Abstract
A human endogenous retrovirus type E was recently found to be selectively expressed in most renal cell carcinomas (RCC). Importantly, antigens derived from this provirus are immunogenic, stimulating cytotoxic T-cells that kill RCC cells in vitro and in vivo. Here we show HERV-E expression is restricted to the clear cell subtype of RCC (ccRCC) characterized by an inactivation of the von Hippel-Lindau (VHL) tumor suppressor gene with subsequent stabilization of hypoxia-inducible transcription factors HIF-1α and -2α. HERV-E expression in ccRCC linearly correlated with HIF-2α levels and could be silenced in tumor cells by either transfection of normal VHL or siRNA inhibition of HIF-2α. Using chromatin immunoprecipitation, we demonstrated that HIF-2α can serve as transcriptional factor for HERV-E by binding with HIF response elements (HRE) localized in the proviral 5′LTR. Remarkably, the LTR was found to be hypomethylated only in HERV-E-expressing ccRCC while other tumors and normal tissues possessed a hypermethylated LTR preventing proviral expression. Taken altogether, these findings provide the first evidence that inactivation of a tumor suppressor gene can result in aberrant proviral expression in a human tumor and give insights needed for translational research aimed at boosting human immunity against antigenic components of this HERV-E.
Keywords: renal cell carcinoma, endogenous retrovirus, VHL, HIF, DNA methylation
Introduction
It is estimated that 1 in 70 men and women will be diagnosed with kidney cancer in their lifetime (SEER data http://seer.cancer.gov/statfacts/html/kidrp.html). Although highly curable when disease is localized, metastatic kidney cancer is associated with an extremely poor prognosis, with median survivals of only 10 months. Despite its poor prognosis, kidney cancer is unusual among solid tumors as it appears to be susceptible to the immune system; sustained regression of tumors in patients treated with interferon alpha, IL-2 and allogeneic stem cell transplantation have established the susceptibility of this tumor to attack by T-cells (Childs et al., 2000; Bregni et al., 2002; Rini et al., 2002). Although some patients achieve long-term disease free survival with immune-based therapies, surprisingly little data is known about the target antigens of T-cell populations that mediate regression of RCC. Using allogeneic T-cells, we recently discovered two novel transcripts (CT-RCC-8 and CT-RCC-9) derived from a HERV-E provirus (herein named CT-RCC HERV-E) located on chromosome 6q that are expressed in kidney cancer cells (Takahashi et al., 2008). CT-RCC HERV-E expression was detected at variable levels in fresh kidney cancer samples and in >50% of cultured RCC cell lines, but was not detected in normal tissues and non-RCC tumors. A 10 amino acid peptide antigen (CT-RCC-1) encoded from the shared common sequence region of CT-RCC-8 and CT-RCC-9 was found to be the target antigen of RCC-reactive T-cells that were isolated from a patient who had prolonged regression of kidney cancer following an allogeneic hematopoietic stem cell transplant. Furthermore, this HERV-E derived peptide is highly immunogenic, stimulating peptide specific CD8+ T-cells in vitro and in vivo in kidney cancer patients that kill RCC cells (Rao et al., 2008). These data suggest antigens derived from the CT-RCC HERV-E could be potential targets for T-cell based immunotherapy directed at RCC.
Endogenous retroviruses represent the ancestral remnants of exogenous retroviral infection into germ line cells. Due to negative selection, the majority of HERVs accumulated mutations or insertions/deletions and thus do not contain uninterrupted ORFs to code full-length proteins, or have had their transcription silenced by promoter methylation. However, there is a growing literature on HERV up-regulation at the transcriptional and protein levels in a variety of cancers (reviewed by Ruprecht et al., 2008; Romanish et al., 2010). An increasing number of tumors have recently been shown to express endogenous retroviral products that elicit antitumor immunity in vivo (Hsiao et al., 2006; Hahn et al., 2008; Takahashi et al., 2008; Wang-Johanning et al., 2008). Furthermore, since retroviral-derived proteins are intrinsically immunogenic, antigens encoded by HERVs potentially represent excellent targets for immune-based cancer therapy.
Here we investigated the mechanisms leading to specific expression of the CT-RCC HERV-E in RCC. HERV-E expression was found to be restricted to the clear cell histological subtype of RCC (ccRCC) and caused by inactivation of the VHL tumor suppressor gene and HIF-2α stabilization. We demonstrated that HIF-2α may serve as transcriptional factor for the CT-RCC HERV-E by binding with HIF response elements (HRE) localized in proviral long terminal repeat (LTR). Remarkably, all ccRCC tumors expressing CT-RCC HERV-E possessed hypomethylated LTR. In contrast, non-ccRCC tumors and normal tissues had a highly methylated proviral LTR and no CT-RCC HERV-E expression. These findings suggest loss of function of the VHL tumor suppressor and HIF-2α stabilization together with epigenetic modifications lead to the selective expression of this endogenous retrovirus in ccRCC that serves as an antigenic target of T-cells in vivo.
Results
Expression of CT-RCC HERV-E in different RCC histological subtypes
The classification for renal neoplasms divides RCC into distinct histological subtypes. CT-RCC HERV-E expression was analyzed from fresh tumor samples representing 5 different histological types of kidney tumors: clear cell kidney cancer (both without and with sarcomatoid features), papillary RCC, chromophobe RCC, collecting duct RCC, and oncocytomas. Quantitative real-time RT-PCR (qRT-PCR) using primers specific for the proviral transcripts CT-RCC-8, CT-RCC-9 and their shared common sequence region (herein named CT-RCC common region) was used for this analysis as described previously (Takahashi et al., 2008). Analysis of 17 freshly isolated clear cell renal cell carcinomas procured at NIH (14 without sarcomatoid features and 3 with) showed 13/17 (76%) expressed the CT-RCC HERV-E (Table 1). In contrast, none of the 17 non-clear cell kidney fresh tumors expressed any CT-RCC transcripts (Table 1).
Table 1.
Expression of CT-RCC HERV-E in fresh RCC tumors.
