Abstract
OBJECTIVE
Insulin resistance associates with chronic inflammation, and participatory elements of the immune system are emerging. We hypothesized that bacterial elements acting on distinct intracellular pattern recognition receptors of the innate immune system, such as bacterial peptidoglycan (PGN) acting on nucleotide oligomerization domain (NOD) proteins, contribute to insulin resistance.
RESEARCH DESIGN AND METHODS
Metabolic and inflammatory properties were assessed in wild-type (WT) and NOD1/2−/− double knockout mice fed a high-fat diet (HFD) for 16 weeks. Insulin resistance was measured by hyperinsulinemic euglycemic clamps in mice injected with mimetics of meso-diaminopimelic acid–containing PGN or the minimal bioactive PGN motif, which activate NOD1 and NOD2, respectively. Systemic and tissue-specific inflammation was assessed using enzyme-linked immunosorbent assays in NOD ligand–injected mice. Cytokine secretion, glucose uptake, and insulin signaling were assessed in adipocytes and primary hepatocytes exposed to NOD ligands in vitro.
RESULTS
NOD1/2−/− mice were protected from HFD-induced inflammation, lipid accumulation, and peripheral insulin intolerance. Conversely, direct activation of NOD1 protein caused insulin resistance. NOD1 ligands induced peripheral and hepatic insulin resistance within 6 h in WT, but not NOD1−/−, mice. NOD2 ligands only modestly reduced peripheral glucose disposal. NOD1 ligand elicited minor changes in circulating proinflammatory mediators, yet caused adipose tissue inflammation and insulin resistance of muscle AS160 and liver FOXO1. Ex vivo, NOD1 ligand caused proinflammatory cytokine secretion and impaired insulin-stimulated glucose uptake directly in adipocytes. NOD1 ligand also caused inflammation and insulin resistance directly in primary hepatocytes from WT, but not NOD1−/−, mice.
CONCLUSIONS
We identify NOD proteins as innate immune components that are involved in diet-induced inflammation and insulin intolerance. Acute activation of NOD proteins by mimetics of bacterial PGNs causes whole-body insulin resistance, bolstering the concept that innate immune responses to distinctive bacterial cues directly lead to insulin resistance. Hence, NOD1 is a plausible, new link between innate immunity and metabolism.
Insulin resistance is a major predictor and leading cause of type 2 diabetes (1). Intricate links between metabolic and immune responses underlie the disease development (2) and a chronic, low-level inflammation has been associated with insulin resistance (3,4). Innate and adaptive immune systems have been implicated in the proinflammatory responses and have emerged as critical factors in the manifestation of insulin resistance (5–7). It is paramount to define the specific immune elements that mediate inflammation-induced metabolic alterations. This may yield therapeutic approaches to combat insulin resistance by divorcing metabolic and immune responses from one another, which could include blocking the activation-specific innate immune receptors.
The integration of nutrient and pathogen-sensing systems has prompted scrutiny of pattern recognition receptors, such as Toll-like receptors (TLRs) and other danger sensors, in the association of inflammation with insulin resistance (6,8). Although bacterial sepsis causes insulin resistance, seminal work revealed that relatively low circulating levels of endotoxin and factors derived from the gut flora are linked to obesity and dietary-induced insulin resistance (9–11). In particular, chronically elevating circulating lipopolysaccharide (LPS) recapitulates metabolic defects associated with a high-fat diet (HFD), including insulin resistance (10).
Microbiota-derived factors in the circulation prime the innate immune system to augment pathogen removal (12). Surprisingly, an intracellular innate immune sensor for the bacterial cell wall component peptidoglycan (PGN), but not LPS, was involved in these augmented innate immune responses. PGN levels are lower in the circulation of germ-free and antibiotic-treated mice (12). Given the known immunoactive nature of PGN and the fact that antibiotic treatment can attenuate HFD-induced insulin resistance (13), these results position PGN as a potential link between innate immunity and insulin resistance. Nucleotide oligomerization domain (NOD) proteins, specifically NOD1 and NOD2, members of the NOD-like receptor (NLR) family in mammals (14), are currently the only proteins known to propagate inflammatory signals in response to PGN, outside of controversial results implicating TLR2 (15,16). NOD1 detects d-glutamyl-meso-diaminopimelic acid (meso-DAP)–containing PGN found principally in Gram-negative bacteria, whereas NOD2 detects muramyl dipeptide (MDP) present in all bacteria, though more abundant in Gram-positive strains (14,17). Although NLRP3 has recently been implicated as an inflammasome component that provides a connection between inflammation and insulin resistance (18–21), any input by NOD1 or NOD2 to glucose metabolism remains unexplored, except for our recent finding linking NOD2 activation to muscle cell–autonomous inflammation and insulin resistance (22). We hypothesize that NOD proteins are involved in diet-induced metabolic alterations and that direct recognition of specific PGN motifs by NOD1 and/or NOD2 would result in insulin resistance. We show that NOD1/NOD2-null mice are protected from obesity-induced inflammation and peripheral insulin resistance. Conversely, NOD1 activators induced profound acute insulin resistance in mice in the absence of major systemic inflammation, potentially mediated by direct action on hepatocytes and adipocytes and indirect action on skeletal muscle. These observations suggest that intracellular innate immune PGN sensors can trigger insulin resistance.
