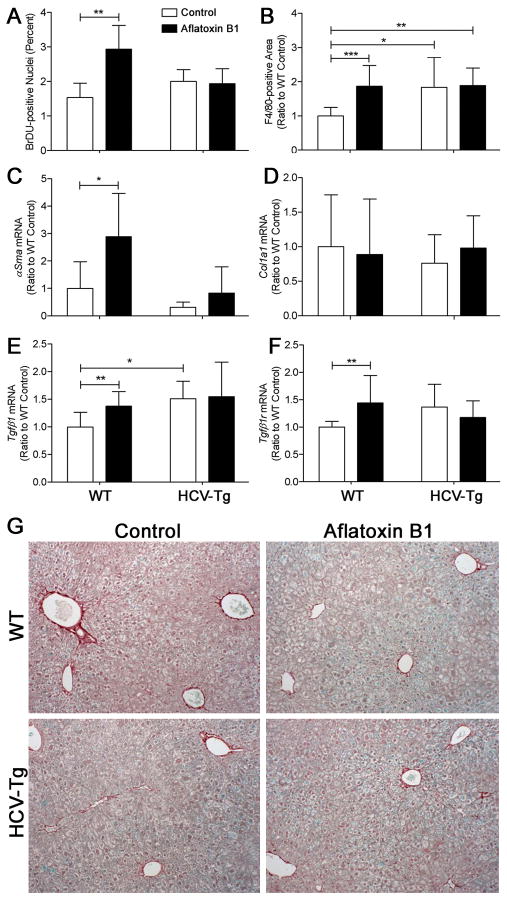
Interestingly, cell proliferation rate in non-tumoral liver tissue was increased in response to AFB1 treatment at 12 months in WT, but not HCV-Tg groups (Figure 2A). An elevation in the number of F4/80-positive macrophages in the liver was also observed in WT, but not HCV-Tg mice in response to AFB1 (Figure 2B). Even though a significant elevation in expression of alpha-smooth muscle actin, transcription growth factor beta 1 or its receptor were observed in WT/AFB1 group, no effect was observed in HCV-Tg mice (Figures 2C, E and F). Furthermore, no evidence of liver fibrosis was found using histological examination, Sirius red staining (Figure 2G), or expression of collagen 1-alpha 1 (Figure 2D).
Figure 2. Hepatocyte proliferation, inflammation and fibrosis markers.
(A) Percent BrDU-positive nuclei (n=4–8/group; mean±standard deviation for 5 randomly selected fields at 200x); (B) area occupied by F4/80-stained cells (n=9–16/group; mean±standard deviation for 10 randomly selected fields at 100x) relative to WT group. Liver expression (evaluated using real-time PCR and normalized to levels in WT group; n=6–16/group) of smooth muscle actin-alpha (C), collagen 1-alpha1 (D), transforming growth factor beta1 (E) and its receptor (F). Asterisks (*, ** and ***) denote statistical significance (p<0.05, p<0.01 and p<0.001, respectively) between the groups as indicated. (G) Representative Sirius red-stained sections (100x) from 12 months old animals of WT/AFB1 and HCV-Tg/AFB1 groups.
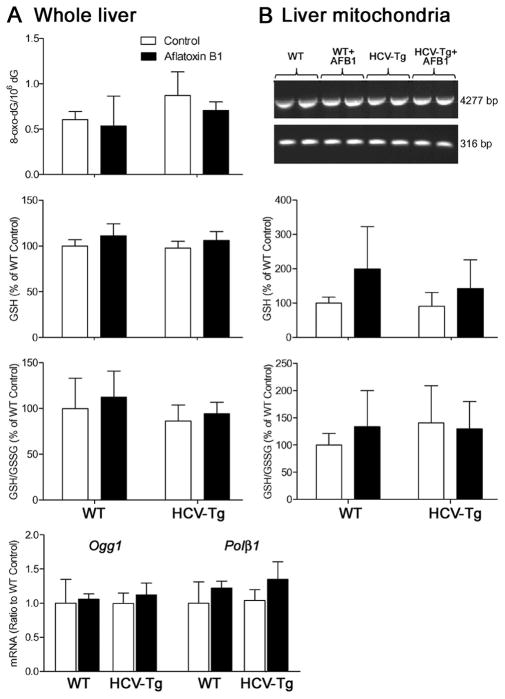
Since an increase in lipid peroxidation was observed in AFB1-treated mice (Figure 1C), we further examined markers of oxidative stress (Figure 3). In whole liver, we examined oxidative damage to genomic DNA (8-oxo-dG and expression of base excision DNA repair genes 8-oxo-dG DNA glycosylase 1 and polymerase beta 1) and amounts of reduced and oxidized glutathione (Figure 3A), yet no effect of either AFB1 or HCV-Tg was observed. Next, we evaluated mitochondrial DNA damage and glutathione content (Figure 3B) and also found no difference between the groups.
