Abstract
Aging is thought to negatively affect multiple cellular processes, including the ability to maintain chromosome stability. Chromosome instability (CIN) is a common property of cancer cells and may be a contributing factor to cellular transformation. The types of DNA aberrations that arise during aging prior to tumor development and that contribute to tumorigenesis are currently unclear. Mdm2, a key regulator of the p53 tumor suppressor and modulator of DNA break repair, is frequently overexpressed in malignancies and contributes to CIN. To determine the relationship between aging and CIN and the role of Mdm2, pre-cancerous wild-type C57Bl/6 and littermate-matched Mdm2 transgenic mice at various ages were evaluated. Metaphase analyses of wild-type cells showed a direct correlation between age and increased chromosome and chromatid breaks, chromosome fusions, and aneuploidy, but the frequency of polyploidy remained stable over time. Elevated levels of Mdm2 in pre-cancerous mice increased both the numerical and the structural chromosomal abnormalities observed. Chromosome and chromatid breaks, chromosome fusions, aneuploidy, and polyploidy were increased in older Mdm2 transgenic mice compared to wild-type littermates. Unexpectedly, chromosome fusions, aneuploidy, and polyploidy rates in Mdm2 transgenic mice, but not chromosome and chromatid breaks, showed cooperation between Mdm2 overexpression and age. Notably, Mdm2 overexpression promoted gains in one or more chromosomes with age, while it did not affect the rate of chromosome loss. Therefore, aging increased specific forms of genomic instability, and elevated Mdm2 expression cooperated with aging to increase the likelihood of gaining certain chromosomal abnormalities of the kind thought to lead to cancer development.
Keywords: aging, Mdm2, p53, chromosome instability, aneuploidy
INTRODUCTION
Genomic instability refers to the accumulation or acquisition of numerical and/or structural abnormalities in chromosomes. It has long been observed that chromosome instability (CIN) is a hallmark of cancer cells and is postulated to be required for tumorigenesis (Lengauer et al., 1998, Negrini et al., 2010). Genomic changes, such as chromosome breaks, translocations, genome rearrangements, aneuploidy, and telomere shortening have been observed in aging organisms (Aubert and Lansdorp, 2008, Dolle and Vijg, 2002, Nisitani et al., 1990, Tucker et al., 1999, Zietkiewicz et al., 2009). Characteristics of genetically unstable cells include structural changes such as insertions, deletions, and translocations, as well as changes in the number of chromosomes (aneuploidy or polyploidy). Cells can gain or lose one or more chromosomes (aneuploidy) or gain part of or an entire genome (polyploidy) (Ganem et al., 2007, Lengauer et al., 1998). Additionally, cells with genomic instability frequently display increased chromosome or chromatid breaks, chromosomal fusions, and centrosome amplification, which itself promotes CIN.
Mechanisms leading to CIN are diverse and incompletely understood. Unrepaired DNA double-strand breaks or eroded telomeres can lead to CIN by serving as substrates for chromosomal fusions and translocations (Morgan et al., 1998). Amplification of centrosomes may lead to the missegregation of chromosomes and result in aneuploidy (Ganem et al., 2007). The uncoupling of DNA replication and mitosis can result in polyploidy, which is postulated to be a precursor to aneuploidy (Fujiwara et al., 2005, Ganem et al., 2007, Thompson et al., 2010, Vitale et al., 2010). Whether this genomic instability, commonly observed in cancer cells, precedes tumorigenesis or is a by-product of transformation is not clear. However, mice with mutations in genes encoding proteins involved in cell cycle control or DNA damage signaling or repair have increased CIN and many spontaneously develop cancer (Garinis et al., 2008, Negrini et al., 2010). Therefore maintaining genomic stability appears essential to limit tumorigenesis.
The oncogene Mdm2 is frequently amplified or overexpressed in many human and murine cancers; approximately 10% of all human cancers harbor MDM2 amplifications (Marine and Lozano, 2010, Rayburn et al., 2005). Elevated levels of Mdm2 are associated with increased transformation in vitro and in vivo (Marine and Lozano, 2010). Mdm2 is an E3 ubiquitin ligase that functions as a critical negative regulator of the tumor suppressor p53, and Mdm2 can also delay DNA repair through association with a DNA repair complex (Bouska and Eischen, 2009, Marine and Lozano, 2010). Mdm2 interferes with the transcriptional activity of p53 by binding to and ubiquitinating p53, targeting it for degradation by the proteosome (Marine and Lozano, 2010). Inactivation of p53 function results in a loss of cell cycle checkpoint control and polyploidy, aneuploidy, and tumorigenesis (Fujiwara et al., 2005, Levine and Oren, 2009). Mdm2 also binds to Nbs1, a protein in the Mre11/Rad50/Nbs1 DNA repair complex, and inhibits DNA double-strand break repair (Alt et al., 2005, Bouska et al., 2008). Cells with reduced levels of Nbs1 or that contain mutated Nbs1 have an altered response to DNA damage and are delayed in repairing DNA damage (Difilippantonio et al., 2005, Williams et al., 2002). Humans with mutations in NBS1 frequently develop malignancies (Demuth and Digweed, 2007). Therefore Mdm2 regulates two proteins essential for monitoring, signaling, and/or repairing DNA damage, which is thought to be a contributing factor to tumorigenesis.
Several studies have linked Mdm2 overexpression to genomic instability through both p53-dependent and p53-independent mechanisms. Overexpression of human Mdm2 in murine fibroblasts that contain wild-type p53 resulted in aneuploidy and centrosome amplification (Carroll et al., 1999). Recently, loss of p53 was demonstrated to be important for the survival and proliferation of aneuploid cells (Fujiwara et al., 2005, Thompson et al., 2010, Vitale et al., 2010). Mammary epithelial cells from mammary specific-Mdm2 transgenic mice have increased polyploidy, likely due to endoreduplication, and this occurred irrespective of p53 status (Lundgren et al., 1997). Additionally, B-cells from juvenile Mdm2 transgenic mice, where Mdm2 expression was driven from its native promoter, have increased chromosome and chromatid breaks and aneuploidy compared to B-cells from wild-type mice (Wang et al., 2008). Furthermore, through interaction with Nbs1, Mdm2 overexpression in wild-type or p53-null mouse embryonic fibroblasts inhibited DNA double-strand break repair, leading to genomic instability and transformation (Alt et al., 2005, Bouska et al., 2008). Thus, elevated expression of Mdm2 is linked to genomic instability, but whether elevated levels of Mdm2 affect chromosomal stability as a mammal ages is unknown. Therefore, we performed an in depth study of the effect of aging alone and together with Mdm2 overexpression on genome stability. We show that specific chromosomal alterations linked to chromosomal instability increase with age, and elevated levels of Mdm2 lead to a greater amount of chromosomal instability that accumulated with age prior to tumor development. The increased numerical and structural chromosome abnormalities in Mdm2 transgenic mice likely contribute to their increased predisposition to cancer.