| Histological subtype | number of samples | mean values and range of copies relative to β-actin (× 105) | ||
|---|---|---|---|---|
| Common Region | CT-RCC-8 | CT-RCC-9 | ||
| Clear cell | 14 | 890.75 (6.5 to 1775) | 356 (7 to 705) | 4051 (9.0 to 8093) |
| Clear cell with sarcomatoid features | 3 | 115 (15 to 215) | 95 (15 to 175) | 290.5 (21 to 560) |
| Papillary | 8 | 10 (3.5 to 16.5) | 1.3 (0.1 to 2.5) | 14.5 (5.5 to 23.5) |
| Chromophobe | 3 | 8.8 (5 to 12.6) | 0.4 (0.2 to 0.6) | 17 (10 to 24) |
| Oncocytoma | 4 | 6.4 (4.3 to 8.5) | 1.1 (0.2 to 2) | 13 (10.5 to 15.5) |
| Collecting duct | 2 | 2 (1.2 to 2.8) | 0.9 (0.1 to 1.7) | 6.9 (3.3 to 10.5) |
We next assessed whether there was any correlation between the size and stage of the primary tumor and the level of proviral expression. Twenty seven additional ccRCC primary tumors at different clinical stages obtained from nephrectomy samples were procured at the Engelhardt Institute of Molecular Biology in Moscow. They included 7 stage I tumors (7 cm or smaller primary tumor confined to kidney), 11 stage II tumors (greater than 7 cm primary tumor confined to kidney), and 9 stage III tumors (primary tumor extended into the renal vein or vena cava, or involved the ipsilateral gland and/or perinephric fat, or had spread to one local lymph node). Tumors were dissected away from the normal part of the kidney and the remaining normal kidney tissue served as a control for each tumor sample analyzed. A two-fold or greater expression ratio of the CT-RCC common region in the tumor was considered significant, as this was the maximum fluctuation range for mRNA level of the reference gene RPN1. Among all the samples analyzed, expression of the HERV-E common region was a median 2.7 fold higher in the tumor samples (median expression 11.5) compared to normal kidney tissues (median expression 4.5). Slightly more than half of the fresh tumor samples (15/27; 55.5%) had significantly higher expression of the CT-RCC common region than normal kidney tissue, including 13/27 samples that had a 3 fold or higher expression and 5/27 that had a 10 fold or higher expression than normal kidney tissue. Ten of the tumor samples (37%) had similar expression levels as the control kidney tissue (Table 2) and two tumors (7.4%) had slightly lower expression in comparison to the normal kidney, although this most likely occurred as a consequence of the low copy number of CT-RCC transcripts (less than 6 copies relative to RPN1 X 100) in both the tumor and normal kidney tissue (Table 2).
Table 2.
Expression of CT-RCC HERV-E in fresh (primary) ccRCC tumors and normal kidney tissue obtained from the same nephrectomy samples*.
| ID Number | ccRCC tumors | Normal Kidney | n-fold Expression Ratio (tumor/normal kidney) |
|---|---|---|---|
| 1012 | 4.5 | 3.6 | 1.2 ± 0.1 |
| 1100 | 11.5 | 6.3 | 1.8 ± 0.1 |
| 1051 | 15.2 | 8.3 | 1.8 ± 0.1 |
| 1040 | 8.9 | 4.4 | 2.0 ± 0.1 |
| 1018 | 16.8 | 3.8 | 4.5 ± 0.1 |
| 1108 | 29.5 | 4.5 | 6.5 ± 0.3 |
| 1027 | 118.8 | 4.5 | 26.5 ± 0.2 |
| 1093 | 1.3 | 5.3 | 0.2 ± 0.4 |
| 1061 | 4.9 | 4.8 | 1.0 ± 0.3 |
| 1096 | 4.9 | 3.9 | 1.3 ± 0.2 |
| 1028 | 9.1 | 6.1 | 1.5 ± 0.3 |
| 1049 | 8.2 | 4.2 | 1.9 ± 0.3 |
| 1005 | 12.2 | 4.5 | 2.7 ± 0.3 |
| 1036 | 13.1 | 4.0 | 3.2 ± 0.2 |
| 1079 | 23.7 | 4.0 | 5.9 ± 0.2 |
| 1129 | 30.2 | 4.5 | 6.7 ± 0.3 |
| 1063 | 459.3 | 3.8 | 121.0 ± 0.3 |
| 1119 | 2825.0 | 4.5 | 630.4 ± 0.2 |
| 1039 | 2.7 | 5.9 | 0.5 ± 0.2 |
| 1147 | 3.6 | 4.5 | 0.8 ± 0.2 |
| 1111 | 4.3 | 4.2 | 1.0 ± 0.1 |
| 1014 | 3.9 | 3.0 | 1.3 ± 0.2 |
| 1103 | 9.5 | 2.8 | 3.4 ± 0.2 |
| 1077 | 9.3 | 2.6 | 3.5 ± 0.2 |
| 1104 | 18.4 | 4.5 | 4.1 ± 0.2 |
| 1110 | 87.8 | 5.2 | 16.7 ± 0.2 |
| 1068 | 436.6 | 3.9 | 111.0 ± 0.3 |
| median | 11.5 | 4.5 | 2.7 |
The median copy numbers of the CT-RCC common region relative to RPN1 × 100 and n-fold expression ratio of values for tumors and matched normal kidney tissues are shown.
When the results were analyzed according to tumor stage, over half of the tumors in each stage had 2-fold or greater expression of the HERV-E compared to normal kidney tissue, including 4/7 (57.1%) stage I tumors, 6/11 (54.5%) stage II tumors and 5/9 (55.6%) stage III tumors. Of the 9 stage III tumors, 3 samples (1103, 1147, and 1110) were known to have metastases to regional lymph nodes. Two of these three tumors had greater than 2-fold expression of the HERV-E common region compared to normal kidney tissue.
In summary, expression of CT-RCC transcripts is limited to clear cell histological subtype of kidney cancer and occurs in the majority of ccRCC tumors, even in the earliest stages of carcinogenesis and is not limited to more advanced stage tumor lesions.
Status of the VHL tumor suppressor gene and CT-RCC HERV-E expression in ccRCC
Inactivation of the VHL tumor suppressor gene has been shown to occur in the vast majority of ccRCC tumors. Therefore, we investigated for genetic and epigenetic alterations in the VHL gene as well as for VHL mRNA expression levels in all 14 ccRCC cell lines used in this study. Sequence analysis of DNA showed 9 of 14 ccRCC cell lines had mutations or deletions in one of three VHL exons resulting in amino acid substitutions or the creation of stop codons, and 3 of 14 tumors had a highly methylated VHL promoter (Table 3). In the remaining two ccRCC lines (lines #13 and 14), no mutations or promoter methylation could be found. Nevertheless, qRT-PCR using primers for all three VHL exons to measure VHL expression showed the two above lines without VHL abnormalities had extremely weak or undetectable levels of VHL mRNA (data not shown). Therefore, all ccRCC cell lines expressing CT-RCC HERV-E were classified as being VHL deficient.
Table 3.
Genetic alterations of the VHL gene in different ccRCC primary tumors and cell lines.
| ccRCC cell lines and tumors | promoter methylation | exon 1 | exon 2 | exon3 |
|---|---|---|---|---|
| 1 | NO | G/A IVS2 +1 | ||
| 2 | NO | 2 bp DEL* AA180 | ||
| 3 | NO | L89P | ||
| 4 | NO | 1 bp DEL AA124 | ||
| 5 | YES | |||
| 6 | NO | 25 bp DEL AA62 | ||
| 7 | YES | |||
| 8 | YES | |||
| 9 | NO | W117S | ||
| 10 | NO | 1 bp DEL AA65 | ||
| 11 | NO | Y98Stop | ||
| 12 | NO | T/A IVS1 +2 | ||
| 13 | NO | |||
| 14 | NO | |||
| UT08-0084 | NO | DEL AA1 | ||
| UT08-0115 | NO | C162F | ||
| UT08-0373 | NO | F136S | ||
| UT09-0294 | NO | P86R | ||
| UT09-0318 | NO | FS** AA144 | ||
| UT09-0320 | NO | R167Q | ||
| UT09-0335 | NO | S65W | ||
| UT09-0416 | NO | FS AA176 | ||
| UT09-0419 | NO | E134X | ||
| UT09-0433 | NO | W117C | ||
| UT09-0540 | NO | DEL AA155 | ||
| UT09-0784 | NO | L158P | ||
| UT10-0134 | NO | R161X | ||
| UT10-0401 | NO | N90I | ||
| UT10-0419 | NO | FS AA197 | ||
| UT10-0465 | NO | DEL AA76 | ||
| UT10-0560 | NO | Y175D | ||
| UT10-0585 | NO | N78S | ||
| UT10-0765 | NO | S111N | ||
| UT10-0927 | NO | FS AA118 | ||
| UT11-0067 | NO | R167W | ||
| UT11-0071 | NO | DEL AA114 | ||
| UT11-0074 | NO | DEL AA155 |
DEL, deletion.