Figure 3. Liver oxidative stress in WT and HCV-Tg mice treated with AFB1.
(A) In whole liver samples, genomic DNA content of 8-oxo-dG adducts, reduced glutathione, reduced to oxidized glutathione ratio, and expression of base-excision DNA repair genes 8-oxo-dG DNA glycosylase and polymerase beta were assessed. (B) In hepatic mitochondria, mtDNA integrity, and levels of reduced glutathione, or reduced to oxidized glutathione ratio were evaluated. No significant differences were observed (n=5–10/group).
Effects of HCV and AFB1 on liver lipid metabolism pathways
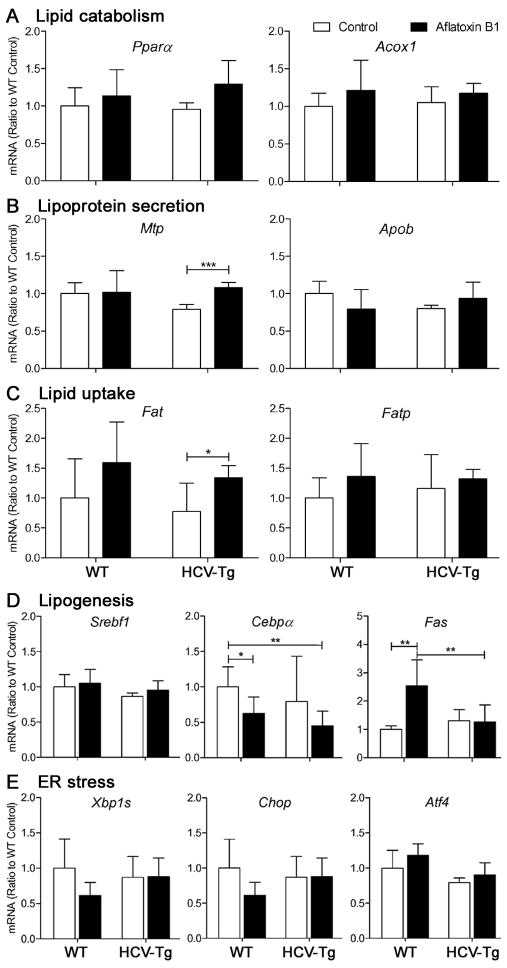
To evaluate the effects of AFB1 and HCV on lipid metabolism pathways, we first examined several factors previously associated with fatty acid and triglyceride metabolism in HCV transgenic mice. We evaluated expression of lipid catabolism, lipoprotein secretion, lipid uptake and lipogenesis genes (Figures 4A–D). Concomitant with little evidence for fatty change in the liver of vehicle-treated HCV-Tg mice, we observed no differences between WT and HCV-Tg groups. Our data shows that the mechanisms for excess fat accumulation may differ between WT/AFB1 and HCV-Tg/AFB1 groups. For example, while AFB1 induced lipogenesis in WT, lipid secretion and uptake were dysregulated in HCV-Tg mice.
Figure 4. Lipid metabolism and UPR response gene expression in livers of WT and HCV-Tg mice treated with AFB1.