RESULTS
Chromosome instability increases as mice age
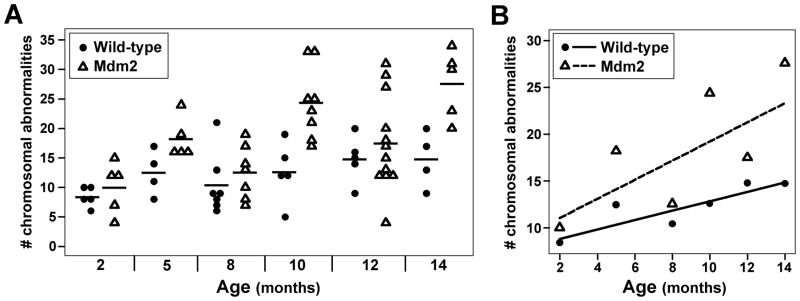
To determine the specific effects of aging on overall genome stability and the contribution of increased Mdm2 levels to this process, approximately 3,600 metaphase spreads of splenocytes from 72 littermate-matched pre-cancerous wild-type C57Bl/6 and Mdm2 hemizygous transgenic mice 2–14 months old were analyzed for chromosomal alterations. (Mice older than 14 months were not evaluated due to the emergence of cancer in the Mdm2 hemizygous transgenic mice.) The number of chromosomal aberrations, including chromosome and chromatid breaks, chromosome fusions, and metaphases with an abnormal number of chromosomes, were quantified in approximately 50 metaphases from each mouse. There was a small, gradual increase in the total number of chromosomal abnormalities in wild-type mice as they aged (Fig. 1A and 1B). In contrast, Mdm2 transgenic mice had a marked increase in CIN with increasing age (p<0.0005). The frequency of chromosomal abnormalities in wild-type and Mdm2 transgenic mice were similar at 2 months of age, but at 14 months old the number of chromosomal abnormalities in Mdm2 transgenic mice was twice that in wild-type mice (Fig. 1B). Therefore, as mice age, the frequency of CIN increases, and this is significantly augmented by elevated levels of Mdm2.
Figure 1.
Increased chromosomal instability with age. Metaphases from splenocytes from wild-type or littermate matched Mdm2 transgenic mice ages 2–14 months old were evaluated for chromosomal abnormalities (e.g., breaks, fusions, altered number of chromosomes). A) The number of chromosomal abnormalities of each mouse at each age is plotted. Each symbol represents an individual mouse, and the mean value for each genotype at each age is denoted by a black line. B) The expected number of chromosomal abnormalities at each age is given by a solid (wild-type) or dashed (transgenic) line. The mean number of chromosomal abnormalities of each genotype at each age is represented by symbols.
Increased chromosome and chromatid breaks in aging mice
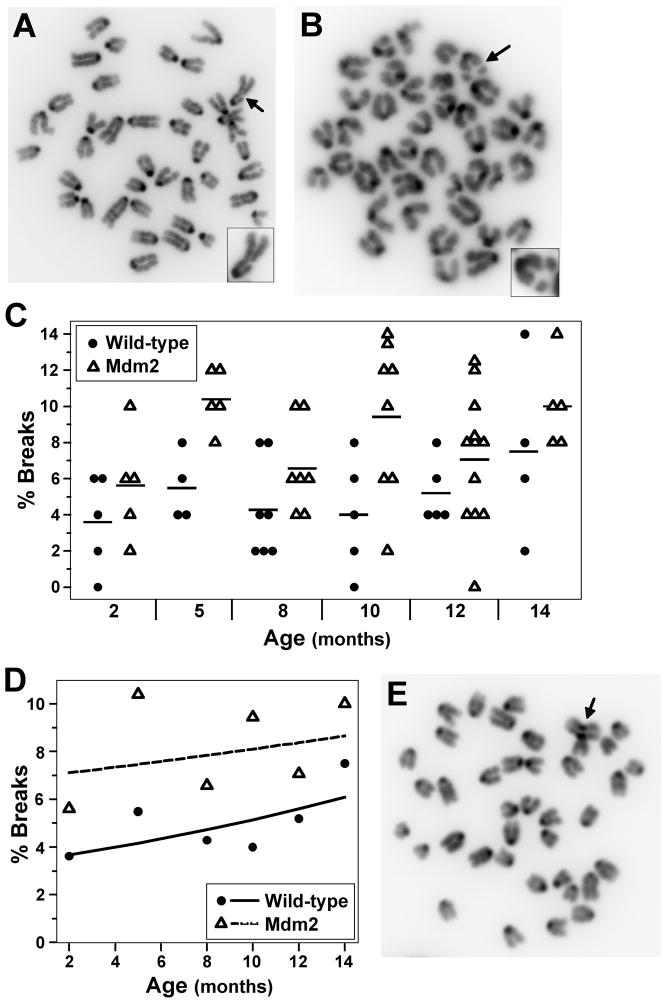
To identify the specific chromosomal alterations that are correlated with aging, and to examine the effects of Mdm2 overexpression on these alterations, we first evaluated chromatid and chromosome breaks from metaphase spreads of splenocytes from pre-cancerous wild-type C57Bl/6 mice and Mdm2 transgenic littermates. Both chromatid and chromosome breaks were detected in wild-type and Mdm2 transgenic mice with chromosome breaks dominating in both genotypes (Fig. 2A–2C). The number of metaphases with breaks gradually increased with age for both wild-type and Mdm2 transgenic mice (Fig. 2D). As we previously reported for splenocytes from mice 3 weeks old (Wang et al., 2008), at 2 months old, a higher percentage of metaphases from Mdm2 transgenic mice had chromosome and chromatid breaks compared to metaphases from wild-type littermates. The mean number of metaphases with DNA breaks was significantly higher for Mdm2 transgenic mice compared to wild-type mice at all ages evaluated (p=0.001) (Fig. 2D). Moreover, tri-radial chromosomes, which lead to DNA breaks, were detected only in Mdm2 transgenic cells (Fig. 2E). Surprisingly, the rate of increase of DNA breaks with age was statistically similar between wild-type and Mdm2 transgenic cells (p=0.49). Therefore, although Mdm2 overexpression increased the frequency of chromosome and chromatid breaks at every age evaluated, Mdm2 overexpression did not accelerate the rate at which breaks occurred with age.