RESEARCH DESIGN AND METHODS
Materials.
Endotoxin-free NOD1 (c12-iEDAP, Tri-DAP) and NOD2 ligands (L18-MDP) and LPS (Escherichia coli 0111:B4 or Salmonella minnesota) were from Invivogen (San Diego, CA). The MDPA-OCH3 (N-AC-Mur-l-ala-D-Glu-OCH3) used in vivo was from D. Mengin-Lecreulx (University Paris-Sud, Paris, France). FK156 (d-lactyl-l-Ala-γ-d-Glu-meso-DAP-Gly) and FK565 (heptanoyl-γ-d-Glu-meso-DAP-Ala) were from Fujisawa Pharmaceutical Company Ltd. (Osaka, Japan). Specificity and purity of all agonists were ensured as previously described (23). 2-Deoxy-d-[3H]glucose (2-[3H]DG) was from Perkin Elmer (Boston, MA). RT-PCR kit was from Qiagen (Valencia, CA). Cytokines and ELISAs were from R&D Systems (Denver, CO).
Mice procedures.
Protocols were approved by the Animal Ethics Committee (University of Toronto and The Hospital for Sick Children). C57BL/6 mice were from Charles River or Jackson Laboratories. NOD1-deficient mice (Millenium Pharmaceuticals) and NOD2-deficent mice (J.-P. Hugot, Hopital Robert Debré, Paris, France) have been back-crossed to the 10th and 8th generation into the C57BL/6 background, respectively. These mice were used to generate NOD1/2−/− double knockout mice with a minimum of 10 generations of backcrossing into the C57BL/6 background. Male wild-type (WT) and NOD1/2−/− mice were on an HFD (TD.93075; Harlan Laboratories Teklad, Montreal, Quebec, Canada) for 16 weeks, and following insulin tolerance tests (ITTs), mice were anesthetized and the entire gonadal adipose tissue depot and livers were surgically excised and weighed. Histological preparation and staining of 8-μm cryosectioned adipose and liver tissues from WT and NOD1/2−/− mice were performed by the Pathology Laboratory Services at The Hospital for Sick Children. Individual adipocyte area was determined using National Institutes of Health (NIH) ImageJ software. Spontaneous activity and food intake were measured in 7–8-week-old chow-fed WT and NOD1/2−/− mice over 3 days using the comprehensive lab animal monitoring system (CLAMS) (Columbus Instruments, Columbus, OH), as previously described (24). All other mice were fed ad libitum with standard diet (5% fat, Harlan Teklad).
Transcript detection.
Total RNA was prepared from mouse tissues using TRIzol (Invitrogen, Carlsbad, CA), and RT-PCR was performed as previously described (25). The primers were inducible nitric oxide synthase (iNOS) 5′-ccttgttcagctacgccttc-3′ and 5′-aaggccaaacacagcatacc-3′ and arginase 5′-gtgaagaacccacggtctgt-3′ and 5′-ctggttgtcaggggagtgtt-3′. Each transcript was corrected to relative levels of 18S (forward, 5′-aaacggctaccacatccaag-3′; reverse, 5′-ccctcttaatcatggcctca-3′). For Fig. 3, iNOS, arginase, and β-actin (housekeeper) were quantified by real-time PCR (Rotorgene 6000; Qiagen, Toronto, Canada) using Assay-on-Demand gene expression kits, as previously described (26).
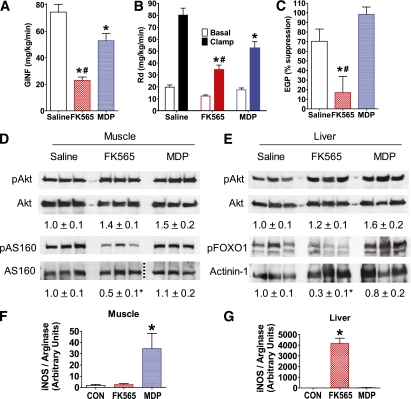
FIG. 3.
