Abstract
Retinoic acid (RA) is the active metabolite of vitamin A (retinol) that controls growth and development. The first step of RA synthesis is controlled by enzymes of the alcohol dehydrogenase (ADH) and retinol dehydrogenase (RDH) families that catalyze oxidation of retinol to retinaldehyde. The second step of RA synthesis is controlled by members of the aldehyde dehydrogenase (ALDH) family also known as retinaldehyde dehydrogenase (RALDH) that further oxidize retinaldehyde to produce RA. RA functions as a ligand for DNA-binding RA receptors that directly regulate transcription of specific target genes. Elucidation of the vitamin A metabolic pathway and investigation of the endogenous function of vitamin A metabolites has been greatly improved by development of mouse ADH, RDH, and RALDH loss-of-function models. ADH knockouts have demonstrated a postnatal role for this enzyme family in clearance of excess retinol to prevent vitamin A toxicity and in generation of RA for postnatal survival during vitamin A deficiency. A point mutation in Rdh10 generated by ethylnitrosourea has demonstrated that RDH10 generates much of the retinaldehyde needed for RA synthesis during embryonic development. Raldh1, Raldh2, and Raldh3 knockouts have demonstrated that RALDH1, RALDH2, and RALDH3 generate most of the RA needed during embryogenesis. These mouse models serve as instrumental tools for providing new insight into retinoid function.
Keywords: Retinol, Retinaldehyde, Retinoic acid, Alcohol dehydrogenase, Retinol dehydrogenase, Aldehyde dehydrogenase, Retinaldehyde dehydrogenase, Retinoid metabolism
1. Introduction
Among the various diffusible cell-cell signaling factors that naturally direct developmental processes, retinoic acid (RA) is unique in that it is a small lipophilic molecule (MW = 300) derived from vitamin A that directly regulates gene expression. Over the years many lessons have been learned about the developmental roles of RA signaling from studies on vitamin A deficiency [1–3] and RA receptor null mice [4, 5]. In addition, recent studies using model organisms carrying genetic defects in enzymes that convert vitamin A to RA (either in the whole embryo or in specific tissues) have provided great insight into the mechanism of RA signaling during development. Such studies are the result of many years of research on retinoid-metabolizing enzymes which finally led to the discovery of enzymes necessary for RA synthesis in vivo [6].
The retinoid metabolic pathway provides mechanisms to store vitamin A, to generate RA for signaling processes, and to degrade excess RA (Fig. 1). Cleavage of beta-carotene (provitamin A) by carotenoid-15,15′-oxygenase (CMO1) generates all-trans-retinaldehyde which can be reduced to all-trans-retinol for storage as vitamin A [7, 8]. For long-term storage of vitamin A, all-trans-retinol is esterified with a fatty acid to form retinyl esters that are stored as lipid droplets; this reaction is catalyzed by lecithin-retinol acyltransferase (LRAT) with the help of cellular retinol-binding protein-1 (CRBP1) [9–11]. Hydrolases that remove the ester group to regenerate retinol have been identified [12, 13].
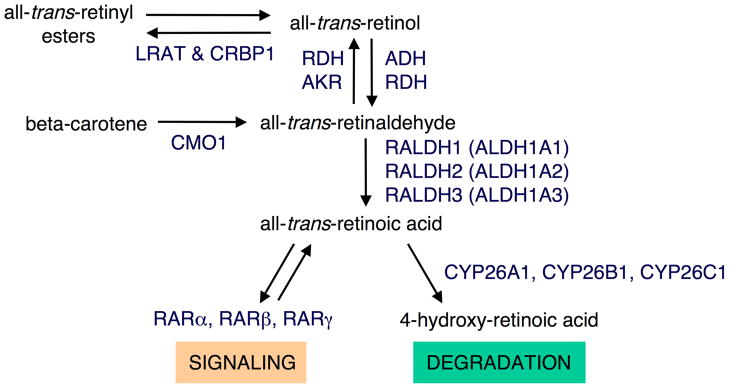
Fig. 1.
Retinoid metabolic pathway. All-trans-retinol (the alcohol form of vitamin A) can be converted to retinyl esters for storage through the action lecithin-retinol acyltransferase (LRAT) and cellular retinol-binding protein-1 (CRBP1). Alternatively, all-trans-retinol can be oxidized to all-trans-retinaldehyde by either alcohol dehydrogenase (ADH) or retinol dehydrogenase (RDH) using NAD as cofactor. All-trans-retinaldehyde can also be generated by cleavage of beta-carotene (provitamin A) by carotenoid-15,15′-oxygenase (CMO1). All-trans-retinaldehyde can be reduced to all-trans-retinol through the action of RDHs that preferentially use NADPH as cofactor or by aldo-keto reductases (AKR). All-trans-retinaldehyde can be further oxidized to all-trans-retinoic acid (RA) by retinaldehyde dehydrogenases (RALDH1, RALDH2, and RALDH3) which are members of the aldehyde dehydrogenase family (i.e. ALDH1A1, ALDH1A2, and ALDH1A3, respectively). RA can be further oxidized to 4-hydroxy-retinoic acid by cytochrome P450s (CYP26A1, CYP26B1, and CYP26C1) which is considered the first step of RA degradation as it leads to more easily excreted metabolites. RA can initiate a signaling event through binding to nuclear RA receptors (RARα, RARβ, and RARγ) that regulate transcription of target genes.
Mobilization of vitamin A for RA synthesis is a two-step metabolic process in which retinol is first oxidized to retinaldehyde in a reversible reaction, followed by an irreversible second oxidation which metabolizes retinaldehyde to RA. Several enzymes have been linked to physiological roles in RA synthesis through genetic loss-of-function studies. In the first step, retinol is oxidized to retinaldehyde by either alcohol dehydrogenases (ADH1, ADH3, and ADH4) [14, 15] or retinol dehydrogenases that are members of the short-chain dehydrogenase/reductase family (RDH1 and RDH10) [16, 17] (Table 1). Due to the reversible nature of the first step, retinaldehyde can be reduced back to retinol through the action of some RDHs such as RDH12 or by some aldo-keto reductases such as AKR1B10 [18–20]. In zebrafish, a retinol dehydrogenase encoded by rdh1l (homolog of mouse Dhrs9) was identified that is required for oxidation of retinol to retinaldehyde for RA synthesis in certain tissues such as intestine [21, 22]. In Xenopus, an rdh10 homolog has been identified which is required for RA synthesis in the hindbrain and other tissues [23]. Also in zebrafish, an rdh10 homolog has been found which presumably oxidizes retinol to retinaldehyde, and another retinol dehydrogenase encoded by dhrs3a was identified that reduces retinaldehyde to retinol [24]. Retinaldehyde can be further oxidized to retinoic acid (RA) by retinaldehyde dehydrogenases (RALDH1, RALDH2, and RALDH3) which are members of the aldehyde dehydrogenase family (i.e. ALDH1A1, ALDH1A2, and ALDH1A3, respectively) [25–29]. In Xenopus, a raldh2 homolog has been identified which generates RA in early embryos [30]. In zebrafish, a raldh2 homolog has been identified that is required for early embryonic RA synthesis [31, 32] and an raldh3 homolog has been found but raldh1 is absent [33, 34].
Table 1.
Mouse mutants for enzymes catalyzing conversion of retinol to retinoic acid (RA).
| Gene | Official gene name | Enzyme name and activity with retinoids | Major defects observed in null mutants |
|---|---|---|---|
| Adh1 | Adh1 | class I alcohol dehydrogenase oxidation of retinol to retinaldehyde | non-lethal; adults fertile; hypersensitive to vitamin A toxicity due to reduced ability to clear excess retinol; increased retinyl esters |
| Adh3 | Adh5 | class III alcohol dehydrogenase oxidation of retinol to retinaldehyde | non-lethal; adults fertile; reduced postnatal survival during vitamin A deficiency |
| Adh4 | Adh7 | class IV alcohol dehydrogenase oxidation of retinol to retinaldehyde | non-lethal; adults fertile; reduced postnatal survival during vitamin A deficiency |
| Rdh1 | Rdh1 | retinol dehydrogenase-1 oxidation of retinol to retinaldehyde | non-lethal; adults fertile; increased body weight, adipose tissue, and retinyl ester levels |
| Rdh10 | Rdh10 | retinol dehydrogenase-10 oxidation of retinol to retinaldehyde | lethal at E13.5; craniofacial defects; small forelimbs |
| Raldh1 | Aldh1a1 | retinaldehyde dehydrogenase-1 oxidation of retinaldehyde to RA | non-lethal; adults fertile; hypersensitive to vitamin A toxicity due to reduced ability to clear excess retinaldehyde; protects against obesity in adult |
| Raldh2 | Aldh1a2 | retinaldehyde dehydrogenase-2 oxidation of retinaldehyde to RA | lethal at E9.5; failure in embryonic turning due to somite defect; enlarged heart; forelimb field absent; posterior hindbrain absent; posterior foregut absent |
| Raldh3 | Aldh1a3 | retinaldehyde dehydrogenase-3 oxidation of retinaldehyde to RA | lethal at birth; blockage of nasal passages; mild perioptic mesenchyme defect (more severe in Raldh1;Raldh3 double knockout); forebrain LGE/striatum defects |
Further oxidation of RA to form 4-hydroxy-RA and 4-oxo-RA is carried out by cytochrome P450s CYP26A1 [35–37], CYP26B1 [38], and CYP26C1 [39]. This reaction is considered the first step of RA degradation as it adds a hydrophilic group which results in the formation of RA metabolites that are more easily excreted than RA itself [40]. Although polar metabolites such as 4-oxo-RA have been found to act similarly to RA as a teratogen when introduced to embryos [41], they are unnecessary for endogenous RA signaling [42]. The unique tissue-specific expression patterns of Cyp26 genes influence where RA signaling is able to occur and provides some level of protection against teratogenic or toxic exposure to retinoids as demonstrated by Cyp26 gene knockout studies [43].