Figure 2.
Chromosome and chromatid breaks increase with age. Metaphases from splenocytes from wild-type or littermate matched Mdm2 transgenic mice ages 2–14 months old were evaluated for chromosome or chromatid breaks. A & B) Representative pictures of chromatid (A) and chromosome (B) breaks. Arrows point to breaks. Insert in the corner of each photograph is the chromosome with the break. C) The percentages of metaphases with chromosome or chromatid breaks from each mouse are shown; each symbol represents an individual mouse. A black line denotes the mean percentage for each genotype at each age. D) The expected percentage of cells with chromosome or chromatid breaks is denoted by a solid (wild-type) or a dashed (transgenic) line. The mean percentages of each genotype at each age are represented by symbols. E) A picture of a metaphase from an Mdm2 transgenic mouse with a tri-radial chromosome (arrow points to defect).
Increased chromosome fusion in cells from Mdm2 transgenic mice
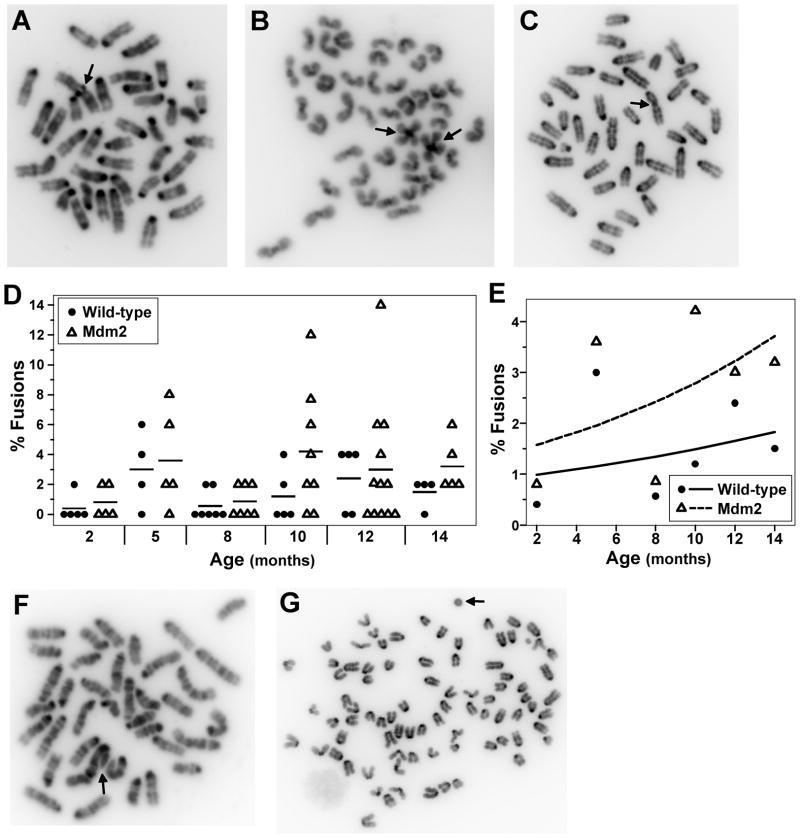
Unrepaired DNA breaks can serve as substrates for genomic rearrangements (Morgan et al., 1998), which have been found to accumulate in aging mice (Dolle and Vijg, 2002). Since aging wild-type and Mdm2 transgenic mice have elevated levels of DNA breaks, we evaluated metaphases for chromosomal fusions from both genotypes of mice at 2–14 months of age. Although fusions were rare, they were identified in both wild-type and Mdm2 transgenic mice (Fig. 3A–3C). Most (66%) of the fusions identified for both genotypes were centromeric-centromeric fusions also known as Robertsonian translocations (Fig. 3A & 3B), which are known to occur in mice (Garagna et al., 2001). Centromeric-telomeric fusions accounted for a quarter of the fusions detected (Fig. 3C), and telomeric-telomeric fusions were rare for both genotypes and accounted for less than 10% of the fusions identified. At 2 months of age, wild-type and Mdm2 transgenic mice had a similar low number of metaphases with fused chromosomes, and both genotypes showed an increase in metaphases with fused chromosomes as the mice aged (Fig. 3D). However, Mdm2 transgenic mice had a greater percentage of metaphases with fused chromosomes than wild-type mice with the greatest difference detected at older ages (p=0.05) (Fig. 3E). Other deleterious chromosomal fusion abnormalities were only observed in cells from Mdm2 transgenic mice. Specifically, three chromosomes fused centromere to telomere and a break in one of the fused chromosomes were detected in a metaphase from a 10 month old Mdm2 transgenic mouse (Fig. 3F). Additionally, a very rare fusion, a ring chromosome, was observed in cells from Mdm2 transgenic mice as young as 5 months old and in 14 month old mice (Fig. 3G). Therefore, fused chromosomes were more common in Mdm2 transgenic mice. The prevalence of these abnormalities increased with age in both wild-type and Mdm2 transgenic mice, but the rate of increase was greater for Mdm2 transgenic mice, suggesting an interaction between Mdm2 and age on chromosomal fusions. Moreover, severe structurally abnormal chromosomes were only detected in pre-cancerous cells in Mdm2 transgenics and not in wild-type cells.
Figure 3.
Mdm2 overexpression increases chromosome fusion frequency as mice age. Metaphases from splenocytes from wild-type or littermate matched Mdm2 transgenic mice ages 2–14 months old were evaluated for chromosome fusions. A–C) Representative pictures of centromere-centromere (A & B) and telomere-centromere (C) fusions. Arrows point to fusions. D) The percentages of metaphases with fusions for each mouse are shown; each symbol represents an individual mouse. The mean values of each genotype at each age are denoted by a black line. E) Plots of the expected percentage of cells with fusions at each age are denoted by a solid (wild-type) or a dashed (transgenic) line. The mean values of each genotype at each age are represented by symbols. F–G) Severe structural chromosomal abnormalities in Mdm2 transgenic cells. F) A picture of a metaphase with three chromosomes fused centromere to telomere and a break in one of the chromosomes. Arrow points to defect. G) Metaphase with a ringed chromosome. Arrow points to ring.