NOD1 activation impairs insulin-mediated glucose disposal, suppression of glucose production, and insulin signaling in liver and muscle. A: Glucose infusion rate (GINF) during a hyperinsulinemic-euglycemic clamp in WT mice 6 h after injection of saline, NOD1 ligand (10 μg FK565 i.p.), or NOD2 ligand (100 μg MDP i.p.). B: Peripheral glucose disposal (Rd) during the basal period before insulin infusion (Basal) and during the hyperinsulinemic-euglycemic clamp (Clamp) in WT mice 6 h after injection of a NOD1 or NOD2 ligand. C: Insulin-mediated suppression of EGP during the hyperinsulinemic-euglycemic clamp in WT mice 6 h after injection of a NOD1 or NOD2 ligand. D: Representative immunoblots of postclamp levels of skeletal muscle phospho-Akt (pAkt), total Akt, phospho-AS160 (pAS160), and total AS160, and densitometric quantification (below) of pAkt/Akt and pAS160/AS160. Aliquots of the same samples used to detect pAkt or pAS160 were used to detect Akt or AS160 on parallel gels; the dotted line indicates splicing of the gel to remove unrelated samples. E: Representative immunoblots of postclamp levels of liver pAkt, total Akt, phospho-FOXO1 (pFOXO1), and actinin-1, and densitometric quantification (below) of pAkt/Akt and pFOXO1/actinin-1. Quantitative PCR analysis of iNOS/arginase transcript ratio in (F) muscle and (G) liver 6 h after NOD ligand injection in WT mice. *P < 0.05, compared with saline injection. #P < 0.05, compared with MDP injection. CON, control. Data are means ± SE, n >3 per group. (A high-quality color representation of this figure is available in the online issue.)
ITTs.
ITTs were performed on 6-h fasted WT and NOD1/2−/− mice 16 weeks after an HFD. Fasting blood glucose was determined from tail vein sampling using a glucometer (LifeScan, Milpitas, CA). Mice were injected with insulin (0.85 IU/kg), and glucose was measured until 2-h postinjection for HFD experiments. In order to test the acute effects of NOD protein activation on insulin sensitivity, NOD ligands or LPS were injected (100 µL i.p.) and food was removed 6 h prior to the ITT. Glucose was measured every 10 min for 1 h for acute NOD ligand activation experiments. Area under the curve (AUC) of blood glucose plotted against time for each experiment was calculated using GraphPad software.
Hyperinsulinemic-euglycemic clamps.
Eleven-week-old male C57BL/6 mice were anesthetized with isofluorane and an indwelling catheter was inserted into the right jugular vein 4–6 days before the experiment, which was performed in conscious animals. Saline, NOD1 ligand (10 µg FK565 i.p.), or NOD2 ligand (100 μg MDP i.p.) were injected at −6 h and mice were fasted at −5 h. At −2 h, [3-3H]glucose was infused with a 2.8-µCi bolus, followed by constant infusion at 0.052 µCi/min for the remainder of the experiment (27). During the basal steady-state period (−30 min to 0 h), blood samples for glucose concentration and specific activity assays were obtained every 15 min. At 0 h, a constant insulin infusion of 5 mU ⋅ kg−1 ⋅ min−1 was initiated. Starting at 0 h, plasma glucose was measured (HemoCue Glucose 201, Ängelholm, Sweden) every 5–10 min via tail vein sampling and the mice were clamped at their basal glucose concentrations (isoglycemic) using a variable infusion of 50% dextrose, which contained 53 µCi/g [3-3H]glucose. Steady state was achieved during the hyperinsulinemic-euglycemic clamp (90 min to 2 h), and blood samples were also obtained every 15 min during this period. Radioactivity from [3-3H]glucose in plasma and in infusates was measured, and Steele non–steady-state equation was used to calculate glucose turnover, taking into account the additional tracer contained in the exogenous glucose infusion (27,28).
Cytokines.
Serum circulating proinflammatory chemokine (C-X-C motif) ligand 1 (CXCL1) was determined in 6-h fasted WT and NOD1/2−/− mice 16 weeks into the HFD. Mice were injected with NOD ligands or LPS and tumor necrosis factor α (TNFα) and CXCL1 in sera (2 h postinjection), and CXCL9 and CXCL10 adipose tissue lysates (6 h postinjection) were determined by ELISA.