(A–D) The expression of lipid metabolism genes subdivided into various categories as shown, as well as (E) genes playing a role in UPR response, was determined by real-time PCR and expression was normalized to WT group. Asterisks (* and **) denote statistical significance (n=5–8/group; p<0.05 and p<0.01, respectively) between the groups as indicated.
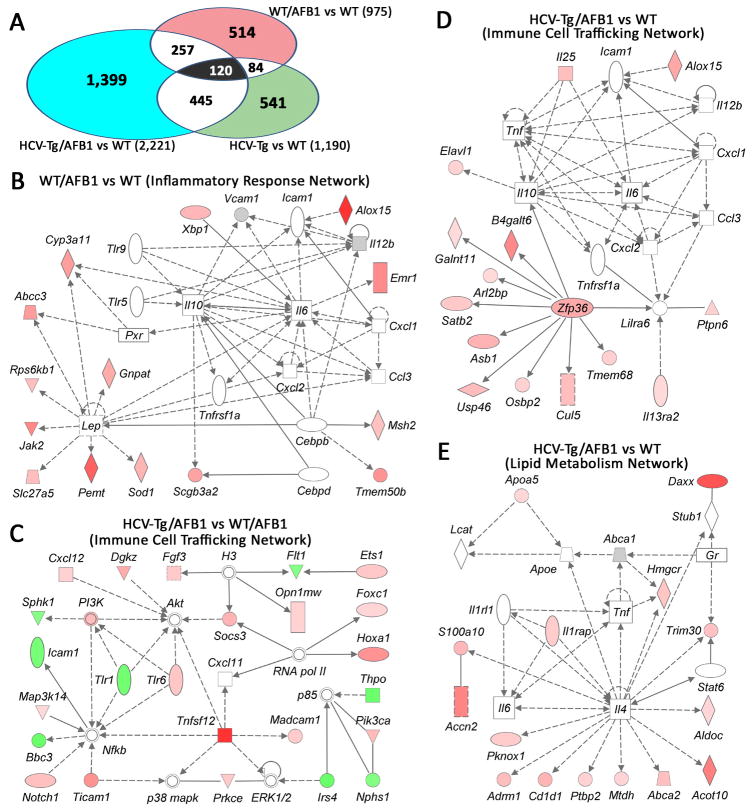
Next, we examined liver gene expression profiles in each experimental group to assess global differences in the molecular pathways that may be affected by each experimental factor (Figure 5). While both AFB1 and HCV transgene had an effect of the liver transcriptome when compared to the WT group (Figure 5A), the overlap between the groups, in terms of the individual genes, was relatively small. Interestingly, the greatest effect of AFB1 treatment in wild-type mice was on the inflammatory response network (Figure 5B), an effect which appears to be synergistic with the impact on immune cell trafficking pathways of HCV-Tg alone (Figure 5C), or a combination of the two (Figure 5D). Lipid metabolism genes, largely related to the inflammatory cytokines, were induced in HCV-Tg/AFB1 mice as compared to WT (Figure 5E).
Figure 5. Microarray analysis of liver tissues from WT and HCV-Tg mice treated with AFB1.
(A) Venn diagram showing genes that were significantly different between the groups as indicated. The number of genes in each comparison is shown. (B–E) Molecular network interactions between the genes significantly different between each group (as indicated) were visualized using Ingenuity® database. Up-regulated genes are identified in red, down-regulated in green.
Unfolded protein response (UPR) is not induced by either HCV or AFB1
It has been shown that HCV envelope proteins induce UPR, a cellular response to accumulation of damaged proteins in the endoplasmatic reticulum, in vitro33. Several in vivo studies using HCV transgenic mouse models, however, showed little evidence for induction of UPR in the liver34 in absence of additional factors, such as iron overload35. Interestingly, a link between the UPR and hepatic steatosis has also been proposed36. Thus, we examined whether AFB1 may have an effect on UPR in WT or HCV-Tg mice. Similar to the observation of others34, we found no evidence that UPR is affected by either HCV, AFB1 or both (Figure 4E).