Increased polyploid DNA content in cells from aging Mdm2 transgenic mice
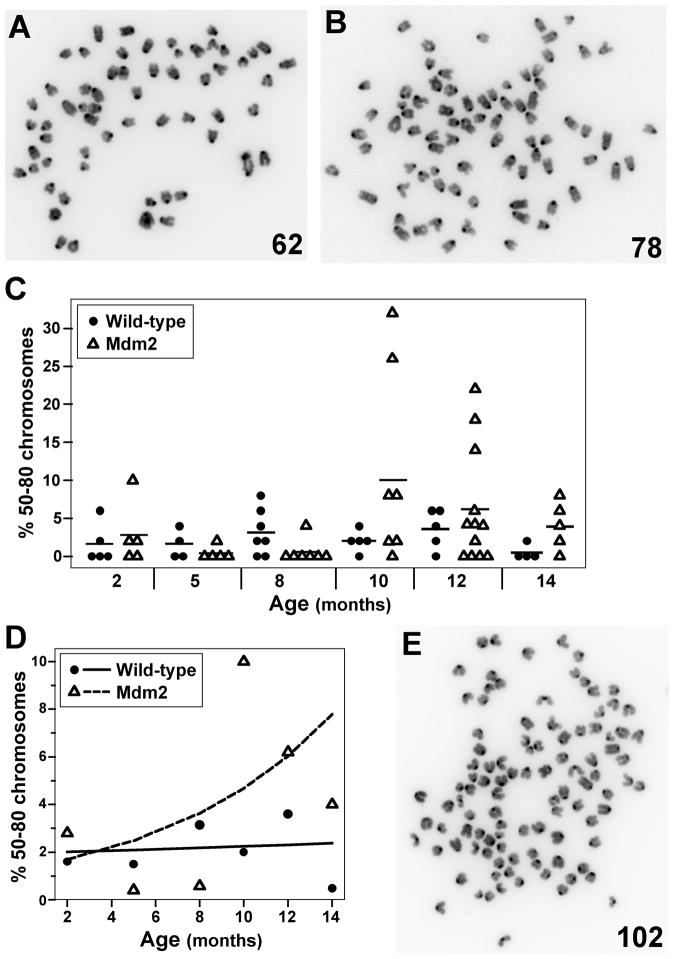
A cell becomes polyploid by gaining whole sets of chromosomes or an entire genome through a variety of incompletely understood mechanisms (Ganem et al., 2007, Lengauer et al., 1998). The number of chromosomes from metaphases of splenocytes from wild-type and Mdm2 transgenic mice were quantified to determine the frequency of polyploid cells (near triploid, triploid, near tetraploid, and tetraploid DNA content). Polyploid metaphases (40 chromosomes is normal) were detected in both wild-type and Mdm2 transgenic mice (Fig. 4A–4C). At two months of age, the number of metaphases with 50–80 chromosomes was similar for wild-type and Mdm2 transgenic littermates (Fig. 4C & 4D). As the mice aged, the number of metaphases with 50–80 chromosomes significantly increased in Mdm2 transgenic mice (p=0.04), but remained relatively constant for wild-type mice (Fig. 4D). Furthermore, Mdm2 transgenic mice also had cells that contained more than 4N DNA (Fig. 4E). Our results indicate that age had little effect on the development of polyploidy in wild-type mice, but elevated levels of Mdm2 significantly increased the rate of emergence of polyploid cells in mice over 14 months. Thus, there was a greater likelihood of gaining partial or whole genomes as mice aged, if Mdm2 was overexpressed.
Figure 4.
Mdm2 overexpression induces polyploidy that increases with age. Chromosome number from metaphases from splenocytes from wild-type or littermate matched Mdm2 transgenic mice ages 2–14 months old were counted. A–B) Representative pictures of metaphases with near triploidy (A) and near tetraploidy (B). Numbers of chromosomes in each metaphase are indicated. C) The percentages of metaphases with 50–80 chromosomes for each mouse are shown; each symbol represents an individual mouse. A black line denotes the mean percentage for each genotype at each age. D) Plots of the expected percentage of polyploidy cells are denoted by a solid (wild-type) or a dashed (transgenic) line. The mean values of each genotype at each age are represented by symbols. E) A representative picture of a metaphase from an Mdm2 transgenic mouse with greater than 4N DNA content.
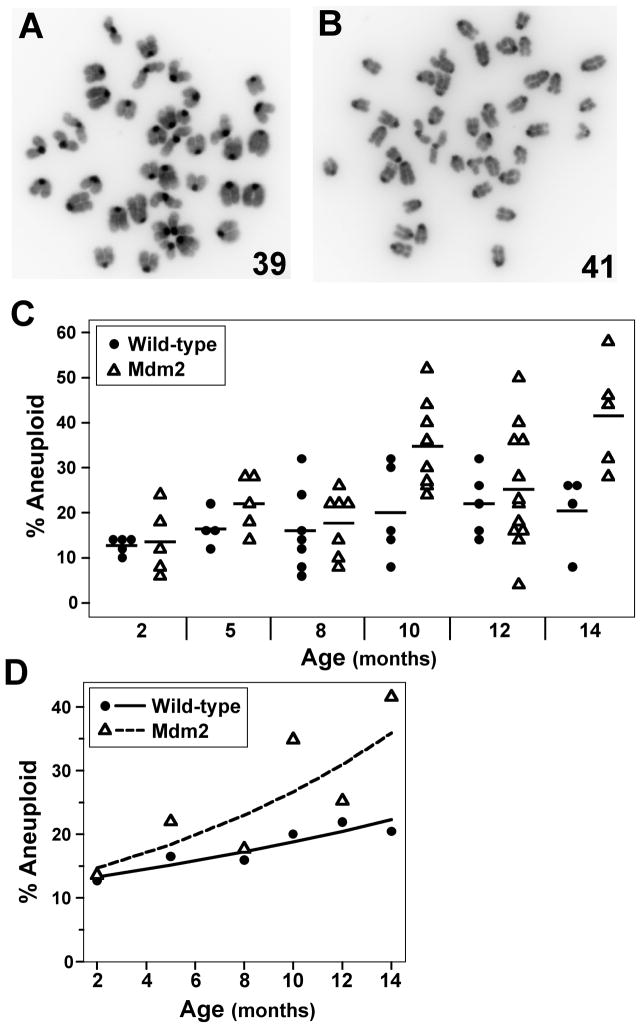
Elevated rates of aneuploidy in aging Mdm2 transgenic mice
Polyploidy is postulated to be a precursor to aneuploidy, which is a loss or gain of whole chromosomes and can result from chromosome missegregation during mitosis (Fujiwara et al., 2005, Ganem et al., 2007, Storchova and Kuffer, 2008, Thompson et al., 2010, Vitale et al., 2010). The number of chromosomes in metaphases of splenocytes from wild-type and Mdm2 transgenic littermates of various ages were counted to assess aneuploidy. Metaphases with greater than or less than the normal number of 40 chromosomes were detected in both wild-type and Mdm2 transgenic mice (Fig. 5A & 5B), but the frequency beyond 2 months of age was different for each (Fig. 5C & 5D). As wild-type mice aged, they showed a small but steady, significant increase in the number of metaphases that were aneuploid (Fig. 5D, p=0.04). Notably, aging Mdm2 transgenic mice had a large significant, non-linear increase in aneuploid cells such that the mean number of aneuploid cells in mice 14 months old was more than twice that in mice 2 months old (p<0.0005) (Fig. 5D). Therefore, Mdm2 overexpression appeared to accelerate the rate of aneuploidy development with age compared to wild-type mice.