The effect of RA signaling on gene regulation is due to its ability to enter the nucleus and activate DNA-bound RA receptors (RARα, RARβ, and RARγ) which directly regulate specific genes via effects on transcriptional co-repressors and co-activators [44]. Each RAR binds to DNA as a heterodimer with one of the retinoid X receptors (RXRα, RXRβ, and RXRγ) [45]. RARs bind the abundant form of RA known as all-trans-RA as well as a very low-abundance isomer known as 9-cis-RA, whereas RXRs bind only 9-cis-RA [46]. However, 9-cis-RA is undetectable [47–51] except under conditions of vitamin A excess [52, 53]. Thus, it appears that 9-cis-RA plays a pharmacological but not physiological role, suggesting that RXR has a different physiological ligand or functions without a ligand. The overall function of RXR transcends its role in retinoid signaling as it forms heterodimers not only with RAR but also with at least ten additional nuclear receptors including thyroid hormone receptor (TR), vitamin D receptor (VDR), peroxisome-proliferator activated receptor (PPAR), and liver X receptor (LXR), demonstrating that RXR is involved in many different signaling pathways [54]. In the case of signaling through RA, thyroid hormone, or vitamin D, in vitro studies have shown that RAR-RXR, TR-RXR, and VDR-RXR heterodimers are stimulated by all-trans-RA, thyroid hormone, or vitamin D3, respectively, but not by RXR ligands due to allosteric RXR subordination which presumably prevents signaling pathway promiscuity [55–58]. Also, binding of a retinoid ligand to RXR is unnecessary for RA signaling in vivo [53, 59]. The polyunsaturated fatty acid docosahexaenoic acid present at high levels in brain has been found to function as an RXR ligand in vitro, but its relevance in vivo is unclear [60]. Thus, whereas RXR is known to facilitate DNA-binding of its heterodimer partners, whether RXR has a physiological ligand required for normal development or adult homeostasis remains unknown [61].
2. Alcohol dehydrogenase and retinol dehydrogenase loss-of function models
All cells have access to retinol via the circulatory system, but the ability of various cell types to oxidize retinol to retinaldehyde varies greatly due to the expression levels of specific retinol-metabolizing enzymes. Mouse genetic loss-of-function studies have led to the identification of several enzymes that play a role in oxidation of retinol to retinaldehyde for RA synthesis (Table 1).
2.1. Adh1 knockout
Class I ADH (ADH1) is expressed in many epithelial tissues and at very high levels in adult liver where it serves as an important enzyme in the clearance of toxic alcohols including ethanol [62, 63]. Adh1−/− mice exhibit normal survival to adulthood and normal fertility, and when maintained on a vitamin A deficient diet during development they display a similar degree of survival compared to wild-type [14]. However, Adh1−/− mice administered a dose of retinol exhibit a marked reduction in metabolism of retinol to RA compared to wild-type, and this failure to clear retinol results in a significant increase in vitamin A toxicity as measured by a reduced LD50 value for retinol and by increased release of cytosolic liver proteins into the serum indicating hepatic cell death [14, 64]. Adh1−/− mice have thus revealed that metabolism of excess retinol to RA through ADH1 is an important mechanism to avoid retinol toxicity in adult tissues, even though this pathway may still generate enough RA to be teratogenic if the mouse is pregnant. Evidently there is less toxicity to adult tissues when retinol is metabolized to RA through the NAD-dependent ADH1 pathway than when retinol is metabolized by other mechanisms such as hydroxylation by cytochrome P450s which can generate reactive oxygen species or other toxic byproducts. Also, other metabolic routes are less efficient allowing high levels of retinol to accumulate in various tissues which likely disrupts many cellular processes through non-specific binding of retinol. This mechanism may be important not only pharmacologically for individuals taking vitamin A supplements, but also physiologically as some foods such as liver contain high amounts of vitamin A which can be toxic if consumed in high quantities.
As liver is the major site of detoxification, the expression patterns of ADHs and their relative activities with retinol provide further evidence that ADH1 has an important function in prevention of retinol toxicity. Thus, the physiological findings on vitamin A toxicity obtained in Adh1−/− mice correlate with the observation that ADH1 exhibits high retinol activity and is found at much higher levels in adult liver than ADH3 which exhibits low retinol activity, and the observation that ADH4 (also with high retinol activity) is not expressed in adult liver [15, 62].
An interesting relationship exists between cellular retinol-binding protein-1 (CRBP1) and ADH1. Crbp1−/− mice are very sensitive to vitamin A deficiency due to greatly reduced stores of liver retinyl esters [9]. Adh1−/− mice have higher than normal levels of liver retinyl esters due to reduced oxidation of retinol, and Crbp1−/−;Adh1−/− double mutant mice have relatively normal levels of liver retinyl esters and reduced sensitivity to vitamin A deficiency [65]. Thus, ADH1 and CRBP1 exhibit opposing roles in the liver, with ADH1 ensuring that excess dietary retinol is metabolized to prevent vitamin A toxicity while CRBP1 ensures that a large amount of retinol is stored as retinyl esters to prevent vitamin A deficiency.
2.2. Adh3 knockout
Class III ADH (ADH3) is expressed ubiquitously from embryogenesis to adulthood [62]. Adh3−/− mice maintained on a normal diet survive to adulthood and are fertile, but they exhibit 15% postnatal lethality and a growth deficiency resulting in adult body weights 30% lower than normal [15]. Dietary retinol supplementation can rescue growth deficiency in Adh3−/−, suggesting that other enzymes can replace ADH3 retinol activity. Although ADH1 and ADH4 in vitro retinol oxidation activities are about 1000 times higher than that of ADH3 [15], the ability of ADH3 to contribute to retinol oxidation in vivo may be due to its ubiquitous expression.
A physiological role for ADH3 in retinol oxidation is also supported by studies showing that developmental vitamin A deficiency conditions that result in 40% postnatal lethality in wild-type mice result in 100% postnatal lethality in Adh3−/− mice [15]. ADH3 also participates in prevention of retinol toxicity likely due to its ubiquitous expression. Adh3−/− mice display reduced metabolism of a dose of retinol to RA and a lower LD50 value for retinol, although these effects are smaller than those observed for Adh1−/− mice [64].
2.3. Adh4 knockout
Class IV ADH (ADH4) is expressed in several epithelial tissues with the notable exception of the liver [62]. Adh4−/− mice survive to adulthood, display normal fertility, and do not exhibit defects in growth or survival when maintained on a normal diet [14, 66]. However, Adh4−/− mice exhibit 100% postnatal lethality when placed on a vitamin A deficient diet under conditions where wild-type exhibits 40% postnatal lethality [14, 66]. As mentioned above, Adh1−/− mice do not show a reduction in postnatal survival on a vitamin A deficient diet, and Adh1−/−;Adh4−/− double mutants exhibit similar survival during vitamin A deficiency compared to Adh4−/− mice [14]. Thus, ADH3 and ADH4 play overlapping roles in retinol oxidation for RA synthesis during vitamin A deficiency, but ADH1 does not share this function. As vitamin A deficiency is likely a common occurrence, especially for terrestrial vertebrates in temperate climates, this function of ADH3 and ADH4 is likely to be an important one for species survival.
ADH4 does not play a significant role in oxidation of excess retinol to prevent retinol toxicity. Following administration of a toxic dose of retinol, Adh4−/− mice metabolize retinol similarly to wild-type whereas Adh1−/− and Adh1−/−;Adh4−/− double mutant mice both exhibit a 10-fold decrease in RA production; also LD50 studies indicate only a small increase in acute retinol toxicity for Adh4−/− mice [14].