Discussion
An interaction between AFB1 exposure and HBV infection on HCC occurrence is a well-established phenomenon confirmed by multiple well-controlled epidemiological studies37, 38. Data from several recent reports from the Middle East and South-East Asia9, 10, 39, where both HCV prevalence and dietary exposure to AFB1 are high, also suggests that AFB1 exposure may be associated with advanced liver disease and increased HCC incidence in HCV-infected patients. Our study shows that HCV proteins core, E1, E2 and p7 are sufficient to reproduce the co-carcinogenic effect of HCV and AFB1 in the mouse, opening a possibility for understanding the molecular mechanisms of the interaction.
Much progress has been made in elucidating the mechanisms that underlie AFB1 and HCV hepatocarcinogenesis. Biotransformation of AFB1 to the putative carcinogenic intermediate, AFB-8,9-exo-epoxide, is a key step in the formation of pro-mutagenic DNA lesions40. Detoxification of AFB-8,9-epoxide by glutathione S-transferases is an important protective mechanism in mice41, species resistant to AFB1 hepato-carcinogenesis if treated post-neonataly11. Chronic inflammation, a result of HCV infection, was suggested to contribute to both DNA damage and cell proliferation in the liver, processes that may accelerate both accumulation of the mutations and clonal expansion of the initiated cells42. Firm molecular evidence in support of this hypothesis from subjects exposed to HCV and AFB1 is, however, lacking.
Several HCV transgenic mouse models have been developed in the past decade, yet liver tumors have been observed only in a couple of strains, usually after 13 to 24 months15, 18. Most of the HCV transgenic mouse models exhibit a limited overt liver phenotype, even late in life17, yet are susceptible to a number of additional hepatotoxic challenges (e.g., iron overload43 or alcohol20). An HCV-Tg mouse model which shows no spontaneous liver tumor development, yet exhibits an oxidative mitochondrial phenotype22, was selected here to study AFB1/HCV co-carcinogenic effect. We posit that the animal which does not have liver disease, yet is susceptible to additional challenges, may be relevant to model the human carriers of HCV who have not manifested with hepatitis but may be at risk for HCC upon exposure to AFB1.
Our data suggest that the molecular pathways of promotion of AFB1-induced carcinogenesis may differ between wild-type and HCV-Tg mice. We observed an increase in cell proliferation and expression of pro-fibrotic cytokines in the WT/AFB1, but not the HCV-Tg/AFB1 group. Conversely, AFB1 treatment in HCV-Tg mice, as compared to WT/AFB1 group, led to a significant increase in hepatic inflammation and expression of immune cell trafficking genes. These differences illustrate that while the gross liver pathology observed in AFB1-treated mice (e.g., steatosis and lipid peroxidation) are similar between the groups, the molecular events leading to steatohepatitis and tumor promotion are impacted greatly by the HCV.
The effects of HCV on lipid metabolism are well documented in human studies29, a phenomenon also observed in some HCV-Tg models28, 34. Steatohepatitis also appears to play a role in promoting HCV replication27. However, not all HCV-infected patients, or HCV transgenic mouse strains, manifest with steato-hepatitis, or even steatosis, and it is also difficult to distinguish between the pathways that may lead to lipid accumulation in the liver. For example, studies using different HCV transgenic mouse models have reported enhanced fatty acid uptake and decreased mitochondrial beta-oxidation28, as well as increased lipogenesis and decreased triglyceride secretion34, all of which may contribute to this phenotype. In the HCV-Tg model used here, lipid accumulation in the liver was minimal in absence of AFB1 treatment, yet HCV transgene sensitized hepatocytes to steatosis induced by AFB1. AFB1 has been reported to cause fatty change in the liver in humans30, rats31 and mice32; however, no mechanism has been proposed. While our study did not address this issue in full, we show that the steatosis-promoting effect of AFB1 may also differ between WT and HCV-Tg mice. This was evidenced by changes in expression of the lipogenesis gene fatty acid synthase in the WT group, but additional transcripts for fatty acid translocase and microsomal transfer protein, involved in lipid uptake and lipoprotein secretion, were induced by AFB1 in HCV-Tg mice.