Figure 5.
Increased aneuploid cells in aging Mdm2 transgenic mice. Chromosome number from metaphases of splenocytes from wild-type or littermate matched Mdm2 transgenic mice ages 2–14 months old were counted. A & B) Representative pictures of cells with less than 40 chromosomes (A) and more than 40 chromosomes (B). Numbers of chromosomes in each metaphase are indicated. C) Plots of the percentage of aneuploid metaphases from each mouse; each symbol represents an individual mouse. The mean values of each genotype at each age are denoted by a black line. D) The expected percentage of cells at each age is denoted by a solid (wild-type) or a dashed (transgenic) line. The mean values of each genotype at each age are represented by symbols.
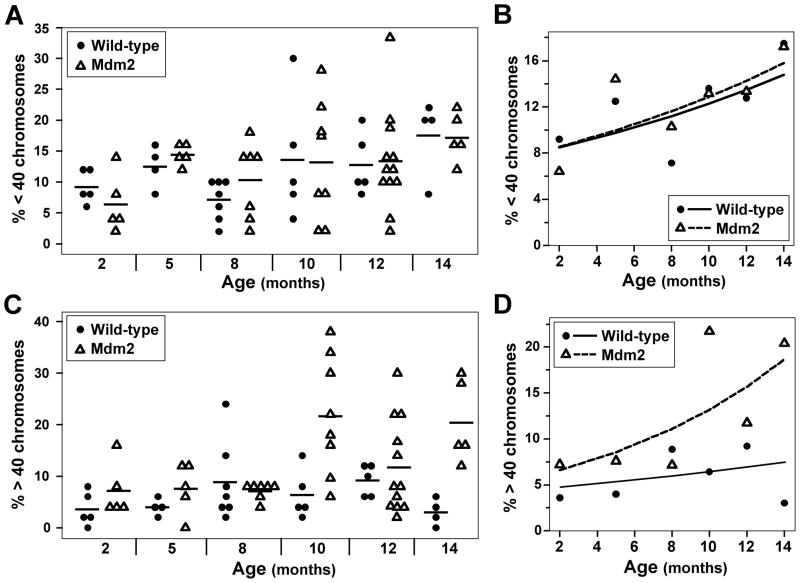
To assess aneuploidy more in depth, metaphases that lost chromosomes were distinguished from metaphases that gained chromosomes. As mice aged, both wild-type and Mdm2 transgenic mice had increased numbers of metaphases with less than 40 chromosomes (p=0.003), doubling the mean frequency between 2 months and 14 months of age (Fig. 6A & 6B). Unexpectedly, wild-type and Mdm2 transgenic mice had a similar number of metaphases with less than 40 chromosomes at all ages evaluated. Consequently, both genotypes of mice showed a similar rate of increase in metaphases with less than 40 chromosomes over 14 months (p=0.88) (Fig. 6B). In contrast, the rate of increase in metaphases with more than 40 chromosomes over 14 months was greater in Mdm2 transgenic mice than wild-type mice (Fig. 6C & 6D). At 2 months of age there were slightly more metaphases from Mdm2 transgenic mice that had greater than 40 chromosomes compared to wild-type mice. However, as the mice aged, the number of metaphases with greater than 40 chromosomes slightly increased in wild-type mice, but this increase was not significant (p=0.34). In contrast, the number of metaphases with greater than 40 chromosomes profoundly increased with age in Mdm2 transgenic mice and was statistically significant (p=0.003) (Fig. 6D). Mdm2 transgenic mice had triple the mean number of metaphases with greater than 40 chromosomes at 14 months old compared to at 2 months of age. These data indicate that as wild-type and Mdm2 transgenic mice age, both gains and losses of chromosomes increased. However, Mdm2 overexpression only resulted in an exacerbation of the rate of gains in chromosomes and not in the loss of chromosomes.
Figure 6.
Mdm2 transgenic mice have a significant gain in chromosomes as they age. Metaphases of splenocytes from wild-type or littermate matched Mdm2 transgenic mice ages 2–14 months old were evaluated for chromosome number. A–D) Data for the loss of chromosomes (A & B) or the gain of chromosomes (C & D) for both genotypes of mice. A & C) Percentages for each mouse are shown; each symbol represents an individual mouse. A black line denotes the mean percentage for each genotype at each age. B & D) Plots of the expected percentage of cells with a loss (B) or a gain (D) of chromosomes are denoted by a solid (wild-type) or a dashed (transgenic) line. The mean values of each genotype at each age are represented by symbols.