FS, frame shift.
To further correlate VHL inactivation with CT-RCC HERV-E expression, 23 primary frozen ccRCC tumors obtained from nephrectomy samples procured at the NCI from patients with VHL disease where specific mutations in the VHL gene were defined by sequencing were analyzed (Table 3). qRT-PCR showed 20/23 of these fresh tumors expressed CT-RCC HERV-E: 13/20 had expression of CT-RCC HERV-E transcripts in a range of 400 up to 5000 copies (relative to GAPDH × 105) and 7/20 demonstrated higher expression levels ranging from 7500 to 25000 copies (relative to GAPDH × 105). In the 20 of these samples that expressed HERV-E, we did not find any significant difference in HERV-E expression level relative to the mutated VHL exon. However, when we performed the same analysis on pooled ccRCC fresh tumors (n=20) and cell lines (n=11) that expressed CT-RCC HERV-E, HERV-E expression was found to be slightly higher in the 10 samples with VHL alterations located in exon 1 (median HERV-E expression is 7231 copies relative to GAPDH ×105) compared to the 21 other samples without exon 1 alterations (median HERV-E expression is 2502 copies relative to GAPDH × 105; p=0.048 in two-sided Wilcoxon rank-sum test).
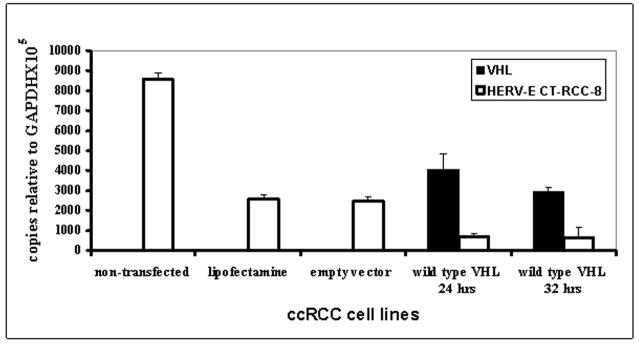
To identify if loss of functional VHL protein is required for HERV-E expression, we analyzed the effect of introducing wt VHL transgenes into VHL-deficient ccRCC cell lines. As shown in Figure 1, transfection of functional VHL significantly reduced expression of CT-RCC HERV-E transcripts. Taken together, these data suggest VHL inactivation is required for CT-RCC HERV-E expression in ccRCC.
Figure 1.
Expression of CT-RCC HERV-E in ccRCC cell line after wt VHL transfection. qRT-PCR analysis of VHL (black bars) and CT-RCC-8 (white bars) expression levels was performed 24 and 32 hours after transfection of a ccRCC tumor line #2 with a plasmid expressing normal wt VHL gene. Empty vector and transfection reagent were used as negative controls at one point (32 hours). The data shown represents mean values of two independent experiments.
Methylation of the 5′LTR affects provirus activity
Although the above data are consistent with VHL inactivation promoting provirus activity, we failed to detect any CT-RCC HERV-E transcripts in some VHL-deficient ccRCC cell lines (lines # 5, 6, 9, and 786-0). Since transcription of HERVs is known to be silenced by promoter methylation, we hypothesized the absence of HERV-E expression might be related to hypermethylation of the proviral LTR. To investigate this further, bisulfite sequencing of the 11 CpGs contained within the HERV-E 5′LTR was performed as described in the ‘Materials and methods section’. Results showed all HERV-E non-expressing ccRCC cell lines as well as non-clear cell kidney tumors, tumors other than kidney cancer, and normal tissues contained CpGs that were hypermethylated in the 5′LTR region (Table 4). In contrast, all 11 CpGs in the HERV-E 5′LTR were hypomethylated in all ccRCC cell lines that expressed HERV-E (Table 4).
Table 4.
Methylation status of the 11 CpGs and the HRE located in 5′LTR of CT-RCC HERV-E in different tumors and normal tissues.
| Type of cancer or normal tissue | Number of samples | % methylation of CpGs | % methylation of the HRE CpG |
|---|---|---|---|
| median (range) | median (range) | ||
| HERV-E expressing ccRCC | 13 | 9.4 (0–32.1) | 9.4 (3.3–17.4) |
| HERV-E non-expressing ccRCC | 5 | 84.4 (47.6–100) | 85.1 (75.3–96) |
| Collecting duct tumor | 2 | 91.7 (57–100) | 96.5 (92.9–100) |
| Papillary kidney cancer | 2 | 91.8 (53.2–100) | 76.6 (53.2–100) |
| Colon carcinoma | 2 | 91.8 (60.7–100) | 94.1 (88.6–99.6) |
| Breast cancer | 1 | 94.8 (88.2–100) | 100 |
| Lung cancer | 3 | 93.7 (81–100) | 95.7 (89.9–99.3) |
| Melanoma | 2 | 93.6 (84.3–100) | 99.8 (99.6–100) |
| Pancreatic carcinoma | 3 | 91.3 (73.5–100) | 86.1 (73.5–100) |
| Normal kidney tissue | 4 | 94.4 (86.2–100) | 99.7 (98.9–100) |
| Normal liver tissue | 2 | 80.5 (46.4–100) | 89 (78.1–100) |
These data show that hypermethylation of the proviral 5′LTR silences proviral expression in normal human tissues, tumors other than ccRCC, and in the minority of ccRCC tumors. Thus, CT-RCC HERV-E was found to be expressed only in VHL-deficient ccRCC tumors that harbored a hypomethylated HERV-E 5′LTR.