Adipose tissue lysates were prepared in ice-cold PBS containing 1% Triton-X100 and protease inhibitor cocktail (1:500; P8340; Sigma, St. Louis, MO) and hand homogenized. Equal amounts of lysate protein were exposed to Proteome Profiler. Duplicate immunoreactive spots were quantified using NIH ImageJ software.
Cell cultures.
Primary hepatocytes were isolated using an in vivo collagenase infusion, as previously described (29). Kupffer cells were essentially absent from our cultures, given that qPCR for specific markers (F4/80 and CD68) only constituted 0.04% of the signal compared with macrophages. Primary hepatocytes were cultured for 18 h in William’s medium containing 0.1% FBS, interleukin (IL)-1β (100 IU), interferon-γ (100 IU), and 200 μmol/L l-glutamine and stimulated with FK565 (5 μg/mL) or iEDAP (120 ng/mL), and a subset were stimulated with insulin (10 nmol/L for 2 min). Nitrite released from hepatocytes into phenol red–free media was quantified using a Greiss reaction kit according to the manufacturer’s instructions (G7921; Invitrogen). 3T3-L1 fibroblasts were differentiated into adipocytes (30) and exposed to NOD ligands for 18 h for measurement of cytokines.
Glucose uptake.
3T3-L1 adipocytes were treated with FK565 (5 μg/mL) or L-18 MDP (1 μg/mL) during serum starvation for 3 h, and a subset of cells was stimulated with 100 nmol/L insulin for 20 min. Glucose uptake was assessed for 5 min in HEPES-buffered saline (140 mmol/L NaCl, 20 mmol/L HEPES, 5 mmol/L KCl, 2.5 mmol/L MgSO4, 1 mmol/L CaCl2 [pH 7.4]) containing 5 μmol/L 2-deoxyglucose (2-DG) (0.25 µCi/ml 2-[3H]DG) at room temperature. Nonspecific uptake was determined with 25 μmol/L cytochalasin B.
Immunoblotting.
Mice were anesthetized after the clamp, and tibialis anterior muscles and liver were rapidly excised and frozen in liquid nitrogen. Tissue homogenates and hepatocyte lysates were prepared as previously described (29,31) and equal amounts of protein were immunoblotted with antibodies to Akt and AS160, phospho-Akt (Ser473), phospho-AS160 (Thr642), and phospho-FOXO1 (Ser256) from Cell Signaling Technology (Danvers, MA) or anti-actinin-1 from Sigma.
Statistical analysis.
Values are mean ± SE, unless specified. Statistical significance was determined by ANOVA with Tukey or Dunnet post hoc test, as appropriate. P < 0.05 was considered significant. Since adipocyte area was not normally distributed, significance was accepted when the geometric mean was outside the 95% CI.
RESULTS
Absence of NOD1 and NOD2 attenuates diet-induced inflammation and insulin intolerance.
We first established a role for NOD proteins in metabolic disease by demonstrating that NOD1/2−/− double knockout mice are more insulin tolerant compared with WT mice after 16 weeks on HFD, evinced by a reduced AUC of blood glucose during an ITT (P < 0.05; Fig. 1A). After the HFD, NOD1/2−/− mice had lower gonadal adipose and liver masses compared with WT mice (P < 0.05; Fig. 1B) and lower levels of CXCL1 (P < 0.05; Fig. 1C). NOD1/2−/− mice had reduced adipocyte size (P < 0.05; Fig. 1D and E) and less adipose tissue inflammation shown by a lower ratio of iNOS to arginase transcripts (P < 0.05; Fig. 1F). NOD1/2−/− mice had a strikingly lower level of steatosis and lipid accumulation in the liver assessed by hematoxylin and eosin (H&E) and Oil-Red-O histology (Fig. 1G) and showed less liver inflammation, given a lower iNOS to arginase transcript ratio (P < 0.05; Fig. 1H). Measurement of a number of transcripts demonstrated that both adipose tissue and liver had attenuated inflammatory and metabolic disturbances after the HFD (Supplementary Fig. 1). These results implicate NOD1 and/or NOD2 in inflammatory and metabolic responses throughout a physiologically relevant model of insulin resistance. However, NOD1/2−/− mice also had increased rates of spontaneous activity and reduced food intake compared with WT mice (Fig. 1I). Hence, examining these mice after an HFD cannot discern between dietary/energy and bacterial factors to cause NOD activation. Therefore, to directly test the hypothesis that NOD1 or NOD2 activation may cause insulin resistance, we took advantage of well-characterized PGN derivatives that are specific for NOD1 or NOD2 (32,33). This further allowed us to explore whether insulin resistance can arise acutely without the long-term phenotypic adaptations that might occur in the absence of NOD proteins. However, a limitation of this approach is that the improved insulin tolerance in NOD1/2−/− mice cannot be directly linked to effects of NOD1 or NOD2 activators, and mechanisms other than PGN-derived NOD-mediated insulin intolerance in muscle, liver, or fat might contribute to the phenotype of these mice.