Overall, studies on ADH knockout mice indicate that ADH1 provides considerable protection against vitamin A toxicity whereas ADH4 promotes survival during vitamin A deficiency, demonstrating largely non-overlapping physiological functions for these two enzymes in retinol metabolism. ADH3 appears to function redundantly both in prevention of retinol toxicity and vitamin A deficiency, a role which is consistent with its ubiquitous expression.
2.4. Rdh1 knockout
RDH1 was the first member of the short-chain dehydrogenase/reductase family found to metabolize retinol to retinaldehyde [67]. The short-chain dehydrogenase/reductase enzyme family is distinct from that of the ADH family, with a different overall structure and enzyme mechanism [68]. Mice carrying a null mutation in Rdh1 survive to adulthood and are fertile, but adult mice were found to have increased size and adiposity [16]. Rdh1−/− mice exhibit altered vitamin A homeostasis, with increased retinyl ester levels in liver and kidney [16]. Thus, retinol metabolism by both RDH1 and ADH1 functions to maintain normal levels of retinyl esters in the liver.
2.5. Rdh10 mutant derived by ethylnitrosourea mutagenesis
A forward genetic screen using ethylnitrosourea-induced mutagenesis resulted in the identification of a mouse strain named trex which displays embryonic lethality at 13.5 days of embryonic development (E13.5) and exhibits forelimb stunting but normal hindlimbs; trex was found to harbor a missense point mutation in Rdh10 that eliminates retinol dehydrogenase activity [17]. Rdh10 had previously been identified as a member of the short-chain dehydrogenase/reductase family that is highly expressed in the adult eye where it presumably plays a role as a retinol dehydrogenase in the visual cycle [69]. However, it is now clear that Rdh10 is expressed in a tissue-specific fashion in several embryonic tissues including the somitic and lateral plate mesoderm where it overlaps with Raldh2 expression and the eye where it overlaps with Raldh1 and Raldh3 expression [17, 70, 71]. The severe defects observed in Rdh10trex mutants have demonstrated conclusively that tissue-specific control of RA synthesis in mouse can be controlled not only by enzymes catalyzing oxidation of retinaldehyde to RA but also by enzymes catalyzing oxidation of retinol to retinaldehyde. Earlier studies in zebrafish employing morpholino knockdown of rdh1l also demonstrated that enzymes catalyzing retinol oxidation can control tissue-specific RA synthesis [21].
Mouse Rdh10trex mutants carrying the RARE-lacZ RA-reporter transgene [72], which detects sites of RA signaling, demonstrated that most RA activity is lost at E9.5 but some RA activity is retained in the neural tube although at a reduced level compared to wild-type [17]. All RA activity in the eye and craniofacial region is lost in Rdh10trex mutants which display severe eye and craniofacial defects by E12.5 [17]. The RA activity remaining in the neural tube of Rdh10trex mutant embryos may be necessary for survival to E13.5 as Raldh2−/− embryos, which completely lack RA activity from E7.5-E8.5, exhibit lethality by approximately E9.5 [25, 26]. Further studies on the relatively normal hindlimbs of Rdh10trex mutants have demonstrated that mutant hindlimb buds completely lack RA activity from initiation of the hindlimb field at E9.5 until E14.5 demonstrating that RA is unnecessary for limb patterning along either the anteroposterior or proximodistal axes [73]. However, the stunted forelimb in this mutant supports the conclusion that RA plays an early role in forelimb bud initiation, a conclusion also supported by studies using Raldh2−/− embryos [74].
2.6. Rdh10 mutants carrying a deletion of the Adh family: Adh-del;Rdh10 mutants
Mice carrying Adh1, Adh3, Adh4, or Rdh10 loss-of-function mutations have shown that oxidation of retinol to retinaldehyde is controlled in vivo by each of these genes postnatally. Whereas ADH plays a role in RA synthesis postnatally, only Rdh10trex mutants display a serious defect in embryonic RA synthesis resulting in embryonic lethality [17]. As Rdh10trex mutant embryos still maintain a reduced level of RA signaling in the neural tube, additional retinol dehydrogenase activity from another enzyme may account for the RA activity detected, or the Rdh10trex mutant may be a severe hypomorph with residual RDH10 activity. Thus, it is possible that Adh1, Adh3, or Adh4, all expressed in mouse embryos [62], may produce retinaldehyde in embryos for RA synthesis.
Our studies of Adh knockout mice have led to the generation of an Adh-del compound knockout lacking all six ADH genes located close together on chromosome 3 (Fig. 2). The rationale for knocking out all ADH genes is based upon the knowledge that not only ADH1, ADH3, and ADH4 catalyze oxidation of retinol to retinaldehyde [15], but also ADH2 (class II ADH) can perform this reaction [75]; ADH5a or ADH5b (class V ADH) protein cannot be detected in any organs and cannot be generated by in vitro translation, thus this form of ADH may be an unstable protein with no function [76]. In order to remove all potential retinol oxidation by ADHs, a 250 kb deletion was generated using Cre-loxP technology. Homozygous Adh-del knockout mice survive postnatally and are fertile (SK and GD, unpublished).
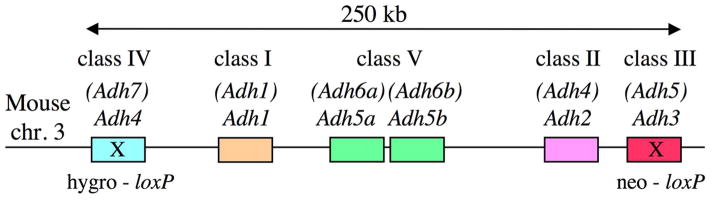
Fig. 2.
Deletion of entire mouse ADH gene family. The Adh-del strain was made possible by the fact that all six ADH genes in the mouse genome are located in a 250 kb stretch on chromosome 3 with no other genes intermixed [111]. Double-targeted embryonic stem (ES) cells were generated that had loxP sites introduced into both Adh4 and Adh3 located on opposite ends of the complex; double-targeted ES cells were selected for resistance to both neomycin (neo) and hygromycin (hygro). Mice generated from double-targeted ES cells were mated to a germ-line Cre to create homozygous Adh-del knockout mice lacking this 250 kb stretch containing the ADH gene family.
After crossing Rdh10trex mutant mice with Adh-del mice, we found that E10.5 homozygous Adh-del;Rdh10 compound mutant embryos appear similar to Rdh10trex mutant embryos, exhibiting similar stunted forelimbs but normal hindlimbs (Fig. 3). The RARE-lacZ RA-reporter transgene was introduced into Adh-del;Rdh10 mutants to monitor RA signaling activity. We found that E10.5 Adh-del;Rdh10 mutants display the same pattern of RA activity as Rdh10trex mutants, particularly loss of RA activity in most mesodermal tissues and eye/nasal pit but retention of RA activity in the neural tube (Fig. 3). Thus, ADH retinol activity is not responsible for the RA activity remaining in Rdh10trex mutants at E10.5. Recent studies have shown that the P450 enzyme CYP1B1 is able to oxidize retinol to retinaldehyde, and Cyp1b1 is expressed in the neural tube of chick and mouse embryos [77, 78]. Thus, RA activity observed in the neural tube of Rdh10trex mutants may be the result of retinol oxidation catalyzed by CYP1B1.
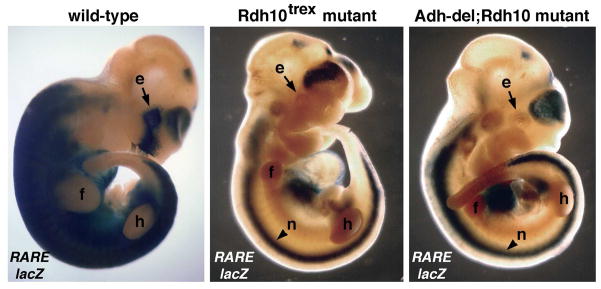
Fig. 3.
Embryogenesis and RA signaling in RDH and ADH mutants. RA activity was visualized in E10.5 embryos carrying the RARE-lacZ RA-reporter transgene. At E10.5, the most obvious phenotype of Rdh10trex mutants is the presence of a stunted forelimb while retaining normal-sized hindlimbs; a very similar phenotype was observed in Adh-del;Rdh10 compound mutant embryos. Both Rdh10trex mutants (n = 4) and Adh-del;Rdh10 compound mutants (n = 4) exhibited loss of most mesodermal RA activity and complete loss of eye/nasal pit RA activity, but retention of neural tube RA activity. These findings demonstrate that the ADH gene family is not responsible for generating retinaldehyde needed for the lower level of RA synthesis still remaining in Rdh10trex mutants. e, eye/nasal pit; f, forelimb bud; h, hindlimb bud; n, neural tube.