HIF-2α is a likely transcription factor for CT-RCC HERV-E
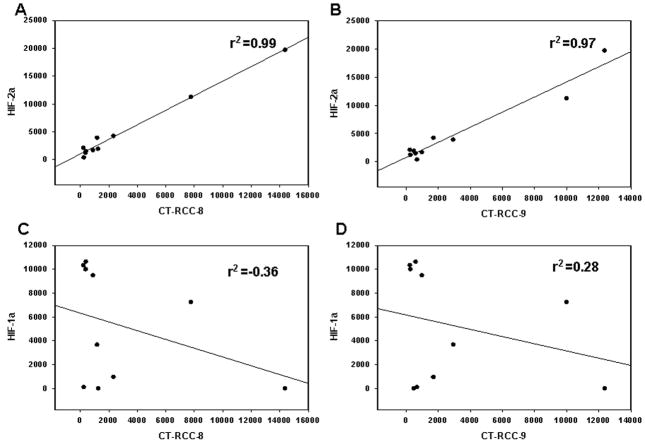
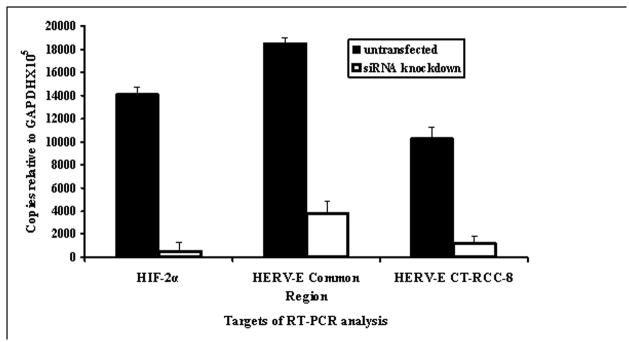
Loss of function of the VHL tumor suppressor or a hypoxic environment ultimately leads to increased expression of hypoxia-inducible transcription factors (HIFs) at mRNA and protein levels (Gnarra et al., 1996; Iliopoulos et al., 1996; Maxwell et al., 1999; Krieg et al., 2000). Because loss of function of VHL appears to be associated with CT-RCC HERV-E expression, we evaluated whether HIF might serve to promote expression of this provirus in ccRCC. qRT-PCR of VHL defective ccRCC lines showed all had elevated mRNA levels of HIF-2α, either alone or in combination with elevated levels of HIF-1α. The HERV-E expressing ccRCC lines #7 and #12 were found to have high HIF-2α expression in the absence of HIF-1α mRNA, establishing that HERV-E transcription was not dependent on HIF-1α. Furthermore, we found no correlation between HIF-1α mRNA levels and expression of either CT-RCC-8 or CT-RCC-9 HERV-E transcripts (Figure 2). In contrast, all HERV-E expressing ccRCC tumors expressed HIF-2α and there appeared to be a significant correlation between HIF-2α expression and CT-RCC HERV-E expression (Figure 2). Based on these observations, we hypothesized that HIF-2α can activate HERV-E transcription in ccRCC. To test this hypothesis, HERV-E expression was measured by qRT-PCR in ccRCC lines following transfection with siRNA specific for HIF-2α. As shown in Figure 3, proviral transcription decreased substantially following targeted knockdown of HIF-2α, confirming this transcription factor is involved in activation of CT-RCC HERV-E.
Figure 2.
Relationship between HIF-2α and HIF-1α expression and CT-RCC HERV-E expression in different ccRCC cell lines.
Expression levels of HIFs and the HERV-E transcripts were measured by qRT-PCR in all studied ccRCC lines. HIF-2α mRNA levels strongly correlate with expression levels of the HERV-E transcripts CT-RCC-8 (A) and CT-RCC-9 (B). In contrast, there was no correlation between HIF-1α mRNA levels and expression of CT-RCC-8 (C) or CT-RCC-9 (D) transcripts. The data shown represents mean values of two independent experiments.
Figure 3.
Expression of CT-RCC HERV-E in ccRCC cell line after siRNA knockdown of HIF-2α. qRT-PCR on RNA isolated from the ccRCC line #2 was performed using primers specific for HIF-2α, as well as primers targeting the common region and CT-RCC-8 sequences. Black bars show data from cells cultured in normal media and white bars show data from cells after siRNA knockdown of HIF-2α. The data shown represents mean values of two independent experiments.
Once stabilized and activated, HIFs bind DNA at the consensus HIF response element (HRE) in the promoter region of the target gene (core HRE sequence is CGTG). We identified a core HRE sequence in the HERV-E 5′LTR (located in Human HERV-E AL133408: nt 79333–79336) that was hypomethylated in all HERV-E expressing ccRCCs but was hypermethylated in all normal tissues, non clear cell RCC cell lines, and HERV-E-negative ccRCC cell lines (Table 4). A chromatin immunoprecipitation (ChIP) assay using HIF-2α antibodies and sonicated chromatin isolated from different HIF-2α expressing ccRCC cell lines was performed to analyze possible HIF-2α interactions with the 5′LTR of CT-RCC HERV-E. The ccRCC cell line 786-wt transfected with wtVHL, which eliminates HIF-2α expression, was used as a negative control for this immunoprecipitation assay. Resulting DNA samples were analyzed by PCR utilizing designed primer set covering the HRE element in the 5′LTR of CT-RCC HERV-E. Primers targeting the HRE element of Egln3/PHD3 promoter region which has previously been shown to be a target for HIF-2α binding were used as a positive control (Wright and Rathmell, 2010). In HERV-E expressing ccRCC cell lines, specific PCR signals utilizing both primer sets targeting the HRE elements of Egln3/PHD3 promoter and provirus LTR were observed (Figure 4). PCR products were subsequently extracted from gels and were shown by sequencing to be derived from corresponding genomic regions of the HERV-E 5′LTR confirming binding of HIF-2α to HRE element of CT-RCC HERV-E.
Figure 4.

HIF-2α interaction with the 5′LTR of CT-RCC HERV-E.
Clear cell RCC cell lines over-expressing HIF-2α were subjected to the ChIP assay using antibodies to HIF-2α. The HERV-E expressing RCC cell lines #2 and #12 contain a hypomethylated proviral 5′LTR; the HERV-E non-expressing RCC lines #6 and 786-0 contain a hypermethylated 5′LTR. The VHL transfected ccRCC cell line 786-wt was used as a negative control for immunoprecipitation with HIF-2α antibodies. Primers targeting HRE in 5′LTR of CT-RCC HERV-E and Egln3/PHD3 promoter were used for PCR; amplification at 30 cycles is shown. Input, DNA extracted from cell lines without immunoprecipitation.
Next, the ChIP assay was used to compare RCC cell lines with high levels of HIF-2α expression but with varying levels of proviral LTR methylation and HERV-E expression (Figure 4). The HERV-E expressing RCC lines #2 and #12 that contain a hypomethylated proviral 5′LTR (mean values of 3.1% and 4.9%, respectively) and the HERV-E non-expressing RCC lines #6 and 786-0 that contain a hypermethylated 5′LTR (mean values of 90% and 88.3%, respectively) were immunoprecipitated with HIF-2α antibodies. We observed strong PCR signals for the HERV-E 5′LTR in the hypomethylated RCC cell lines; in contrast, PCR signals for the HERV-E 5′LTR were substantially weaker in the hypermethylated RCC cell lines (Figure 4). PCR products from HERV-E expressing RCC cell lines #2 and #12 reached plateau by cycle 27, whereas HERV-E-negative RCC cell lines #6 and 786-0 did not achieve saturation levels until 38–40 cycles, respectively (data not shown). Importantly, the PCR signal intensity for the Egln3/PHD3 promoter was similar among all the RCC tumor lines, both HERV-E expressing and HERV-E non-expressing. Based on these findings, it is plausible that HIF-2α directly activates CT-RCC HERV-E expression in ccRCC tumors that have a hypomethylated provirus LTR.