DISCUSSION
Genomic instability is characterized by chromosomal alterations, including changes in the number of chromosomes (aneuploidy or polyploidy) and/or in the structure of chromosomes (breaks, fusion, insertions, deletions, and translocations) (Lengauer et al., 1998, Negrini et al., 2010). Limited published studies suggest that genome instability increases with age in mammals (Aubert and Lansdorp, 2008, Nisitani et al., 1990, Tucker et al., 1999, Zietkiewicz et al., 2009). The incidence of cancer increases with age, and cancer cells commonly have genome instability, but the contribution chromosomal aberrations make to the development of malignancies is incompletely understood. This study shows that specific numerical and structural chromosomal changes increased with age in wild-type C57Bl/6 mice and were exacerbated with Mdm2 overexpression. Importantly, all chromosomal alterations in both wild-type and Mdm2 transgenic mice occurred prior to overt tumor development, indicating genome instability precedes tumorigenesis and likely contributes to the development of tumors. Moreover, Mdm2 transgenic mice had greatly increased and more severe chromosomal alterations than wild-type mice that correlated to age; they also develop cancer significantly earlier in life than wild-type mice, which may never develop a malignancy. Therefore, our data from approximately 3,600 metaphases from 72 littermate-matched mice from 2 to 14 months old provide new insight into aging and the types of chromosomal alterations associated with it that may lead to tumorigenesis.
Polyploidy is considered a prerequisite to aneuploidy (Fujiwara et al., 2005, Ganem et al., 2007, Storchova and Kuffer, 2008, Thompson et al., 2010, Vitale et al., 2010), which did increase and was correlated with age in wild-type mice. Therefore, it was expected that polyploid frequency would increase with age in wild-type cells. However, we did not detect an increase in polyploid cells in aging wild-type mice, suggesting that this genetic abnormality is not linked to age up to 14 months old. Alternatively, many of the polyploid cells that did emerge in the wild-type mice may not have survived and were, therefore, not detected, since other genetic alterations, such as loss of functional p53, appear required for polyploid cell survival (Lin et al., 2001, Storchova and Kuffer, 2008). Notably, Mdm2 transgenic mice had both increased polyploid and aneuploid cells that correlated with age. Mdm2 transgenic mice possessed cells with elevated levels of near triploid to true tetraploid DNA content and also had cells that had greater than 4N DNA. Since Mdm2 negatively regulates p53 (Marine and Lozano, 2010), and p53 inhibits the development of tetraploidy (Fujiwara et al., 2005, Ganem et al., 2007, Storchova and Kuffer, 2008, Thompson et al., 2010, Vitale et al., 2010), the polyploidy we observed in Mdm2 transgenic cells is likely due at least in part to inhibition of p53 by Mdm2. Moreover, Mdm2 expression can lead to the degradation of the p53 transcriptional target gene and cell cycle inhibitor p21, and loss of p21 results in endoreduplication and polyploid cells (Jin et al., 2003, Stewart et al., 1999). In addition, Mdm2 also negatively regulates the DNA repair protein, Nbs1 (Alt et al., 2005, Bouska et al., 2008). Nbs1 has a role in preventing endoreduplication (Wu et al., 2004), which leads to polyploidy, suggesting that Mdm2 may influence polyploidy through the regulation of Nbs1 as well. Therefore, there are multiple pathways to tetraploidy mediated by Mdm2 expression to explain the increased polyploidy in Mdm2 transgenic mice.
Aneuploidy frequently arises due to defects in chromosome segregation during mitosis (Thompson et al., 2010). Recently, loss of p53 function was found to be important for the proliferation of aneuploid cells (Thompson and Compton, 2010), which provides an explanation for the increased number of aneuploid cells in Mdm2 transgenic mice over that in wild-type mice. Interestingly, overexpression of Mdm2 resulted in an increase in all measures of genome instability, except the loss of chromosomes. Surprisingly, the number of cells losing one or more chromosomes increased at a similar rate with age in both wild-type and Mdm2 transgenic mice. In contrast, Mdm2 overexpression led to a greater frequency of gains of chromosomes compared to the rate in wild-type mice, and this increased with age. It is currently unknown why Mdm2 overexpression would influence a gain but not a loss in chromosome number; however, it is possible that Mdm2 preferentially affects endoreduplication events rather than chromosome missegregation events. Alternatively, a cell may only be able to sustain the loss of certain chromosomes, or require the acquisition of additional genetic alterations to survive the loss of chromosomes.
DNA breaks can be substrates for fusions and translocations of chromosomes (Morgan et al., 1998), which can accumulate in aging mice (Dolle and Vijg, 2002). Previously we reported that Mdm2 overexpression in primary fibroblasts results in an Nbs1-dependent delay in DNA double-strand break repair, resulting in DNA breaks persisting longer and an increase in chromatid and chromosome breaks (Alt et al., 2005, Bouska et al., 2008). Unexpectedly, the rate of increase in chromatid and chromosome breaks in aging mice was similar between wild-type and Mdm2 transgenics, although Mdm2 transgenic cells had more DNA breaks than their wild-type littermates at every age evaluated. Moreover, tri-radial chromosomes, which are usually due to increased mitotic recombination, were observed in Mdm2 transgenic mice, but not in wild-type mice. Tri-radial chromosomes frequently lead to DNA breaks during the next round of cell cycle. In accordance with the rate of breaks, there was an increase in chromosomal fusions that correlated with age in both the wild-type and the Mdm2 transgenic mice. Notably, Mdm2 overexpression resulted in severe structural chromosome fusion abnormalities that were not observed in wild-type mice. Metaphases with multiple chromosomes fused end-to-end and ring chromosomes were detected in Mdm2 transgenic mice as young as 5 months old. The chromosomal structure of three chromosomes fused centromere to telomere is typical of a breakage-fusion-bridge cycle that can lead to gene amplifications or translocations and is associated with tumorigenesis (DePinho and Polyak, 2004). Thus, elevated expression of Mdm2 over time increased fusion events, which may be due to the increase in total overall breaks in the cells of these mice, but may also be due to other uncharacterized mechanisms.