3. Aldehyde dehydrogenase loss-of function models
Loss-of-function studies in mice have identified several ALDH enzymes that play a role in oxidation of retinaldehyde to RA (Table 1). Oxidation of retinaldehyde to RA is irreversible and all three RALDHs identified as catalysts for this reaction (RALDH1, RALDH2, and RALDH3) are expressed in non-overlapping dynamic spatiotemporal patterns during development indicating that this step is tissue-restricted and time-restricted [26, 79]. All three RALDHs contribute to physiological generation of RA activity as monitored using a transgenic RA-reporter mouse strain (RARE-lacZ) which contains lacZ linked to a RA response element [72].
3.1. Raldh1 knockout
During mouse embryonic development, Raldh1 expression is first observed at E9.5 in the dorsal retina where it remains until adulthood [27, 80]. Later in development a few other sites of Raldh1 expression exist [79], and by adulthood Raldh1 is expressed in many epithelial tissues and at very high levels in liver similar to ADH1 [81]. Raldh1−/− mice exhibit normal survival to adulthood, they are fertile, and they exhibit no obvious defects [27]. Although Raldh1−/− mutants do not exhibit an ocular phenotype, Raldh1−/−;Raldh3−/− double mutant embryos do exhibit a severe eye defect as described below.
As mentioned above, ADH1 clears excess retinol in adult liver by oxidizing it to retinaldehyde which is further oxidized to RA. Treatment of Raldh1−/− mice with a toxic dose of retinol results in an increase in retinaldehyde in the serum, a large reduction in the appearance of RA, and a reduced LD50 for retinol toxicity [82]. Thus, RALDH1 is the primary enzyme which further oxidizes retinaldehyde to RA in adult liver, demonstrating that ADH1 and RALDH1 cooperate to eliminate excess vitamin A.
Adult Raldh1−/− mice were found to exhibit resistance to diet-induced obesity suggesting that accumulation of retinaldehyde in adipose tissue protects against weight increase [83]. Further studies on Raldh1−/− mice or other retinoid-deficient model organisms may be useful to dissect the underlying mechanism behind this relationship between retinoids and obesity.
3.2. Raldh2 knockout
RARE-lacZ expression has demonstrated that RA signaling activity is first observed in mouse embryos at E7.5 (late primitive streak stage) and RA activity increases to very high levels in the developing trunk from E7.5-E8.5 [72]. This observation suggests that endogenous RA synthesis initiates in the mouse at E7.5. Raldh2 is first expressed at E7.5 in trunk presomitic mesoderm and by E8.5 displays expression in somites and lateral plate mesoderm that appears quite similar to the pattern of RA localization using the RARE-lacZ RA-reporter [26, 84]. Studies on Raldh2−/− mice carrying RARE-lacZ have shown that RALDH2 is responsible for all RA signaling activity seen in the embryo from E7.5 to E8.5 [25, 26]. Loss of all RA signaling at this early stage results in a failure to undergo embryonic turning to achieve the fetal position, a failure to develop beyond E8.75, and lethality by E9.5-E10.5 [25, 26].
Raldh2−/− embryos have been quite useful for elucidating the mechanism of RA signaling during early development as they completely lack RA activity in mesoderm, ectoderm, and endoderm from E7.5 when RA is first detectable until E8.5 (after E8.5 RA is generated in the eye and a few other tissues by Raldh1 and Raldh3). Thus, during early mouse development the only source of RA is the trunk mesoderm which expresses Raldh2. Key to understanding the mechanism has been the observation that RA synthesized in trunk mesoderm is secreted and acts in a paracrine fashion on nearby cells [6]. Raldh2−/− embryos exhibit defects in posterior neuroectodermal tissues including the hindbrain [85, 86] and spinal cord [87]; endodermal tissues including the lung [88, 89] and pancreas [90, 91]; and mesodermal tissues including the heart [92, 93], somites [94, 95], and forelimb bud [74].
Early embryonic lethality in Raldh2−/− embryos can be rescued by maternal dietary RA supplementation at a low dose below the teratogenic range which allows survival to E10.5–E14.5 depending on the treatment plan [26, 92]. Interestingly, RA-rescued Raldh2−/− embryos appear similar to Rdh10 mutants in that they have undergone embryonic turning and they exhibit stunted forelimbs but normal hindlimbs [17, 26, 92]. By treating Raldh2−/− embryos with RA for only a short period to rescue early embryonic lethality (i.e. E7.5–E8.5 or E7.5–E9.5), then examining them several days later when the administered RA has been cleared, one creates a conditional rescue that allows the later functions of Raldh2 to be examined. This method has been used to provide mechanistic insight into how RA controls limb interdigital development [96], kidney formation [97], and lung development [98], and has shown that RA signaling acts in the liver rather than the heart to stimulate ventricular myocardial expansion via an RA-EPO-IGF2 signaling axis [99, 100], and contrary to previous findings Raldh2 mutants have demonstrated that RA is unnecessary for both limb patterning [74] and meiotic initiation in embryonic ovary [101]. Use of these genetic and dietary tools to manipulate RA availability and to visualize the locations of RA signaling, makes it possible to determine which tissues require RA signaling and to examine in detail the mechanism of RA action during organogenesis.
3.3. Raldh3 knockout
Raldh3 expression in mouse is first observed from E8.75–E9.5 in the optic vesicle and nasal placode, then at E10.5 expression is seen in the ventral retina, nasal pit, otic vesicle, and mesonephros, and at E12.5 the ventral telencephalon begins to express Raldh3 [102, 103]. Raldh3−/− mice exhibit lethality at birth due to a blockage of the nasal passages which prevents efficient respiration [28]. Raldh3−/− embryos do not display obvious external deformities and appear relatively healthy until birth. Therefore, Raldh3−/− mice have been useful to examine the mechanism of RA signaling during development of the nasal pit [28], eye [104, 105], forebrain [29], and kidney [97]. Raldh3−/− embryos carrying the RARE-lacZ RA-reporter transgene exhibit a complete loss of nasal pit RA activity and a significant reduction in eye RA activity, but some ocular RA activity remains due to Raldh1 expression in the dorsal retina; these observations are consistent with Raldh3−/− embryos exhibiting a lethal nasal defect but a relatively mild eye defect [28].
Initial studies with Raldh1−/− mice demonstrated no ocular defects, suggesting that RALDH1 is unnecessary for eye development despite its high level of expression in the dorsal retina [27]. However, later studies demonstrated that Raldh1 (expressed in dorsal retina) and Raldh3 (expressed in ventral retina) have redundant roles in RA synthesis needed for embryonic eye development as Raldh1−/−;Raldh3−/− double mutants carrying RARE-lacZ exhibit complete loss of RA activity whereas the single mutants do not [104, 105]. Studies on Raldh1−/−;Raldh3−/− double mutants revealed that RA synthesized in the retina by either RALDH1 or RALDH3 does not control dorsoventral patterning of the retina as originally proposed, but instead RA is secreted and travels to the surrounding neural crest-derived perioptic mesenchyme where it prevents overgrowth of this tissue during formation of the cornea and eyelids [104, 105]. This observation provides a great example of how RA regulates developmental processes using a paracrine signaling pathway. Further studies on Raldh1−/−;Raldh3−/− double mutants demonstrated that RA synthesized in the retina functions in a paracrine fashion on the perioptic mesenchyme to induce Pitx2 [104] which was found to contain a functional RA response element [106]. Interestingly, other studies have shown that Pitx2 induces expression of Dkk2, a Wnt antagonist, in perioptic mesenchyme [107]. Raldh1−/−;Raldh3−/− double mutants were found to have decreased expression of Dkk2 and increased Wnt signaling in the perioptic mesenchyme, suggesting that the ocular defects observed in these double mutants are due to a failure of RA to restrict Wnt signaling in the perioptic mesenchyme [106].
Raldh3−/− mice have also been quite useful to examine the mechanism of RA action during forebrain development. Raldh3 is expressed from E12.5 to birth in the lateral ganglionic eminence (LGE), a neural progenitor zone located in the ventral telencephalon. RARE-lacZ is not expressed in this region of the forebrain in mouse embryos, but RA activity has been detected in the LGE using an in vitro assay in which tissue explants are grown on a lawn of RARE-lacZ RA-reporter cells [108, 109]. Using this assay, Raldh3−/− LGE explants were shown to completely lack RA activity [29]. Raldh3−/− embryos were found to have reduced expression of dopamine receptor DRD2 in a restricted region of the basal ganglia that forms the nucleus accumbens [29]. Raldh3−/− embryos were recently found to lack GABAergic differentiation throughout all LGE-derived striatal projection neurons and interneurons that migrate to the forebrain and olfactory bulb [110]. Further studies using Raldh3−/− mice should shed more light on the normal physiological role of RA signaling in forebrain neural differentiation.