Demethylating agents enhance CT-RCC HERV-E expression in ccRCC
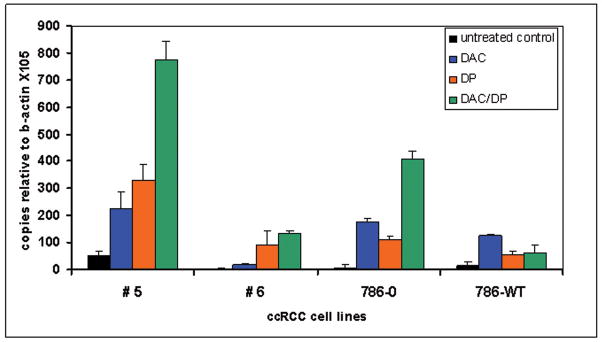
Having observed promoter hypermethylation correlates with absence of HERV-E expression, we assessed whether the demethylating agent DAC and the histone deacetylase inhibitor DP could enhance the provirus expression. HERV-E-negative ccRCC lines containing a hypermethylated 5′LTR region were exposed to normal media, 0.1 uM DAC, DP (25 ng/ml), or combination DAC/DP treatment. qRT-PCR of RNA extracted from tumor lines exposed to DAC or DP alone or sequential DAC/DP showed HERV-E expression was enhanced relative to untreated controls (Figure 5). In the ccRCC cell line #5, expression of the HERV-E common region increased 4.5-fold (range 50 to 225 copies relative to β-actin × 105) with DAC alone, 6.6-fold (50 to 330) with DP alone, and 15.5-fold (50 to 774) with sequential DAC/DP exposure relative to untreated controls.
Figure 5.
Expression of CT-RCC HERV-E in ccRCC cell lines after DAC and DP treatment. qRT-PCR of RNA isolated from ccRCC cell lines with primers specific for CT-RCC common region. Cell lines were treated with DAC alone (blue), DP alone (orange), and combination of DAC/DP (green bars). Black bars show normal media controls.
Remarkably, upregulation of the HERV-E expression by DAC/DP was substantially higher in the ccRCC cell line 786-0 with inactivated VHL compared to the same cell line that was transfected with wt VHL: Relative to untreated controls, HERV-E common region copies following DAC/DP treatment increased 50 fold in the VHL deficient 786-0 line compared to only a 4.2-fold increase in the 786-wt line which expresses VHL. Since both 786 cell lines have a high level of 5′LTR methylation (mean values of 88.3% and 91% for 786-0 and 786-wt, respectively), the smaller increase in proviral expression observed after demethylation of the 786-wt line can be attributed to functional VHL reducing levels of HIF-2α, the proposed CT-RCC HERV-E transcriptional factor.
Discussion
In this study, we provide insight into the mechanisms controlling expression of the CT-RCC HERV-E in kidney cancer. In our prior analysis, we found this HERV-E was expressed in the majority of RCC cell lines and fresh tumors and was highly immunogenic (Takahashi et al., 2008). The CT-RCC-1 peptide derived form this provirus was found to encode an antigen that stimulates cytotoxic T-cells that kill RCC cells in vitro and in vivo in a patient with metastatic kidney cancer who had durable tumor regression following an allogeneic hematopoietic stem cell transplant. Remarkably, this provirus was found to have expression restricted to kidney cancer tumors, with no detectable expression in normal tissues or non kidney cancer tumors. Renal neoplasms are divided into distinct histological subtypes and include clear cell carcinoma, papillary RCC, chromophobe tumors, collecting duct tumors and medullary RCC (Cohen and McGovern, 2005). Here we show expression of this HERV-E is restricted to clear cell carcinoma, which, interestingly, appears to be the subtype of kidney cancer that is immuno-responsive. Although expression levels varied among tumors, a majority of ccRCCs (80%) were found to express CT-RCC transcripts. Furthermore, proviral expression occurred at the earliest stages of carcinogenesis, with CT-RCC transcripts being detected in small (<3cm) primary tumors with no correlation between tumor lesion size and level of expression.
Clear cell carcinoma is the most common subtype of RCC, accounting for approximately 70–80% of all RCC tumors. In most clear cell cancers, the tumor suppressor gene VHL is inactivated from either gene mutation or promoter hypermethylation. In accordance, we found VHL was deficient in all the ccRCC cell lines used in this analysis. Sequencing analysis showed 12/14 ccRCC lines contained VHL mutations or promoter methylation. The remaining two ccRCC lines did not contain any genetic or epigenetic alterations in the VHL gene. Nevertheless, both had strongly reduced levels of VHL, which may have occurred as a consequence of other non-genetic alterations which lead to loss of VHL expression (i.e. micro-RNA reducing VHL expression in CLL B cells; Ghosh et al., 2009). Among tumors where VHL was found to be mutated, we found that the majority (31/37; 84%) expressed the CT-RCC HERV-E including 11/14 (79%) ccRCC cell lines and 20/23 (87%) primary tumors obtained from patients with VHL disease. Importantly, transfection of wt VHL into VHL deficient ccRCC cell lines dramatically reduced the proviral expression. Taken together, these data suggest inactivation of VHL is an essential event required for the CT-RCC HERV-E expression in RCC.
Genetic mutations that disrupt the function of VHL protein in clear cell tumors induce the activation of HIF-mediated hypoxia-induced gene pathways during normoxia (reviewed by Kaelin, 2009). HIF is a basic heterodimeric transcription factor consisting of a constitutively expressed beta subunit and one of three oxygen sensitive alpha subunits (HIF-1α, HIF-2α, or HIF-3α). The HIF-1α and HIF-2α are known to be widely expressed in different types of cancer and play a role in angiogenesis and tumor apoptosis resistance. HIF-2α was reported to be expressed in ccRCC, but its expression was variable in non-clear cell RCC tumors and absent or present at very low levels in the normal kidney cortex (Turner et al., 2002; Kim et al., 2006; Sandlund et al., 2009). Furthermore, VHL-defective RCC cells seem to show a bias toward HIF-2α rather than HIF-1α expression (Krieg et al., 2000; Turner et al., 2002; Kim et al., 2006; Sandlund et al., 2009). In accordance with these findings, all the ccRCC lines studied in this work demonstrated elevated expression of HIF-2α subunits, either alone or in combination with HIF-1α. Because VHL inactivation was found to be associated with HERV-E expression, we evaluated whether HIF promotes proviral expression in ccRCC. HIF-1α was found to be absent in some ccRCC cell lines that had high levels of the HERV-E expression. In contrast, HIF-2α subunits were detected in all HERV-E expressing ccRCC tumors, with HIF-2α mRNA levels linearly correlating with the levels of proviral transcripts. The observation that siRNA silencing of HIF-2α dramatically decreased proviral expression firmly establishes that HIF-2α plays a role in promoting expression of CT-RCC HERV-E.