We also observed an increase in lipid peroxidation in AFB1-treated mice even though no changes in genomic or mitochondrial oxidative DNA damage or intracellular or mitochondrial glutathione were found. Moriya et al.44 previously showed that no lipid peroxidation is found in HCV-Tg mice until 16 months of age when the mice begin to develop HCC. Moreover, they also observed lipid peroxidation in young HCV mice if treated with alcohol or carbon tetrachloride indicating that the HCV core protein predisposes hepatocytes to oxidative stress44. Thus, our study suggests that AFB1 challenge in HCV-Tg and WT mice promotes lipid peroxidation which is most pronounced when tumors are observed.
It should be noted that an alternative explanation for the observed differences in the incidence of AFB1-induced neoplastic and pre-neoplastic liver lesions between WT and HCV-Tg mice is that AFB-DNA adduct formation or removal in neonatal mice could have been effected by the transgene. Even though a comparison of liver gene expression data between WT and HCV-Tg mice at 12 months of age showed no difference in transcript levels for AFB1 metabolizing enzymes, additional experiments examining the time course of AFB1 DNA damage in neonates are necessary to address this possible mechanism.
In conclusion, while HCV mice exhibit a phenotype similar to wild-type mice at 12 months, they are susceptible to AFB1 exposure. We show that the HCV structural proteins potentiate carcinogenesis by AFB1 via increases in steatosis and tumor incidence; however, HCV and AFB1 may operate to increase liver cancer incidence through different mechanisms. In addition, while tumor occurrence may be also linked to lipid peroxidation consequently to AFB1 exposure, we posit that the conventional explanations (e.g., oxidative stress, lipid accumulation) do not appear to be the cause of the observed effect and other more specific, but as yet unidentified effects are likely to be responsible.
Supplementary Material
(A) Representative image of a hepatocellular carcinoma in WT/AFB1 and HCV/AFB1 mouse. (B) Different subtypes of adenomas observed in both WT/AFB1 and HCV/AFB1 groups, representative microphotographs (100x).
Acknowledgments
Supported in part by grants from the National Institutes of Health: ES005948, AA016258, ES010126 and AA012863.
List of Abbreviations
- AFB1
aflatoxin B1
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HCV-Tg
HCV transgenic mouse
- HBV
hepatitis B virus
- Tbp
TATA box binding protein
- UPR
unfolded protein response
Footnotes
Note: The views expressed herein do not necessarily represent those of the U.S. Food and Drug Administration.
References
- 1.El Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Bosch FX, Ribes J, Diaz M, Cleries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127:S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Jepsen P, Vilstrup H, Tarone RE, Friis S, Sorensen HT. Incidence rates of hepatocellular carcinoma in the U.S. and Denmark: recent trends. Int J Cancer. 2007;121:1624–6. doi: 10.1002/ijc.22860. [DOI] [PubMed] [Google Scholar]
- 4.International Agency for Research on Cancer. World Cancer Report 2008. Lyon, France: IARC Press; 2008. [Google Scholar]
- 5.London WT, Evans AA, McGlynn K, Buetow K, An P, Gao L, Lustbader E, Ross E, Chen G, Shen F. Viral, host and environmental risk factors for hepatocellular carcinoma: a prospective study in Haimen City, China. Intervirology. 1995;38:155–61. doi: 10.1159/000150426. [DOI] [PubMed] [Google Scholar]