4. Conclusions
A requirement for vitamin A in growth and development has been known for several decades, but we are still actively engaged in research to understand the mechanisms involved. A major clue was revealed with the discovery that the vitamin A metabolite RA directly regulates gene expression by functioning as a ligand for nuclear RA receptors. Since then, there has been a great deal of interest in determining the physiological processes controlled by RA and elucidating the target genes directly controlled by RA. Many conclusions in the retinoid field have been drawn from experiments employing treatment with non-physiological levels of RA or high levels of RA receptor antagonists which can both result in non-specific effects. Thus, a major advantage of the genetic models described here is that they result in loss of endogenous RA, thus providing a method which does not suffer from the non-specific effects of adding RA. Mice lacking the ability to metabolize vitamin A to RA in specific tissues are now a valuable resource for current studies designed to understand the mechanisms related to vitamin A function in mammalian organisms. Studies on RDH and RALDH mutant mice have revealed that RA activity can be reduced or completely eliminated in certain tissues, thus providing very useful genetic loss-of-function models to decipher the mechanism of RA signaling. It is now clear that vitamin A functions mostly (or maybe only) through a paracrine RA signaling pathway rather than an endocrine or autocrine pathway during embryonic development. Using this paradigm, it will be interesting to see if paracrine signaling is also the preferred mechanism used by RA when it acts in postnatal and adult tissues. In order to completely understand the RA signaling pathway, future studies need to be designed to determine the sources of RA synthesis and the actual target tissues/cells that respond to physiological levels of RA, i.e. the tissues/cells where direct RA target genes are expressed. The observations made so far with ADH, RDH, and RALDH mutant mice will thus guide future studies designed to understand vitamin A function in embryonic, postnatal, and adult tissues.
Acknowledgments
Sources of Funding: Research was supported by grants from the National Institutes of Health (GM062848 and RR032045 to GD) and (DE016082 to PAT), and by the Stowers Institute for Medical Research (PAT).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dersch H, Zile MH. Induction of normal cardiovascular development in the vitamin A-deprived quail embryo by natural retinoids. Dev Biol. 1993;160:424–433. doi: 10.1006/dbio.1993.1318. [DOI] [PubMed] [Google Scholar]
- 2.Maden M, Gale E, Kostetskii I, Zile MH. Vitamin A-deficient quail embryos have half a hindbrain and other neural defects. Curr Biol. 1996;6:417–426. doi: 10.1016/s0960-9822(02)00509-2. [DOI] [PubMed] [Google Scholar]
- 3.Clagett-Dame M, DeLuca HF. The role of vitamin A in mammalian reproduction and embryonic development. Annu Rev Nutr. 2002;22:347–381. doi: 10.1146/annurev.nutr.22.010402.102745E. [DOI] [PubMed] [Google Scholar]
- 4.Kastner P, Mark M, Chambon P. Nonsteroid nuclear receptors: What are genetic studies telling us about their role in real life? Cell. 1995;83:859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- 5.Niederreither K, Dolle P. Retinoic acid in development: towards an integrated view. Nature Rev Genet. 2008;9:541–553. doi: 10.1038/nrg2340. [DOI] [PubMed] [Google Scholar]
- 6.Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134:921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lampert JM, Holzschuh J, Hessel S, Driever W, Vogt K, Von Lintig J. Provitamin A conversion to retinal via the β,β-carotene-15,15′-oxygenase (bcox) is essential for pattern formation and differentiation during zebrafish embryogenesis. Development. 2003;130:2173–2186. doi: 10.1242/dev.00437. [DOI] [PubMed] [Google Scholar]
- 8.Hessel S, Eichinger A, Isken A, Amengual J, Hunzelmann S, Hoeller U, Elste V, Hunziker W, Goralczyk R, Oberhauser V, von Lintig J, Wyss A. CMO1 deficiency abolishes vitamin A production from beta-carotene and alters lipid metabolism in mice. J Biol Chem. 2007;282:33553–33561. doi: 10.1074/jbc.M706763200. [DOI] [PubMed] [Google Scholar]
- 9.Ghyselinck NB, Båvik C, Sapin V, Mark M, Bonnier D, Hindelang C, Dierich A, Nilsson CB, Håkansson H, Sauvant P, Azaïs-Braesco V, Frasson M, Picaud S, Chambon P. Cellular retinol-binding protein I is essential for vitamin A homeostasis. EMBO J. 1999;18:4903–4914. doi: 10.1093/emboj/18.18.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batten ML, Imanishi Y, Maeda T, Tu DC, Moise AR, Bronson D, Possin D, Van Gelder RN, Baehr W, Palczewski K. Lecithin-retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver. J Biol Chem. 2004;279:10422–10432. doi: 10.1074/jbc.M312410200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Byrne SM, Wongsiriroj N, Libien J, Vogel S, Goldberg IJ, Baehr W, Palczewski K, Blaner WS. Retinoid absorption and storage is impaired in mice lacking lecithin:retinol acyltransferase (LRAT) J Biol Chem. 2005;280:35647–35657. doi: 10.1074/jbc.M507924200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison EH. Enzymes catalysing the hydrolysis of retinyl esters. Biochim Biophys Acta Lipids Lipid Metab. 1993;1170:99–108. doi: 10.1016/0005-2760(93)90058-h. [DOI] [PubMed] [Google Scholar]
- 13.Strom K, Gundersen TE, Hansson O, Lucas S, Fernandez C, Blomhoff R, Holm C. Hormone-sensitive lipase (HSL) is also a retinyl ester hydrolase: evidence from mice lacking HSL. FASEB J. 2009;23:2307–2316. doi: 10.1096/fj.08-120923. [DOI] [PubMed] [Google Scholar]
- 14.Molotkov A, Deltour L, Foglio MH, Cuenca AE, Duester G. Distinct retinoid metabolic functions for alcohol dehydrogenase genes Adh1 and Adh4 in protection against vitamin A toxicity or deficiency revealed in double null mutant mice. J Biol Chem. 2002;277:13804–13811. doi: 10.1074/jbc.M112039200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molotkov A, Fan X, Deltour L, Foglio MH, Martras S, Farrés J, Parés X, Duester G. Stimulation of retinoic acid production and growth by ubiquitously-expressed alcohol dehydrogenase Adh3. Proc Natl Acad Sci USA. 2002;99:5337–5342. doi: 10.1073/pnas.082093299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M, Hu P, Krois CR, Kane MA, Napoli JL. Altered vitamin A homeostasis and increased size and adiposity in the rdh1-null mouse. FASEB J. 2007;21:2886–2896. doi: 10.1096/fj.06-7964com. [DOI] [PubMed] [Google Scholar]
- 17.Sandell LL, Sanderson BW, Moiseyev G, Johnson T, Mushegian A, Young K, Rey JP, Ma JX, Staehling-Hampton K, Trainor PA. RDH10 is essential for synthesis of embryonic retinoic acid and is required for limb, craniofacial, and organ development. Genes Dev. 2007;21:1113–1124. doi: 10.1101/gad.1533407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallego O, Belyaeva OV, Porte S, Ruiz FX, Stetsenko AV, Shabrova EV, Kostereva NV, Farres J, Pares X, Kedishvili NY. Comparative functional analysis of human medium-chain dehydrogenases, short-chain dehydrogenases/reductases and aldo-keto reductases with retinoids. Biochem J. 2006;399:101–109. doi: 10.1042/BJ20051988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SA, Belyaeva OV, Popov IK, Kedishvili NY. Overproduction of Bioactive Retinoic Acid in Cells Expressing Disease-associated Mutants of Retinol Dehydrogenase 12. J Biol Chem. 2007;282:35621–35628. doi: 10.1074/jbc.M706372200. [DOI] [PubMed] [Google Scholar]
- 20.Gallego O, Ruiz FX, Ardevol A, Dominguez M, Alvarez R, de Lera AR, Rovira C, Farres J, Fita I, Pares X. Structural basis for the high all-trans-retinaldehyde reductase activity of the tumor marker AKR1B10. Proc Natl Acad Sci USA. 2007;104:20764–20769. doi: 10.1073/pnas.0705659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nadauld LD, Sandoval IT, Chidester S, Yost HJ, Jones DA. Adenomatous polyposis coli control of retinoic acid biosynthesis is critical for zebrafish intestinal development and differentiation. J Biol Chem. 2004;279:51581–51589. doi: 10.1074/jbc.M408830200. [DOI] [PubMed] [Google Scholar]