HERV LTRs are known to function as promoters, with some being enriched with binding sites for transcription factors such as p53, CTCF, Pou5F1-Sox2, and ESR1 (Cohen et al., 2009). We identified core HRE sequences in the 5′LTR of the CT-RCC HERV-E, leading us to hypothesize that HIF-2α binds directly to the 5′LTR promoting proviral expression. Indeed, chromatin immunoprecipitation using HIF-2α antibodies demonstrated an interaction between HIF-2α and the proviral LTR in HERV-E expressing ccRCC cell lines. We found the HRE motif as well as other surrounding CpGs in the HERV-E 5′LTR were hypomethylated in all HERV-E expressing ccRCCs. In contrast, this genomic region was highly methylated in normal tissues, non-kidney tumors, non clear cell RCC cell lines, and the small subset of ccRCC cell lines that were found to be HERV-E negative. Using ChIP analysis we observed substantially lower levels of HIF-2α/5′LTR interaction in HERV-E-negative ccRCC cell lines that possessed hypermethylated HERV-E LTRs. These data are consistent with prior data showing that methylation of a cytosine nucleotide in the core HRE blocks binding of HIF alpha subunits inhibiting transcription (Wenger et al., 1998). Taken together, these data suggest methylation of CpGs in the HERV-E 5′LTR silences proviral expression, even when the transcriptional factor HIF-2α is present, potentially explaining why a minority of ccRCC tumors had no CT-RCC HERV-E expression. Consistent with this observation, we found treatment of ccRCC tumors with the demethylating agent DAC or the histone deacetylase inhibitor depsipeptide, given alone or together, increased proviral expression in ccRCC cells that possessed hypermethylated HERV-E 5′LTRs. Remarkably, the upregulation in proviral expression by combined DAC/depsipeptide treatment decreased substantially when wt VHL was transfected into the 786-0 ccRCC line, providing further evidence that functional pVHL suppresses activation of CT-RCC HERV-E by destabilization of the transcriptional factor HIF-2α.
Although the mechanisms involved are not totally understood, deregulation of DNA methylation is thought to contribute to genomic instability in tumors. Global hypomethylation of CpG dinucleotides is characteristic of many cancers and there is evidence for reactivation of a number of different types of HERVs in a variety of cancer histologies due to the liberation of their LTRs from epigenetic constraints (Florl et al., 1999; Menendez et al., 2004; Lavie et al., 2005; Szpakowski et al., 2009; Gimenez et al., 2010). However, limited data exists on the factors controlling locus-specific transcription of HERVs that are selectively expressed in specific histological subtypes of malignancies. Hsiao et al. recently characterized the mechanism through which HERV-K18 transactivation occurs in EBV-infected B lymphocytes (Hsiao et al., 2006, 2009). Our study provides the first insights into the mechanisms accounting for the selective expression of a HERV in a solid tumor, demonstrating that inactivation of a tumor suppressor gene can result in aberrant expression of a highly immunogenic provirus in ccRCC. The transcriptional up-regulation of HERV-E appears to be related to three critical events: (i) VHL inactivation, (ii) HIF-2α overexpression, and (iii) hypomethylation of the HERV-E 5′LTR. This work provides insights needed for research investigating a potential oncogenic role of CT-RCC HERV-E in kidney cancer.
Materials and methods
Tumor samples and cell lines
A total of 67 fresh (primary) ccRCC tumors were analyzed in this study. Forty of these tumors were procured by the Urologic Oncology Branch, NCI from nephrectomy samples obtained from 17 patients with sporadic ccRCC and 23 patients with VHL disease. Twenty seven of 67 fresh tumors were procured from patients with sporadic ccRCC by nephrectomy at the Engelhardt Institute of Molecular Biology in Moscow; these samples were used to analyze CT-RCC HERV-E expression in tumors compared to matched normal kidney tissue, and to correlate the size and stage of the primary tumor with the level of proviral expression. All fresh tumor samples were snap frozen in liquid nitrogen. The histology of tumors was confirmed by reviewing H&E stained slides made from the tumors. Seventeen non ccRCC tumor samples including papillary (n=8), collecting duct (n=2), chromophobe (n=3), and oncocytoma (n=4) tumors were provided by companies ILSbio (Chestertown, MD, USA) and Asterand (Detroit, MI, USA). The 14 human ccRCC cell lines were established from surgically resected tumors procured at the NIH by the Urologic Oncology Branch of the NCI, the Surgery Branch of the NCI or the Hematology Branch of the NHLBI on IRB approved protocols 97-C-0147, 97-H-0196 and 04-H-0012 as described previously (Takahashi et al., 2008).
VHL gene sequencing and promoter methylation analysis
Analysis of VHL was conducted as previously described (Nickerson et al., 2008). PCR amplification was carried out in 50 μL reactions with ~15 ng tumor DNA. PCR primer sequences were the following: exon 1 forward 5′-CTACGGAGGTCGACTCGGGAG-3′ and reverse 5′-GGGCTTCAGACCGTGCTATCG-3′; exon 2 forward 5′-CCGTGCCCAGCCACCGGTGTG-3′ and reverse 5′-GGATAACGTGCCTGACATCAG-3′; exon 3 forward 5′-CGTTCCTTGTACTGAGACCCTAG-3′ and reverse 5′-GAACCAGTCCTGTATCTAGATCAAG-3′ primers. PCR reactions were analyzed by electrophoresis in 2% agarose gel. Heteroduplexed PCR samples were analyzed using Surveyor Nuclease (Transgenomic, Omaha, NE, USA) and standard non-denaturing HPLC conditions appropriate for DNA fragment sizing. PCR reactions for sequencing were prepared using the AMPure PCR Purification system (Agencourt Bioscience, Beverly, MA, USA). Sequencing products were purified using Cleanseq reagents (Agencourt Bioscience). A standard protocol was used for bisulfite modification of ~250 ng of tumor DNA (Zymo Research, Irvine, CA). Next primers were designed to amplify both methylated and unmethylated alleles of the VHL promoter: forward 5′-TTAYGGAGGTYGATTYGGGAG-3′ and reverse 5′-RCRATTRCARAARATRACCTR-3′ primer, where R = G/A, and Y = C/T. Nested PCR included 1 μL of a 1:10 dilution of first-round product using following primers: forward 5′ YGGGTGGTTTGGATYG-3′ and reverse 5′-ARTTCACCRARCRCARCA-3′. Primer sequences for PCR of untreated (wild type) DNA were the following: forward 5′-CTACGGAGGTCGACTCGGGAG-3′ and reverse 5′-GCGATTGCAGAAGATGACCTG-3′; primer sequences for nested PCR were forward 5′-CGGGTGGTCTGGATCG-3′ and reverse 5′-AGTTCACCGAGCGCAGCA-3′. PCR products were bi-directionally sequenced. Cytosine positions in CpGs were inspected for thymine or cytosine signals in chromatograms, and scoring was conducted as follows: T only, not methylated; both cytosine and thymine, partially methylated; C only, fully methylated. Tumor samples with at least four methylated CpGs (>36%) were considered as hypermethylated. All analyses were run in duplicate, blinded to VHL mutation status, and with positive (CpGenome Universal Methylated DNA, Millipore, Temecula, CA) and negative (K562 Human Genomic DNA, Promega, Madison, WI, USA) controls.
Plasmids
The VHL expressing plasmid and control vector pReceiver-M02 with neomycin selection marker were obtained from Genecopoeia (Rockville, MD, USA).