The mechanisms of how genome instability increases with age are unclear, although a decrease in the efficiency of DNA repair and apoptotic pathways, which lead to cells surviving DNA damage and possibly producing progeny, are likely causes. Studies using a variety of techniques have shown age-related changes in DNA repair mechanisms (Gorbunova et al., 2007). Specifically, the expression and activity of DNA repair enzymes decrease with age. For example, signaling from the DNA damage kinase, ATM, and the response of other DNA repair proteins declines with age (Feng et al., 2007, Gorbunova et al., 2007). Similarly, there is a decrease in p53 function with age (Feng et al., 2007). A reduction in ATM and p53 function results in decreased DNA repair, reduced apoptosis, and an increase in CIN and tumorigenesis (Negrini et al., 2010). Mdm2 negatively regulates both p53 and mediators of ATM activation, the Mre11/Rad50/Nbs1 DNA repair complex (Alt et al., 2005, Bouska et al., 2008, Marine and Lozano, 2010). Thus, Mdm2 overexpression should both induce and allow chromosomal aberrations to accumulate in cells over the course of a lifetime, ultimately leading to cellular transformation and tumorigenesis. We previously showed that in primary B cells in young mice, Mdm2 overexpression leads to an increase in DNA breaks and aneuploidy (Wang et al., 2008). Here our data reveal that prolonged Mdm2 overexpression worsens the genomic abnormalities observed over time, and that chromosomal alterations accumulate with age prior to tumor development. Moreover, the increased CIN correlates to an increased tumor incidence in the Mdm2 transgenic mice. It will be important in the future to further characterize the genomic alterations that arise as an organism ages and determine the contribution of specific alterations to tumorigenesis.
METERIALS AND METHODS
Mice
Mdm2 transgenic mice (from Dr. Steve Jones; U Mass) that express the Mdm2 transgene under the control of its native promotor (Jones et al., 1998) were backcrossed over ten generations to generate congenic C57Bl/6 mice. Experimental mice were generated by breeding hemizygous Mdm2 transgenic males to C57Bl/6 females to generate hemizygous Mdm2 transgenic mice and wild-type littermate controls. Mdm2 transgenic mice and non-transgenic littermate matched controls were sacrificed at specific ages. Mice greater than 14 moths old were excluded from the study due to tumor development in Mdm2 transgenic mice. Tissues from each mouse were carefully evaluated for malignancies; any mouse with overt cancer or suspected of having a malignancy were excluded from the study. All mice in this study conformed to institutional, state and federal rules and regulations.
Chromosome analysis
Spleens were harvested from Mdm2 transgenic mice and littermate matched wild-type controls at 2, 5, 8, 10, 12, and 14 months old. Splenocytes were stimulated to mitosis by cultivation for 48 hours in RPMI-1640 supplemented with L-glutamine, 10% fetal bovine serum, 0.5% β-mercaptoethanol, 3μg/ml lipopolysaccharide (LPS), and 5μg/ml concavalin A. Colcemid (1:100 dilution, KaryoMAX, Gibco) was added for an additional 4 hours. Cells were harvested and metaphase spreads were prepared by standard procedures. Chromosomes were stained with propidium iodide (PI) (Sigma, St. Louis, MO) and 4′, 6′-diamidino-2-phenyl-indo-dihydrochloride (DAPI) (Sigma). Slides were mounted with Vectashield (Vector Laboratories, Burlingame, CA), blinded, and 48–52 metaphases for each mouse for each genotype for each age were visualized and then photographed with a CCD camera coupled with fluorescent microscopy (Nikon, Eclipse 80i, Melville, NY, USA) using MetaMorph software (Molecular Devices Corp., Sunnyvale, CA). The type and number of chromosomal abnormalities were counted in each image. Only breaks visible by both PI and DAPI staining were scored as a break. The number of chromosomes was quantified in each image with Image Tool (UTHSCSA, TX).
Statistical Methods
The effects of age and genotype on the total number of chromosome abnormalities were assessed using multiple linear regression. The number of abnormalities per mouse was regressed on their genotype and age at sacrifice. An interaction term between genotype and age at sacrifice was included in this model. Negative binomial regression was used to model the event rates (the proportion of metaphase cells with the genetic abnormality of interest) in terms of genotype and age at sacrificed was performed (Agresti, 2002). The logarithm of the number of evaluated cells in metaphase from each mouse was included as an offset in our models. An interaction term between genotype and age was also included in these models whenever there was evidence that the multiplicative model might be false. Likelihood ratio tests of over-dispersion were performed for all models. Wald tests were used to evaluate how event rates varied with age and genotype. Estimates of the expected event rates shown in Figures 1B, 2D, 3E, 4D, 5D, 6B, and 6D were derived from these models. These data were also analyzed using general estimating equations (GEE) models with a logarithmic link function and a binomial random component (Diggle et al., 2002). The Huber-White sandwich estimator was used in these analyses to adjust the variance-covariance matrix of the models’ parameter estimates. These models gave results that were very similar to those obtained from our negative binomial regression models. The results given in this manuscript are from our negative binomial models.
Acknowledgments
We thank the members of the Eischen lab for thoughtful discussions. This work was supported by NIH/NCI grants: R01 CA117935 (CME) and the Vanderbilt Cancer Center grant P30 CA068485.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- Agresti A. Categroical Data Analysis. 2. Hoboken NJ: John Wiley & Sons; 2002. [Google Scholar]
- Alt JR, Bouska A, Fernandez MR, Cerny RL, Xiao H, Eischen CM. Mdm2 binds to Nbs1 at sites of DNA damage and regulates double strand break repair. J Biol Chem. 2005;280:18771–18781. doi: 10.1074/jbc.M413387200. [DOI] [PubMed] [Google Scholar]
- Aubert G, Lansdorp PM. Telomeres and aging. Physiol Rev. 2008;88:557–579. doi: 10.1152/physrev.00026.2007. [DOI] [PubMed] [Google Scholar]
- Bouska A, Eischen CM. Mdm2 affects genome stability independent of p53. Cancer Res. 2009;69:1697–1701. doi: 10.1158/0008-5472.CAN-08-3732. [DOI] [PubMed] [Google Scholar]