- 22.Rai K, Sarkar S, Broadbent TJ, Voas M, Grossmann KF, Nadauld LD, Dehghanizadeh S, Hagos FT, Li Y, Toth RK, Chidester S, Bahr TM, Johnson WE, Sklow B, Burt R, Cairns BR, Jones DA. DNA demethylase activity maintains intestinal cells in an undifferentiated state following loss of APC. Cell. 2010;142:930–942. doi: 10.1016/j.cell.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strate I, Min TH, Iliev D, Pera EM. Retinol dehydrogenase 10 is a feedback regulator of retinoic acid signalling during axis formation and patterning of the central nervous system. Development. 2009;136:461–472. doi: 10.1242/dev.024901. [DOI] [PubMed] [Google Scholar]
- 24.Feng L, Hernandez RE, Waxman JS, Yelon D, Moens CB. Dhrs3a regulates retinoic acid biosynthesis through a feedback inhibition mechanism. Dev Biol. 2010;338:1–14. doi: 10.1016/j.ydbio.2009.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niederreither K, Subbarayan V, Dollé P, Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nature Genet. 1999;21:444–448. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- 26.Mic FA, Haselbeck RJ, Cuenca AE, Duester G. Novel retinoic acid generating activities in the neural tube and heart identified by conditional rescue of Raldh2 null mutant mice. Development. 2002;129:2271–2282. doi: 10.1242/dev.129.9.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan X, Molotkov A, Manabe S-I, Donmoyer CM, Deltour L, Foglio MH, Cuenca AE, Blaner WS, Lipton SA, Duester G. Targeted disruption of Aldh1a1 (Raldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina. Mol Cell Biol. 2003;23:4637–4648. doi: 10.1128/MCB.23.13.4637-4648.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dupé V, Matt N, Garnier J-M, Chambon P, Mark M, Ghyselinck NB. A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment. Proc Natl Acad Sci USA. 2003;100:14036–14041. doi: 10.1073/pnas.2336223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molotkova N, Molotkov A, Duester G. Role of retinoic acid during forebrain development begins late when Raldh3 generates retinoic acid in the ventral subventricular zone. Dev Biol. 2007;303:601–610. doi: 10.1016/j.ydbio.2006.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen YL, Pollet N, Niehrs C, Pieler T. Increased XRALDH2 activity has a posteriorizing effect on the central nervous system of Xenopus embryos. Mech Dev. 2001;101:91–103. doi: 10.1016/s0925-4773(00)00558-x. [DOI] [PubMed] [Google Scholar]
- 31.Begemann G, Schilling TF, Rauch GJ, Geisler R, Ingham PW. The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development. 2001;128:3081–3094. doi: 10.1242/dev.128.16.3081. [DOI] [PubMed] [Google Scholar]
- 32.Grandel H, Lun K, Rauch GJ, Rhinn M, Piotrowski T, Houart C, Sordino P, Küchler AM, Schulte-Merker S, Geisler R, Holder N, Wilson SW, Brand M. Retinoic acid signalling in the zebrafish embryo is necessary during pre-segmentation stages to pattern the anterior-posterior axis of the CNS and to induce a pectoral fin bud. Development. 2002;129:2851–2865. doi: 10.1242/dev.129.12.2851. [DOI] [PubMed] [Google Scholar]
- 33.Pittlik S, Domingues S, Meyer A, Begemann G. Expression of zebrafish aldh1a3 (raldh3) and absence of aldh1a1 in teleosts. Gene Exp Patt. 2008;8:141–147. doi: 10.1016/j.gep.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 34.Canestro C, Catchen JM, Rodriguez-Mari A, Yokoi H, Postlethwait JH. Consequences of Lineage-Specific Gene Loss on Functional Evolution of Surviving Paralogs: ALDH1A and Retinoic Acid Signaling in Vertebrate Genomes. PLoS Genet. 2009;5:e1000496. doi: 10.1371/journal.pgen.1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White JA, Guo YD, Baetz K, Beckett-Jones B, Bonasoro J, Hsu KE, Dilworth FJ, Jones G, Petkovich M. Identification of the retinoic acid-inducible all-trans-retinoic acid 4-hydroxylase. J Biol Chem. 1996;271:29922–29927. doi: 10.1074/jbc.271.47.29922. [DOI] [PubMed] [Google Scholar]
- 36.Sakai Y, Meno C, Fujii H, Nishino J, Shiratori H, Saijoh Y, Rossant J, Hamada H. The retinoic acid-inactivating enzyme CYP26 is essential for establishing an uneven distribution of retinoic acid along the anterio-posterior axis within the mouse embryo. Genes Dev. 2001;15:213–225. doi: 10.1101/gad.851501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abu-Abed S, Dollé P, Metzger D, Beckett B, Chambon P, Petkovich M. The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures. Genes Dev. 2001;15:226–240. doi: 10.1101/gad.855001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yashiro K, Zhao X, Uehara M, Yamashita K, Nishijima M, Nishino J, Saijoh Y, Sakai Y, Hamada H. Regulation of retinoic acid distribution is required for proximodistal patterning and outgrowth of the developing limb. Dev Cell. 2004;6:411–422. doi: 10.1016/s1534-5807(04)00062-0. [DOI] [PubMed] [Google Scholar]
- 39.Uehara M, Yashiro K, Mamiya S, Nishino J, Chambon P, Dolle P, Sakai Y. CYP26A1 and CYP26C1 cooperatively regulate anterior-posterior patterning of the developing brain and the production of migratory cranial neural crest cells in the mouse. Dev Biol. 2007;302:399–411. doi: 10.1016/j.ydbio.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 40.Frolik CA, Roberts AB, Tavela TE, Roller PP, Newton DL, Sporn MB. Isolation and identification of 4-hydroxy- and 4-oxoretinoic acid. In vitro metabolites of all-trans-retinoic acid in hamster trachea and liver. Biochemistry. 1979;18:2092–2097. doi: 10.1021/bi00577a039. [DOI] [PubMed] [Google Scholar]
- 41.Pijnappel WWM, Hendriks HFJ, Folkers GE, Van den Brink CE, Dekker EJ, Edelenbosch C, Van der Saag PT, Durston AJ. The retinoid ligand 4-oxo-retinoic acid is a highly active modulator of positional specification. Nature. 1993;366:340–344. doi: 10.1038/366340a0. [DOI] [PubMed] [Google Scholar]
- 42.Niederreither K, Abu-Abed S, Schuhbaur B, Petkovich M, Chambon P, Dollé P. Genetic evidence that oxidative derivatives of retinoic acid are not involved in retinoid signaling during mouse development. Nature Genet. 2002;31:84–88. doi: 10.1038/ng876. [DOI] [PubMed] [Google Scholar]
- 43.Pennimpede T, Cameron DA, MacLean GA, Li H, Abu-Abed S, Petkovich M. The role of CYP26 enzymes in defining appropriate retinoic acid exposure during embryogenesis. Birth Defects Res. 2010;88:883–894. doi: 10.1002/bdra.20709. [DOI] [PubMed] [Google Scholar]
- 44.Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 45.Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 46.Heyman RA, Mangelsdorf DJ, Dyck JA, Stein RB, Eichele G, Evans RM, Thaller C. 9-Cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell. 1992;68:397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- 47.Dong D, Zile MH. Endogenous retinoids in the early avian embryo. Biochem Biophys Res Comm. 1995;217:1026–1031. doi: 10.1006/bbrc.1995.2872. [DOI] [PubMed] [Google Scholar]
- 48.Kurlandsky SB, Gamble MV, Ramakrishnan R, Blaner WS. Plasma delivery of retinoic acid to tissues in the rat. J Biol Chem. 1995;270:17850–17857. doi: 10.1074/jbc.270.30.17850. [DOI] [PubMed] [Google Scholar]
- 49.Costaridis P, Horton C, Zeitlinger J, Holder N, Maden M. Endogenous retinoids in the zebrafish embryo and adult. Dev Dyn. 1996;205:41–51. doi: 10.1002/(SICI)1097-0177(199601)205:1<41::AID-AJA4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 50.Ulven SM, Gundersen TE, Weedon MS, Landaas VO, Sakhi AK, Fromm SH, Geronimo BA, Moskaug JO, Blomhoff R. Identification of endogenous retinoids, enzymes, binding proteins, and receptors during early postimplantation development in mouse: Important role of retinal dehydrogenase type 2 in synthesis of all-trans-retinoic acid. Dev Biol. 2000;220:379–391. doi: 10.1006/dbio.2000.9634. [DOI] [PubMed] [Google Scholar]