- 6.International Agency for Research on Cancer. Some Naturally Occurring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins. Lyon, France: IARC Press; 1993. [Google Scholar]
- 7.Garner RC, Miller EC, Miller JA. Liver microsomal metabolism of aflatoxin B1 to a reactive derivative toxic to Salmonella typhimurium TA 1530. Cancer Res. 1972;32:2058–66. [PubMed] [Google Scholar]
- 8.Foster PL, Eisenstadt E, Miller JH. Base substitution mutations induced by metabolically activated aflatoxin B1. Proc Natl Acad Sci U S A. 1983;80:2695–8. doi: 10.1073/pnas.80.9.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen CH, Wang MH, Wang JH, Hung CH, Hu TH, Lee SC, Tung HD, Lee CM, Changchien CS, Chen PF, Hsu MC, Lu SN. Aflatoxin exposure and hepatitis C virus in advanced liver disease in a hepatitis C virus endemic area in Taiwan. Am J Trop Med Hyg. 2007;77:747–52. [PubMed] [Google Scholar]
- 10.Sayed HA, El Ayyat A, El Dusoki H, Zoheiry M, Mohamed S, Hassan M, El Assaly N, Awad A, El Ansary M, Saad A, El Karim AA. A cross sectional study of hepatitis B, C, some trace elements, heavy metals, aflatoxin B1 and schistosomiasis in a rural population, Egypt. J Egypt Public Health Assoc. 2005;80:355–88. [PubMed] [Google Scholar]
- 11.Vesselinovitch SD, Mihailovich N, Wogan GN, Lombard LS, Rao KV. Aflatoxin B 1, a hepatocarcinogen in the infant mouse. Cancer Res. 1972;32:2289–91. [PubMed] [Google Scholar]
- 12.McGlynn KA, Hunter K, LeVoyer T, Roush J, Wise P, Michielli RA, Shen FM, Evans AA, London WT, Buetow KH. Susceptibility to Aflatoxin B1-related Primary Hepatocellular Carcinoma in Mice and Humans. Cancer Res. 2003;63:4594–601. [PubMed] [Google Scholar]
- 13.Liu SP, Li YS, Lee CM, Yen CH, Liao YJ, Huang SF, Chien CH, Chen YM. Higher susceptibility to aflatoxin B(1)-related hepatocellular carcinoma in glycine N-methyltransferase knockout mice. Int J Cancer. 2011;128:511–23. doi: 10.1002/ijc.25386. [DOI] [PubMed] [Google Scholar]
- 14.Bradley DW. Studies of non-A, non-B hepatitis and characterization of the hepatitis C virus in chimpanzees. Curr Top Microbiol Immunol. 2000;242:1–23. doi: 10.1007/978-3-642-59605-6_1. [DOI] [PubMed] [Google Scholar]
- 15.Moriya K, Fujie H, Shintani Y, Yotsuyanagi H, Tsutsumi T, Ishibashi K, Matsuura Y, Kimura S, Miyamura T, Koike K. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat Med. 1998;4:1065–7. doi: 10.1038/2053. [DOI] [PubMed] [Google Scholar]
- 16.Lerat H, Honda M, Beard MR, Loesch K, Sun J, Yang Y, Okuda M, Gosert R, Xiao SY, Weinman SA, Lemon SM. Steatosis and liver cancer in transgenic mice expressing the structural and nonstructural proteins of hepatitis C virus. Gastroenterology. 2002;122:352–65. doi: 10.1053/gast.2002.31001. [DOI] [PubMed] [Google Scholar]
- 17.Koike K, Moriya K, Matsuura Y. Animal models for hepatitis C and related liver disease. Hepatol Res. 2010;40:69–82. doi: 10.1111/j.1872-034X.2009.00593.x. [DOI] [PubMed] [Google Scholar]
- 18.Liang TJ, Heller T. Pathogenesis of hepatitis C-associated hepatocellular carcinoma. Gastroenterology. 2004;127:S62–S71. doi: 10.1053/j.gastro.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 19.Naas T, Ghorbani M, Alvarez-Maya I, Lapner M, Kothary R, De Repentigny Y, Gomes S, Babiuk L, Giulivi A, Soare C, Azizi A, Diaz-Mitoma F. Characterization of liver histopathology in a transgenic mouse model expressing genotype 1a hepatitis C virus core and envelope proteins 1 and 2. J Gen Virol. 2005;86:2185–96. doi: 10.1099/vir.0.80969-0. [DOI] [PubMed] [Google Scholar]