- Bouska A, Lushnikova T, Plaza S, Eischen CM. Mdm2 promotes genetic instability and transformation independent of p53. Mol Cell Biol. 2008;28:4862–4874. doi: 10.1128/MCB.01584-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll PE, Okuda M, Horn HF, Biddinger P, Stambrook PJ, Gleich LL, Li YQ, Tarapore P, Fukasawa K. Centrosome hyperamplification in human cancer: chromosome instability induced by p53 mutation and/or Mdm2 overexpression. Oncogene. 1999;18:1935–1944. doi: 10.1038/sj.onc.1202515. [DOI] [PubMed] [Google Scholar]
- Demuth I, Digweed M. The clinical manifestation of a defective response to DNA double-strand breaks as exemplified by Nijmegen breakage syndrome. Oncogene. 2007;26:7792–7798. doi: 10.1038/sj.onc.1210876. [DOI] [PubMed] [Google Scholar]
- DePinho RA, Polyak K. Cancer chromosomes in crisis. Nat Genet. 2004;36:932–934. doi: 10.1038/ng0904-932. [DOI] [PubMed] [Google Scholar]
- Difilippantonio S, Celeste A, Fernandez-Capetillo O, Chen HT, Reina San Martin B, Van Laethem F, Yang YP, Petukhova GV, Eckhaus M, Feigenbaum L, Manova K, Kruhlak M, Camerini-Otero RD, Sharan S, Nussenzweig M, Nussenzweig A. Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models. Nat Cell Biol. 2005;7:675–685. doi: 10.1038/ncb1270. [DOI] [PubMed] [Google Scholar]
- Diggle P, Heagerty P, Liang K-Y, Zeger S. Analysis of Longitudinal Data. 2. Oxford: Oxford University Press; 2002. [Google Scholar]
- Dolle ME, Vijg J. Genome dynamics in aging mice. Genome Res. 2002;12:1732–1738. doi: 10.1101/gr.125502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Hu W, Teresky AK, Hernando E, Cordon-Cardo C, Levine AJ. Declining p53 function in the aging process: a possible mechanism for the increased tumor incidence in older populations. Proc Natl Acad Sci U S A. 2007;104:16633–16638. doi: 10.1073/pnas.0708043104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17:157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Garagna S, Marziliano N, Zuccotti M, Searle JB, Capanna E, Redi CA. Pericentromeric organization at the fusion point of mouse Robertsonian translocation chromosomes. Proc Natl Acad Sci U S A. 2001;98:171–175. doi: 10.1073/pnas.98.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garinis GA, van der Horst GT, Vijg J, Hoeijmakers JH. DNA damage and ageing: new-age ideas for an age-old problem. Nat Cell Biol. 2008;10:1241–1247. doi: 10.1038/ncb1108-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V, Seluanov A, Mao Z, Hine C. Changes in DNA repair during aging. Nucleic Acids Res. 2007;35:7466–7474. doi: 10.1093/nar/gkm756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Lee H, Zeng SX, Dai MS, Lu H. MDM2 promotes p21waf1/cip1 proteasomal turnover independently of ubiquitylation. EMBO J. 2003;22:6365–6377. doi: 10.1093/emboj/cdg600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SN, Hancock AR, Vogel H, Donehower LA, Bradley A. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc Natl Acad Sci U S A. 1998;95:15608–15612. doi: 10.1073/pnas.95.26.15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, de Carvalho P, Kho D, Tai CY, Pierre P, Fink GR, Pellman D. Polyploids require Bik1 for kinetochore-microtubule attachment. J Cell Biol. 2001;155:1173–1184. doi: 10.1083/jcb.200108119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren K, Montes de Oca Luna R, McNeill YB, Emerick EP, Spencer B, Barfield CR, Lozano G, Rosenberg MP, Finlay CA. Targeted expression of MDM2 uncouples S phase from mitosis and inhibits mammary gland development independent of p53. Genes Dev. 1997;11:714–725. doi: 10.1101/gad.11.6.714. [DOI] [PubMed] [Google Scholar]
- Marine JC, Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2010;17:93–102. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- Morgan WF, Corcoran J, Hartmann A, Kaplan MI, Limoli CL, Ponnaiya B. DNA double-strand breaks, chromosomal rearrangements, and genomic instability. Mutat Res. 1998;404:125–128. doi: 10.1016/s0027-5107(98)00104-3. [DOI] [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- Nisitani S, Hosokawa M, Sasaki MS, Yasuoka K, Naiki H, Matsushita T, Takeda T. Acceleration of chromosome aberrations in senescence-accelerated strains of mice. Mutat Res. 1990;237:221–228. doi: 10.1016/0921-8734(90)90003-a. [DOI] [PubMed] [Google Scholar]
- Rayburn E, Zhang R, He J, Wang H. MDM2 and human malignancies: expression, clinical pathology, prognostic markers, and implications for chemotherapy. Curr Cancer Drug Targets. 2005;5:27–41. doi: 10.2174/1568009053332636. [DOI] [PubMed] [Google Scholar]
- Stewart ZA, Leach SD, Pietenpol JA. p21(Waf1/Cip1) inhibition of cyclin E/Cdk2 activity prevents endoreduplication after mitotic spindle disruption. Mol Cell Biol. 1999;19:205–215. doi: 10.1128/mcb.19.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchova Z, Kuffer C. The consequences of tetraploidy and aneuploidy. J Cell Sci. 2008;121:3859–3866. doi: 10.1242/jcs.039537. [DOI] [PubMed] [Google Scholar]
- Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability. Curr Biol. 2010;20:R285–295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker JD, Spruill MD, Ramsey MJ, Director AD, Nath J. Frequency of spontaneous chromosome aberrations in mice: effects of age. Mutat Res. 1999;425:135–141. doi: 10.1016/s0027-5107(99)00036-6. [DOI] [PubMed] [Google Scholar]
- Vitale I, Senovilla L, Jemaa M, Michaud M, Galluzzi L, Kepp O, Nanty L, Criollo A, Rello-Varona S, Manic G, Metivier D, Vivet S, Tajeddine N, Joza N, Valent A, Castedo M, Kroemer G. Multipolar mitosis of tetraploid cells: inhibition by p53 and dependency on Mos. EMBO J. 2010;29:1272–1284. doi: 10.1038/emboj.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Lushnikova T, Odvody J, Greiner TC, Jones SN, Eischen CM. Elevated Mdm2 expression induces chromosomal instability and confers a survival and growth advantage to B cells. Oncogene. 2008;27:1590–1598. doi: 10.1038/sj.onc.1210788. [DOI] [PubMed] [Google Scholar]
- Williams BR, Mirzoeva OK, Morgan WF, Lin J, Dunnick W, Petrini JH. A murine model of Nijmegen breakage syndrome. Curr Biol. 2002;12:648–653. doi: 10.1016/s0960-9822(02)00763-7. [DOI] [PubMed] [Google Scholar]
- Wu X, Avni D, Chiba T, Yan F, Zhao Q, Lin Y, Heng H, Livingston D. SV40 T antigen interacts with Nbs1 to disrupt DNA replication control. Genes Dev. 2004;18:1305–1316. doi: 10.1101/gad.1182804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietkiewicz E, Wojda A, Witt M. Cytogenetic perspective of ageing and longevity in men and women. J Appl Genet. 2009;50:261–273. doi: 10.1007/BF03195682. [DOI] [PubMed] [Google Scholar]