- 51.Kane MA, Chen N, Sparks S, Napoli JL. Quantification of endogenous retinoic acid in limited biological samples by LC/MS/MS. Biochem J. 2005;388:363–369. doi: 10.1042/BJ20041867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arnhold T, Tzimas G, Wittfoht W, Plonait S, Nau H. Identification of 9-cis-retinoic acid, 9,13-di-cis-retinoic acid, and 14-hydroxy-4,14-retro-retinol in human plasma after liver consumption. Life Sci. 1996;59:169–177. doi: 10.1016/0024-3205(96)00408-0. [DOI] [PubMed] [Google Scholar]
- 53.Mic FA, Molotkov A, Benbrook DM, Duester G. Retinoid activation of retinoic acid receptor but not retinoid X receptor is sufficient to rescue lethal defect in retinoic acid synthesis. Proc Natl Acad Sci USA. 2003;100:7135–7140. doi: 10.1073/pnas.1231422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: Opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 55.Kurokawa R, DiRenzo J, Boehm M, Sugarman J, Gloss B, Rosenfeld MG, Heyman RA, Glass CK. Regulation of retinoid signalling by receptor polarity and allosteric control of ligand binding. Nature. 1994;371:528–531. doi: 10.1038/371528a0. [DOI] [PubMed] [Google Scholar]
- 56.Forman BM, Umesono K, Chen J, Evans RM. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell. 1995;81:541–550. doi: 10.1016/0092-8674(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 57.Vivat V, Zechel C, Wurtz JM, Bourguet W, Kagechika H, Umemiya H, Shudo K, Moras D, Gronemeyer H, Chambon P. A mutation mimicking ligand-induced conformational change yields a constitutive RXR that senses allosteric effects in heterodimers. EMBO J. 1997;16:5697–5709. doi: 10.1093/emboj/16.18.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson PD, Jurutka PW, Haussler CA, Whitfield CK, Haussler MR. Heterodimeric DNA binding by the vitamin D receptor and retinoid X receptors is enhanced by 1,25-dihydroxyvitamin D3 and inhibited by 9-cis-retinoic acid - Evidence for allosteric receptor interactions. J Biol Chem. 1998;273:8483–8491. doi: 10.1074/jbc.273.14.8483. [DOI] [PubMed] [Google Scholar]
- 59.Calleja C, Messaddeq N, Chapellier B, Yang H, Krezel W, Li M, Metzger D, Mascrez B, Ohta K, Kagechika H, Endo Y, Mark M, Ghyselinck NB, Chambon P. Genetic and pharmacological evidence that a retinoic acid cannot be the RXR-activating ligand in mouse epidermis keratinocytes. Genes Dev. 2006;20:1525–1538. doi: 10.1101/gad.368706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Urquiza AM, Liu SY, Sjöberg M, Zetterström RH, Griffiths W, Sjovall J, Perlmann T. Docosahexaenoic acid, a ligand for the retinoid X receptor in mouse brain. Science. 2000;290:2140–2144. doi: 10.1126/science.290.5499.2140. [DOI] [PubMed] [Google Scholar]
- 61.Mascrez B, Ghyselinck NB, Chambon P, Mark M. A transcriptionally silent RXRalpha supports early embryonic morphogenesis and heart development. Proc Natl Acad Sci USA. 2009;106:4272–4277. doi: 10.1073/pnas.0813143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ang HL, Deltour L, Zgombic-Knight M, Wagner MA, Duester G. Expression patterns of class I and class IV alcohol dehydrogenase genes in developing epithelia suggest a role for alcohol dehydrogenase in local retinoic acid synthesis. Alcohol Clin Exp Res. 1996;20:1050–1064. doi: 10.1111/j.1530-0277.1996.tb01946.x. [DOI] [PubMed] [Google Scholar]
- 63.Deltour L, Foglio MH, Duester G. Metabolic deficiencies in alcohol dehydrogenase Adh1, Adh3and Adh4 null mutant mice: Overlapping roles of Adh1 and Adh4 in ethanol clearance and metabolism of retinol to retinoic acid. J Biol Chem. 1999;274:16796–16801. doi: 10.1074/jbc.274.24.16796. [DOI] [PubMed] [Google Scholar]
- 64.Molotkov A, Fan X, Duester G. Excessive vitamin A toxicity in mice genetically deficient in either alcohol dehydrogenase Adh1 or Adh3. FEBS J. 2002;269:2607–2612. doi: 10.1046/j.1432-1033.2002.02935.x. [DOI] [PubMed] [Google Scholar]
- 65.Molotkov A, Ghyselinck NB, Chambon P, Duester G. Opposing actions of cellular retinol-binding protein and alcohol dehydrogenase control the balance between retinol storage and degradation. Biochem J. 2004;383:295–302. doi: 10.1042/BJ20040621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deltour L, Foglio MH, Duester G. Impaired retinol utilization in Adh4 alcohol dehydrogenase mutant mice. Dev Genet. 1999;25:1–10. doi: 10.1002/(SICI)1520-6408(1999)25:1<1::AID-DVG1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 67.Zhang M, Chen WG, Smith SM, Napoli JL. Molecular characterization of a mouse short chain dehydrogenase/reductase active with all-trans-retinol in intact cells, mRDH1. J Biol Chem. 2001;276:44083–44090. doi: 10.1074/jbc.M105748200. [DOI] [PubMed] [Google Scholar]
- 68.Kallberg Y, Oppermann U, Jörnvall H, Persson B. Short-chain dehydrogenase/reductase (SDR) relationships: A large family with eight clusters common to human, animal, and plant genomes. Protein Sci. 2002;11:636–641. doi: 10.1110/ps.26902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu BX, Moiseyev G, Chen Y, Rohrer B, Crouch RK, Ma JX. Identification of RDH10, an all-trans retinol dehydrogenase, in retinal Muller cells. Invest Ophthalmol Vis Sci. 2004;45:3857–3862. doi: 10.1167/iovs.03-1302. [DOI] [PubMed] [Google Scholar]
- 70.Cammas L, Romand R, Fraulob V, Mura C, Dolle P. Expression of the murine retinol dehydrogenase 10 (Rdh10) gene correlates with many sites of retinoid signalling during embryogenesis and organ differentiation. Dev Dyn. 2007;236:2899–2908. doi: 10.1002/dvdy.21312. [DOI] [PubMed] [Google Scholar]
- 71.Romand R, Kondo T, Cammas L, Hashino E, Dolle P. Dynamic expression of the retinoic acid-synthesizing enzyme retinol dehydrogenase 10 (Rdh10) in the developing mouse brain and sensory organs. J Comp Neurol. 2008;508:879–892. doi: 10.1002/cne.21707. [DOI] [PubMed] [Google Scholar]
- 72.Rossant J, Zirngibl R, Cado D, Shago M, Giguère V. Expression of a retinoic acid response element-hsplacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes Dev. 1991;5:1333–1344. doi: 10.1101/gad.5.8.1333. [DOI] [PubMed] [Google Scholar]
- 73.Cunningham TJ, Chatzi C, Sandell LL, Trainor PA, Duester G. Rdh10 mutants deficient in limb field retinoic acid signaling exhibit normal limb patterning but display interdigital webbing. Dev Dyn. 2011;240:1142–1150. doi: 10.1002/dvdy.22583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao X, Sirbu IO, Mic FA, Molotkova N, Molotkov A, Kumar S, Duester G. Retinoic acid promotes limb induction through effects on body axis extension but is unnecessary for limb patterning. Curr Biol. 2009;19:1050–1057. doi: 10.1016/j.cub.2009.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hellgren M, Stromberg P, Gallego O, Martras S, Farres J, Persson B, Pares X, Hoog JO. Alcohol dehydrogenase 2 is a major hepatic enzyme for human retinol metabolism. Cell Mol Life Sci. 2007;64:498–505. doi: 10.1007/s00018-007-6449-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Strömberg P, Höög JO. Human class V alcohol dehydrogenase (ADH5): A complex transcription unit generates C-terminal multiplicity. Biochem Biophys Res Commun. 2000;278:544–549. doi: 10.1006/bbrc.2000.3837. [DOI] [PubMed] [Google Scholar]
- 77.Chambers D, Wilson L, Maden M, Lumsden A. RALDH-independent generation of retinoic acid during vertebrate embryogenesis by CYP1B1. Development. 2007;134:1369–1383. doi: 10.1242/dev.02815. [DOI] [PubMed] [Google Scholar]
- 78.Chambers D, Wilson LJ, Alfonsi F, Hunter E, Saxena U, Blanc E, Lumsden A. Rhombomere-specific analysis reveals the repertoire of genetic cues expressed across the developing hindbrain. Neural Dev. 2009;4 doi: 10.1186/1749-8104-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Niederreither K, Fraulob V, Garnier JM, Chambon P, Dollé P. Differential expression of retinoic acid-synthesizing (RALDH) enzymes during fetal development and organ differentiation in the mouse. Mech Dev. 2002;110:165–171. doi: 10.1016/s0925-4773(01)00561-5. [DOI] [PubMed] [Google Scholar]
- 80.McCaffery P, Tempst P, Lara G, Dräger UC. Aldehyde dehydrogenase is a positional marker in the retina. Development. 1991;112:693–702. doi: 10.1242/dev.112.3.693. [DOI] [PubMed] [Google Scholar]