- 20.Machida K, Tsukamoto H, Mkrtchyan H, Duan L, Dynnyk A, Liu HM, Asahina K, Govindarajan S, Ray R, Ou JH, Seki E, Deshaies R, et al. Toll-like receptor 4 mediates synergism between alcohol and HCV in hepatic oncogenesis involving stem cell marker Nanog. Proc Natl Acad Sci U S A. 2009;106:1548–53. doi: 10.1073/pnas.0807390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fox JG, Feng Y, Theve EJ, Raczynski AR, Fiala JL, Doernte AL, Williams M, McFaline JL, Essigmann JM, Schauer DB, Tannenbaum SR, Dedon PC, et al. Gut microbes define liver cancer risk in mice exposed to chemical and viral transgenic hepatocarcinogens. Gut. 2010;59:88–97. doi: 10.1136/gut.2009.183749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korenaga M, Wang T, Li Y, Showalter LA, Chan T, Sun J, Weinman SA. Hepatitis C Virus Core Protein Inhibits Mitochondrial Electron Transport and Increases Reactive Oxygen Species (ROS) Production. J Biol Chem. 2005;280:37481–8. doi: 10.1074/jbc.M506412200. [DOI] [PubMed] [Google Scholar]
- 23.James SJ, Rose S, Melnyk S, Jernigan S, Blossom S, Pavliv O, Gaylor DW. Cellular and mitochondrial glutathione redox imbalance in lymphoblastoid cells derived from children with autism. FASEB J. 2009;23:2374–83. doi: 10.1096/fj.08-128926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milano J, Day BJ. A catalytic antioxidant metalloporphyrin blocks hydrogen peroxide-induced mitochondrial DNA damage. Nucleic Acids Res. 2000;28:968–73. doi: 10.1093/nar/28.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Demeilliers C, Maisonneuve C, Grodet A, Mansouri A, Nguyen R, Tinel M, Letteron P, Degott C, Feldmann G, Pessayre D, Fromenty B. Impaired adaptive resynthesis and prolonged depletion of hepatic mitochondrial DNA after repeated alcohol binges in mice. Gastroenterology. 2002;123:1278–90. doi: 10.1053/gast.2002.35952. [DOI] [PubMed] [Google Scholar]
- 26.Powell CL, Kosyk O, Ross PK, Schoonhoven R, Boysen G, Swenberg JA, Heinloth AN, Boorman GA, Cunningham ML, Paules RS, Rusyn I. Phenotypic anchoring of acetaminophen-induced oxidative stress with gene expression profiles in rat liver. Toxicol Sci. 2006;93:213–22. doi: 10.1093/toxsci/kfl030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piver E, Roingeard P, Pages JC. The cell biology of hepatitis C virus (HCV) lipid addiction: molecular mechanisms and its potential importance in the clinic. Int J Biochem Cell Biol. 2010;42:869–79. doi: 10.1016/j.biocel.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka N, Moriya K, Kiyosawa K, Koike K, Gonzalez FJ, Aoyama T. PPARalpha activation is essential for HCV core protein-induced hepatic steatosis and hepatocellular carcinoma in mice. J Clin Invest. 2008;118:683–94. doi: 10.1172/JCI33594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Negro F, Sanyal AJ. Hepatitis C virus, steatosis and lipid abnormalities: clinical and pathogenic data. Liver Int. 2009;29 (Suppl 2):26–37. doi: 10.1111/j.1478-3231.2008.01950.x. [DOI] [PubMed] [Google Scholar]
- 30.Colon AR. Hepatic steatosis in children. Am J Gastroenterol. 1977;68:260–9. [PubMed] [Google Scholar]
- 31.Sachan DS, Yatim AM. Suppression of aflatoxin B1-induced lipid abnormalities and macromolecule-adduct formation by L-carnitine. J Environ Pathol Toxicol Oncol. 1992;11:205–10. [PubMed] [Google Scholar]
- 32.Ankrah NA, Addo PG, Abrahams CA, Ekuban FA, Addae MM. Comparative effects of aflatoxins G1 and B1 at levels within human exposure limits on mouse liver and kidney. West Afr J Med. 1993;12:105–9. [PubMed] [Google Scholar]