Total RNA extraction
Tumor samples frozen in OCT were sliced on a cryotome (Leica CM 3050S) into five to seven 30 μm sections, while flash frozen samples were cut with a scalpel into small pieces approximately 300 mg each. Each sample was placed in 3 ml Trizol (Invitrogen, Carlsbad, CA, USA), homogenized using 7 × 110 mm disposable generators on a GLH homogenizer (Omni International, Kennesaw, GA, USA), and centrifuged at 3000 rpm for 5 min. Chloroform (600 μl) was added to the upper phase and incubated at room temperature for 15 min, followed by centrifugation at 10,000 × g for 15 min at 4°C. To precipitate RNA, 1.5 mL of isopropanol was added to supernatant, incubated at room temperature for 10 min, and centrifuged at 10,000 × g for 10 min. The pellets were washed with 1.4 mL of 70% ethanol and centrifuged at 8500 × g for 5 min at 4 °C. The RNA pellets were dried and suspended in 100 μl of DEPC-treated water. The RNA was treated with Amplification Grade DNase I (Invitrogen) to remove residual genomic DNA. RNA integrity was verified on the Agilent 2100 bioanalyzer using RNA Nano Chips (Agilent Technologies, Santa Clara, CA, USA).
Quantitative RT-PCR
cDNA was synthesized following the protocol of the SuperScript III First-Strand Synthesis System (Invitrogen) using the oligo(dT)20 primer and 1.5 μg total RNA. qRT-PCR using the TaqMan Universal PCR Master Mix was carried out in the 7500 Fast Real-Time PCR System (Applied Biosystems, Austin, TX, USA). Primers specific for HIF-1α and HIF-2α (SABiosciences, Frederick, MD, USA) were used for measuring HIFs expression levels. TaqMan Gene Expression assay for VHL gene (Applied Biosystems) was used to measure expression of all 3 exons of VHL gene. Primers specific for the CT-RCC transcripts were described before (Takahashi et al., 2008). Copies relative to β-actin (× 105) were measured to determine HERV-E expression where less than 20 copies was designated “no expression,” 20–100 copies as “low level”, and greater than 100 copies as “high level”.
siRNA transfection
siRNA targeting HIF-2α and control siRNA were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Cells were transfected with siRNA using transfection reagent and medium provided by Santa Cruz Biotechnology according to the manufacturer’s directions. After 3 days cells were lysed and analyzed by western blot to verify knockdown of HIF-2α.
Quantitative CpG methylation analysis
Quantitative CpG methylation analysis of 5′LTR of HERV-E (Human HERV AL133408: nt 79153–79409) was provided by EpigenDx (Worcester, MA, USA). The following primers were used for PCR of target region: forward primer 5′-GAGGTGATTAGTGGATAGTTGAGG-3′; biotin labeled reverse primer 5′-CCAAAACCAATCTTACCATATTCC-3′. HotStar Taq Polymerase kit (Qiagen, Valencia, CA) was used for PCR with following cycling conditions: 95°C 15 min; 45 × (95°C 30 s; 64°C 30 s; 72°C 30 s); 72°C 5 min. The PSQ™96HS system was used for pyrosequencing analysis according to standard procedures. Following forward primers were used for pyrosequencing: 5′-GGATAGTTGAGGTAGTTTTTTA-3′; 5′-AAGTAAAGTAGATTTTGAGAGT-3′; and 5′-TTGGTTAAATGTTTTGTTAT-3′. 500 ng of DNA and EZ DNA methylation kit (Zymo Research, Orange, CA, USA) were used for bisulfite modification. Low methylated DNA control and in vitro methylated DNA were mixed at different ratios followed by bisulfite modification, PCR, and Pyrosequencing analysis to perform PCR Bias Testing.
Chromatin immunoprecipitation analysis
Cells were grown up to 80–90% confluence before being cross-linked with 11% formaldehyde; extracted chromatin was then sonicated. ChIP assay was performed using Abcam ChIP kit and HIF-2α antibodies according to protocol provided by the company (Abcam Inc, Cambridge, MA, USA). The DNA regions of interest were analyzed by PCR and confirmed by sequencing. 100 ng of DNA and Platinum PCR SuperMix High Fidelity kit (Invitrogen) were used for PCR with following conditions: 95°C 5 min; 25–45 cycles series × (95°C 30 s; 56°C 30 s; 68°C 30 s); 68°C 5 min. Next primers targeting HRE in 5′LTR of the CT-RCC HERV-E were designed: forward 5′-GGAGCACTCTCGCGTAGG-3′, reverse 5′-GATCAGCCCACACTTCCACT-3′. PCR products were analyzed by 2% agarose gel electrophoresis.
DAC and DP treatment of cells
5-aza-2′ deoxycytidine (DAC) or/and depsipeptide (DP) were used along or in combination to treat cell lines. Cell lines with a media containing DAC (0.1uM) were cultured for 72 hrs in a CO2 incubator, followed by incubation in normal media for an additional 24 hrs. Cell lines with media containing DP (25ng/mL) were cultured for 6 hours in a CO2 incubator, followed by incubation in normal media for 18 hours. Combinational DAC/DP treatment was started with DAC treatment for 72 hours, followed by DP treatment for 6 hours, followed by incubation in normal media for 18 hours.