- 81.Haselbeck RJ, Hoffmann I, Duester G. Distinct functions for Aldh1 and Raldh2 in the control of ligand production for embryonic retinoid signaling pathways. Dev Genet. 1999;25:353–364. doi: 10.1002/(SICI)1520-6408(1999)25:4<353::AID-DVG9>3.0.CO;2-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Molotkov A, Duester G. Genetic evidence that retinaldehyde dehydrogenase Raldh1 (Aldh1a1) functions downstream of alcohol dehydrogenase Adh1 in metabolism of retinol to retinoic acid. J Biol Chem. 2003;278:36085–36090. doi: 10.1074/jbc.M303709200. [DOI] [PubMed] [Google Scholar]
- 83.Ziouzenkova O, Orasanu G, Sharlach M, Akiyama TE, Berger JP, Viereck J, Hamilton JA, Tang G, Dolnikowski GG, Vogel S, Duester G, Plutzky J. Retinaldehyde represses adipogenesis and diet-induced obesity. Nature Med. 2007;13:695–702. doi: 10.1038/nm1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Niederreither K, McCaffery P, Dräger UC, Chambon P, Dollé P. Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech Dev. 1997;62:67–78. doi: 10.1016/s0925-4773(96)00653-3. [DOI] [PubMed] [Google Scholar]
- 85.Niederreither K, Vermot J, Schuhbaur B, Chambon P, Dollé P. Retinoic acid synthesis and hindbrain patterning in the mouse embryo. Development. 2000;127:75–85. doi: 10.1242/dev.127.1.75. [DOI] [PubMed] [Google Scholar]
- 86.Sirbu IO, Gresh L, Barra J, Duester G. Shifting boundaries of retinoic acid activity control hindbrain segmental gene expression. Development. 2005;132:2611–2622. doi: 10.1242/dev.01845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Molotkova N, Molotkov A, Sirbu IO, Duester G. Requirement of mesodermal retinoic acid generated by Raldh2 for posterior neural transformation. Mech Dev. 2005;122:145–155. doi: 10.1016/j.mod.2004.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Z, Dolle P, Cardoso WV, Niederreither K. Retinoic acid regulates morphogenesis and patterning of posterior foregut derivatives. Dev Biol. 2006;297:433–445. doi: 10.1016/j.ydbio.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 89.Chen F, Desai TJ, Qian J, Niederreither K, Lu J, Cardoso WV. Inhibition of Tgfβ signaling by endogenous retinoic acid is essential for primary lung bud induction. Development. 2007;134:2969–2979. doi: 10.1242/dev.006221. [DOI] [PubMed] [Google Scholar]
- 90.Molotkov A, Molotkova N, Duester G. Retinoic acid generated by Raldh2 in mesoderm is required for mouse dorsal endodermal pancreas development. Dev Dyn. 2005;232:950–957. doi: 10.1002/dvdy.20256. [DOI] [PubMed] [Google Scholar]
- 91.Martín M, Gallego-Llamas J, Ribes V, Kedinger M, Niederreither K, Chambon P, Dollé P, Gradwohl G. Dorsal pancreas agenesis in retinoic acid-deficient Raldh2 mutant mice. Dev Biol. 2005;284:399–411. doi: 10.1016/j.ydbio.2005.05.035. [DOI] [PubMed] [Google Scholar]
- 92.Niederreither K, Vermot J, Messaddeq N, Schuhbaur B, Chambon P, Dollé P. Embryonic retinoic acid synthesis is essential for heart morphogenesis in the mouse. Development. 2001;128:1019–1031. doi: 10.1242/dev.128.7.1019. [DOI] [PubMed] [Google Scholar]
- 93.Sirbu IO, Zhao X, Duester G. Retinoic acid controls heart anteroposterior patterning by down-regulating Isl1 through the Fgf8 pathway. Dev Dyn. 2008;237:1627–1635. doi: 10.1002/dvdy.21570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vermot J, Llamas JG, Fraulob V, Niederreither K, Chambon P, Dollé P. Retinoic acid controls the bilateral symmetry of somite formation in the mouse embryo. Science. 2005;308:563–566. doi: 10.1126/science.1108363. [DOI] [PubMed] [Google Scholar]
- 95.Sirbu IO, Duester G. Retinoic acid signaling in node ectoderm and posterior neural plate directs left-right patterning of somitic mesoderm. Nature Cell Biol. 2006;8:271–277. doi: 10.1038/ncb1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhao X, Brade T, Cunningham TJ, Duester G. Retinoic acid controls expression of tissue remodeling genes Hmgn1 and Fgf18 at the digit-interdigit junction. Dev Dyn. 2010;239:665–671. doi: 10.1002/dvdy.22188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rosselot C, Spraggon L, Chia I, Batourina E, Riccio P, Lu B, Niederreither K, Dolle P, Duester G, Chambon P, Costantini F, Gilbert T, Molotkov A, Mendelsohn C. Non-cell-autonomous retinoid signaling is crucial for renal development. Development. 2010;137:283–292. doi: 10.1242/dev.040287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen F, Cao Y, Qian J, Shao F, Niederreither K, Cardoso WV. A retinoic acid-dependent network in the foregut controls formation of the mouse lung primordium. J Clin Invest. 2010;120:2040–2048. doi: 10.1172/JCI40253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lin SC, Dolle P, Ryckebusch L, Noseda M, Zaffran S, Schneider MD, Niederreither K. Endogenous retinoic acid regulates cardiac progenitor differentiation. Proc Natl Acad Sci USA. 2010;107:9234–9239. doi: 10.1073/pnas.0910430107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brade T, Kumar S, Cunningham TJ, Chatzi C, Zhao X, Cavallero S, Li P, Sucov HM, Ruiz-Lozano P, Duester G. Retinoic acid stimulates myocardial expansion by induction of hepatic erythropoietin which activates epicardial Igf2. Development. 2011;138:139–148. doi: 10.1242/dev.054239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kumar S, Chatzi C, Brade T, Cunningham TJ, Zhao X, Duester G. Sex-specific timing of meiotic initiation is regulated by Cyp26b1 independent of retinoic acid signaling. Nature Commun. 2011;2:151. doi: 10.1038/ncomms1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li H, Wagner E, McCaffery P, Smith D, Andreadis A, Dräger UC. A retinoic acid synthesizing enzyme in ventral retina and telencephalon of the embryonic mouse. Mech Dev. 2000;95:283–289. doi: 10.1016/s0925-4773(00)00352-x. [DOI] [PubMed] [Google Scholar]
- 103.Mic FA, Molotkov A, Fan X, Cuenca AE, Duester G. RALDH3, a retinaldehyde dehydrogenase that generates retinoic acid, is expressed in the ventral retina, otic vesicle and olfactory pit during mouse development. Mech Dev. 2000;97:227–230. doi: 10.1016/s0925-4773(00)00434-2. [DOI] [PubMed] [Google Scholar]
- 104.Matt N, Dupé V, Garnier J-M, Dennefeld C, Chambon P, Mark M, Ghyselinck NB. Retinoic acid-dependent eye morphogenesis is orchestrated by neural crest cells. Development. 2005;132:4789–4800. doi: 10.1242/dev.02031. [DOI] [PubMed] [Google Scholar]
- 105.Molotkov A, Molotkova N, Duester G. Retinoic acid guides eye morphogenetic movements via paracrine signaling but is unnecessary for retinal dorsoventral patterning. Development. 2006;133:1901–1910. doi: 10.1242/dev.02328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kumar S, Duester G. Retinoic acid signaling in perioptic mesenchyme represses Wnt signaling via induction of Pitx2 and Dkk2. Dev Biol. 2010;340:67–74. doi: 10.1016/j.ydbio.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gage PJ, Qian M, Wu D, Rosenberg KI. The canonical Wnt signaling antagonist DKK2 is an essential effector of PITX2 function during normal eye development. Dev Biol. 2008;317:310–324. doi: 10.1016/j.ydbio.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Luo TL, Wagner E, Grün F, Dräger UC. Retinoic acid signaling in the brain marks formation of optic projections, maturation of the dorsal telencephalon, and function of limbic sites. J Comp Neurol. 2004;470:297–316. doi: 10.1002/cne.20013. [DOI] [PubMed] [Google Scholar]
- 109.Wagner M, Han B, Jessell TM. Regional differences in retinoid release from embryonic neural tissue detected by an in vitro reporter assay. Development. 1992;116:55–66. doi: 10.1242/dev.116.1.55. [DOI] [PubMed] [Google Scholar]
- 110.Chatzi C, Brade T, Duester G. Retinoic acid functions as a key GABAergic differentiation signal in the basal ganglia. PLoS Biol. 2011;9:e1000609. doi: 10.1371/journal.pbio.1000609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Szalai G, Duester G, Friedman R, Jia H, Lin S, Roe BA, Felder MR. Organization of six functional mouse alcohol dehydrogenase genes on two overlapping bacterial artificial chromosomes. FEBS J. 2002;269:224–232. doi: 10.1046/j.0014-2956.2001.02642.x. [DOI] [PubMed] [Google Scholar]