- 33.Chan SW, Egan PA. Hepatitis C virus envelope proteins regulate CHOP via induction of the unfolded protein response. FASEB J. 2005;19:1510–2. doi: 10.1096/fj.04-3455fje. [DOI] [PubMed] [Google Scholar]
- 34.Lerat H, Kammoun HL, Hainault I, Merour E, Higgs MR, Callens C, Lemon SM, Foufelle F, Pawlotsky JM. Hepatitis C virus proteins induce lipogenesis and defective triglyceride secretion in transgenic mice. J Biol Chem. 2009;284:33466–74. doi: 10.1074/jbc.M109.019810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishina S, Korenaga M, Hidaka I, Shinozaki A, Sakai A, Gondo T, Tabuchi M, Kishi F, Hino K. Hepatitis C virus protein and iron overload induce hepatic steatosis through the unfolded protein response in mice. Liver Int. 2010;30:683–92. doi: 10.1111/j.1478-3231.2010.02210.x. [DOI] [PubMed] [Google Scholar]
- 36.Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, Clark R, Miao H, Hassler JR, Fornek J, Katze MG, Hussain MM, et al. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15:829–40. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun Z, Lu P, Gail MH, Pee D, Zhang Q, Ming L, Wang J, Wu Y, Liu G, Wu Y, Zhu Y. Increased risk of hepatocellular carcinoma in male hepatitis B surface antigen carriers with chronic hepatitis who have detectable urinary aflatoxin metabolite M1. Hepatology. 1999;30:379–83. doi: 10.1002/hep.510300204. [DOI] [PubMed] [Google Scholar]
- 38.Yu MW, Lien JP, Chiu YH, Santella RM, Liaw YF, Chen CJ. Effect of aflatoxin metabolism and DNA adduct formation on hepatocellular carcinoma among chronic hepatitis B carriers in Taiwan. J Hepatol. 1997;27:320–30. doi: 10.1016/s0168-8278(97)80178-x. [DOI] [PubMed] [Google Scholar]
- 39.Anwar WA, Khaled HM, Amra HA, El Nezami H, Loffredo CA. Changing pattern of hepatocellular carcinoma (HCC) and its risk factors in Egypt: possibilities for prevention. Mutat Res. 2008;659:176–84. doi: 10.1016/j.mrrev.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 40.Eaton DL, Gallagher EP. Mechanisms of aflatoxin carcinogenesis. Annu Rev Pharmacol Toxicol. 1994;34:135–72. doi: 10.1146/annurev.pa.34.040194.001031. [DOI] [PubMed] [Google Scholar]
- 41.Ilic Z, Crawford D, Vakharia D, Egner PA, Sell S. Glutathione-S-transferase A3 knockout mice are sensitive to acute cytotoxic and genotoxic effects of aflatoxin B1. Toxicol Appl Pharmacol. 2010;242:241–6. doi: 10.1016/j.taap.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kensler TW, Roebuck BD, Wogan GN, Groopman JD. Aflatoxin: A 50 Year Odyssey of Mechanistic and Translational Toxicology. Toxicol Sci. 2011 doi: 10.1093/toxsci/kfq283. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furutani T, Hino K, Okuda M, Gondo T, Nishina S, Kitase A, Korenaga M, Xiao SY, Weinman SA, Lemon SM, Sakaida I, Okita K. Hepatic iron overload induces hepatocellular carcinoma in transgenic mice expressing the hepatitis C virus polyprotein. Gastroenterology. 2006;130:2087–98. doi: 10.1053/j.gastro.2006.02.060. [DOI] [PubMed] [Google Scholar]
- 44.Moriya K, Nakagawa K, Santa T, Shintani Y, Fujie H, Miyoshi H, Tsutsumi T, Miyazawa T, Ishibashi K, Horie T, Imai K, Todoroki T, et al. Oxidative stress in the absence of inflammation in a mouse model for hepatitis C virus-associated hepatocarcinogenesis. Cancer Res. 2001;61:4365–70. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Representative image of a hepatocellular carcinoma in WT/AFB1 and HCV/AFB1 mouse. (B) Different subtypes of adenomas observed in both WT/AFB1 and HCV/AFB1 groups, representative microphotographs (100x).