Acknowledgments
We thank Robert Worrell for his assistance in providing tissue samples. This work was supported by the intramural research program of NIH, National Heart, Lung, and Blood Institute, Hematology Branch. We wish to acknowledge ACKC (Action to Cure Kidney Cancer) for their support of the ACKC fellow who conducted this research and The Dean R. O’Neill Memorial Fellowship for generous contributions supporting this research. We also thank Bill and Giuliana Rancic for their contributions supporting kidney cancer research.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Bregni M, Dodero A, Peccatori J, Pescarollo A, Bernardi M, Sassi I, et al. Nonmyeloablative conditioning followed by hematopoietic cell allografting and donor lymphocyte infusions for patients with metastatic renal and breast cancer. Blood. 2002;99:4234–4236. doi: 10.1182/blood.v99.11.4234. [DOI] [PubMed] [Google Scholar]
- Childs R, Chernoff A, Contentin N, Bahceci E, Schrump D, Leitman S, et al. Regression of metastatic renal-cell carcinoma after nonmyeloablative allogeneic peripheral-blood stem-cell transplantation. N Engl J Med. 2000;343:750–758. doi: 10.1056/NEJM200009143431101. [DOI] [PubMed] [Google Scholar]
- Cohen HT, McGovern FJ. Renal-cell carcinoma. N Engl J Med. 2005;353:2477–2490. doi: 10.1056/NEJMra043172. [DOI] [PubMed] [Google Scholar]
- Cohen C, Lock WM, Mager DL. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene. 2009;448:105–114. doi: 10.1016/j.gene.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Florl A, Lower R, Schmitz-Drager BJ, Schulz WA. DNA methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. Br J Cancer. 1999;80:1312–1321. doi: 10.1038/sj.bjc.6690524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Shanafelt T, Cimmino A, Taccioli C, Volinia S, Liu C, et al. Aberrant regulation of pVHL levels by microRNA promotes the HIF/VEGF axis in CLL B cells. Blood. 2009;113:5568–5574. doi: 10.1182/blood-2008-10-185686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez J, Montgiraud C, Pichon JP, Bonnaud B, Arsac M, Ruel K, et al. Custom human endogenous retroviruses dedicated microarray identifies self-induced HERV-W family elements reactivated in testicular cancer upon methylation control. Nucleic Acid Res. 2010;38:2229–2246. doi: 10.1093/nar/gkp1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnarra JR, Zhou S, Merrill MJ, Wagner JR, Krumm A, Papavassiliou E, et al. Post-transcriptional regulation of vascular endothelial growth factor mRNA by the product of the VHL tumor suppressor gene. Proc Natl Acad Sci USA. 1996;93:10589–10594. doi: 10.1073/pnas.93.20.10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S, Ugurel S, Hanschmann KM, Strobel H, Tondera C, Schadendorf D, et al. Serological response to human endogenous retrovirus K in melanoma patients correlates with survival probability. AIDS Research and Human Retroviruses. 2008;24:717–723. doi: 10.1089/aid.2007.0286. [DOI] [PubMed] [Google Scholar]
- Hsiao F, Tai A, Deglon A, Sutkowski N, Longnecker R, Huber B. EBV LMP-2A employs a novel mechanism to transactivate the HERV-K18 superantigen through its ITAM. Virology. 2009;385:261–266. doi: 10.1016/j.virol.2008.11.025. [DOI] [PubMed] [Google Scholar]
- Hsiao F, Lin M, Tai A, Chen G, Huber B. Epstein-Barr virus transactivates the HERV-K18 superantigen by docking to the human complement receptor 2 (CD21) on primary B-cells. J Immunol. 2006;177:2056–2060. doi: 10.4049/jimmunol.177.4.2056. [DOI] [PubMed] [Google Scholar]
- Iliopoulos O, Levy AP, Jiang C, Kaelin WG, Goldberg MA. Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc Natl Acad Sci USA. 1996;93:10595–10599. doi: 10.1073/pnas.93.20.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG. Treatment of kidney cancer: insights provided by the VHL tumor-suppressor protein. Cancer. 2009;115:2262–2272. doi: 10.1002/cncr.24232. [DOI] [PubMed] [Google Scholar]
- Kim CM, Vocke CD, Torres-Cabala C, Yang Y, Schmidt LS, Walter MM, et al. Expression of hypoxia inducible factor-1alpha and 2alpha in genetically distinct early renal cortical tumors. J Urol. 2006;175:1908–1914. doi: 10.1016/S0022-5347(05)00890-6. [DOI] [PubMed] [Google Scholar]
- Krieg M, Haas R, Brauch H, Acker T, Flamme I, Plate KH. Up-regulation of hypoxia-inducible factors HIF-1alpha and HIF-2alpha under normoxic conditions in renal carcinoma cells by von Hippel-Lindau tumor suppressor gene loss of function. Oncogene. 2000;19:5435–5443. doi: 10.1038/sj.onc.1203938. [DOI] [PubMed] [Google Scholar]
- Lavie L, Kitova M, Maldener E, Meese E, Mayer J. CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K (HML-2) J Virol. 2005;79:876–883. doi: 10.1128/JVI.79.2.876-883.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, et al. The tumor suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Menendez L, Benigno BB, McDonald JF. LI and HERV-W retrotransposons are hypomethylated in human ovarian carcinomas. Mol Cancer. 2004;3:12–17. doi: 10.1186/1476-4598-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson M, Jaeger E, Shi Y, Durocher J, Mahurkar S, Zaridze D, et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clinical Cancer Res. 2008;14:4726–4734. doi: 10.1158/1078-0432.CCR-07-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Abdul T, Kurlander R, Harashima N, Lundqvist A, Hong J, et al. The human endogenous retrovirus (HERV) derived kidney cancer antigen CT-RCC1 induces proliferation of CD8+ antigen-specific T-cells in vitro that kill renal cell carcinoma (RCC) and is up-regulated by inhibiting histone deacetylase. Proceedings of the 99th Annual Meeting of the American Association for Cancer Research; San Diego, CA. 2008. [Google Scholar]
- Rini BI, Zimmerman T, Stadler WM, Gajewski TF, Vogelzang NJ. Allogeneic stem-cell transplantation of renal cell cancer after nonmyeloablative chemotherapy: feasibility, engraftment, and clinical results. J Clin Oncol. 2002;20:2017–2024. doi: 10.1200/JCO.2002.08.068. [DOI] [PubMed] [Google Scholar]
- Romanish MT, Cohen CJ, Mager DL. Potential mechanisms of endogenous retroviral-mediated genomic instability in human cancer. Semin Cancer Biol. 2010;20:246–253. doi: 10.1016/j.semcancer.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Ruprecht K, Mayer J, Sauter M, Roemer K, Mueller-Lantzsch N. Endogenous retroviruses and cancer. Cell Mol Life Sci. 2008;65:3366–3382. doi: 10.1007/s00018-008-8496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandlund J, Ljungberg B, Wikstrom P, Grankvist K, Lindh G, Rasmuson T. Hypoxia-inducible factor-2α mRNA expression in human renal cell carcinoma. Acta Oncol. 2009;48:909–914. doi: 10.1080/02841860902824891. [DOI] [PubMed] [Google Scholar]
- Szpakowski S, Sun X, Lage JM, Dyer A, Rubinstein J, Kowalski D, et al. Loss of epigenetic silencing in tumors preferentially affects primate-specific retroelements. Gene. 2009;448:151–167. doi: 10.1016/j.gene.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Harashima N, Kajigaya S, Yokoyama H, Cherkasova E, McCoy J, et al. Regression of kidney cancer following allogeneic stem-cell transplantation associated with T-cells recognizing a HERV-E antigen. J Clin Invest. 2008;118:1099–1109. doi: 10.1172/JCI34409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner KJ, Moore JW, Jones A, Taylor CF, Cuthbert-Heavens D, Han C, et al. Expression of hypoxia-inducible factors in human renal cell cancer: relationship to angiogenesis and to the von Hippel-Lindau gene mutation. Cancer Res. 2002;62:2957–2961. [PubMed] [Google Scholar]
- Wang-Johanning F, Radvanyi L, Rycaj K, Plummer J, Yan P, Sastry KJ, et al. Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res. 2008;68:5869–5877. doi: 10.1158/0008-5472.CAN-07-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger RH, Kvietikova I, Rolfs A, Camenisch G, Gassmann M. Oxygen-regulated erythropoietin gene expression is dependent on a CpG methylation-free hypoxia-inducible factor-1 DNA-binding site. Eur J Biochem. 1998;253:771–777. doi: 10.1046/j.1432-1327.1998.2530771.x. [DOI] [PubMed] [Google Scholar]
- Wright TM, Rathmell WK. Identification of Ror2 as a hypoxia-inducible factor target in von Hippel-Lindau-associated renal cell carcinoma. J Biol Chem. 2010;285:12916–12924. doi: 10.1074/jbc.M109.073924. [DOI] [PMC free article] [PubMed] [Google Scholar]