FIG. 1.
NOD1/2−/− mice are protected from HFD-induced inflammation, lipid accumulation, and insulin intolerance in peripheral tissues. WT and NOD1/2−/− double knockout mice were fed an HFD for 16 weeks, upon which the following parameters were measured. Relative changes in (A) blood glucose and (inset) AUC during an ITT. B: Mass of gonadal adipose and liver tissue. C: Serum levels of CXCL1. D and E: Gonadal adipose tissue staining with H&E and quantification of adipocyte size. F: iNOS/arginase transcript ratio in gonadal adipose tissue. G: Oil-Red-O and H&E stained liver. H: iNOS/arginase transcript ratio in liver. In addition, (I) spontaneous activity levels and food intake were measured in 7–8-week-old WT and NOD1/2−/− chow-fed mice, and results from the 12-h dark cycle are shown. *P < 0.05, compared with WT mice. Data are means ± SE, n >4 per group, except for adipocyte area, where individual data points and the median are shown. See also Supplementary Fig. 1. (A high-quality digital representation of this figure is available in the online issue.)
NOD1 ligands induce whole-body insulin resistance.
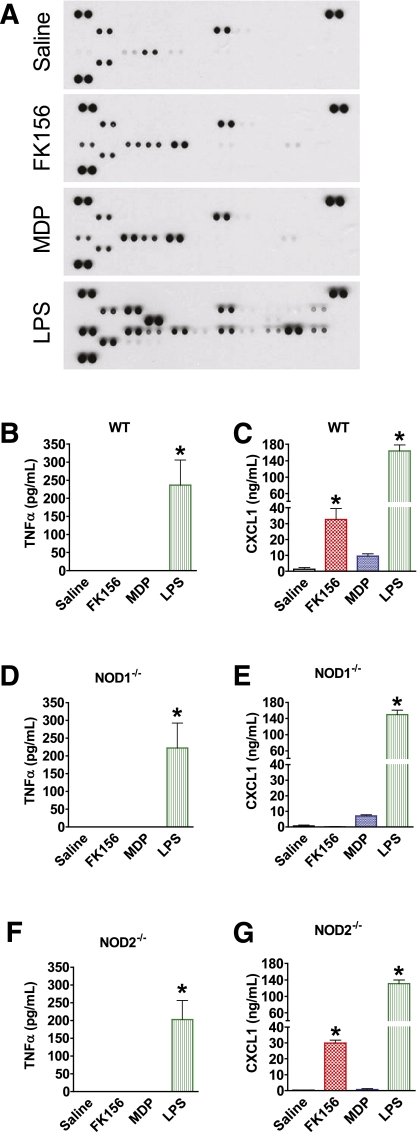
Injection of the NOD1 ligand FK156 into mice provoked insulin intolerance, revealed by a 39% increase in the AUC during the ITT (P < 0.05; Supplementary Fig. 2). NOD1 activation also caused a 45% drop in fasting blood glucose 6 h after injection, an effect that was similar to that of either low (0.025 mg/kg) or high (2.0 mg/kg) doses of LPS (P < 0.05; Supplementary Fig. 2), yet the low dose of LPS did not cause insulin intolerance (Supplementary Fig. 2). The findings using low-dose LPS illustrate that NOD1 ligand-mediated insulin intolerance was likely not driven by this reduction in blood glucose. In addition, the fasting blood glucose concentration after NOD1 ligand injection remained above the threshold for enhanced counterregulatory hormone release in the mouse (34). We confirmed the dose of a NOD1 ligand required to induce insulin intolerance in vivo and found that several NOD1 ligands caused insulin intolerance, which waned by 24-h postinjection (Supplementary Figs. 3 and 4). In contrast, NOD2 activation by MDP failed to alter fasting blood glucose or insulin tolerance when assessed by ITT (Supplementary Fig. 2), even when tested at doses up to 500 μg (data not shown).
Given that NOD1 (and NOD2) transcripts were found in metabolic and immune cells and tissues (Supplementary Fig. 2), we explored if the PGN-induced metabolic effects were mediated by NOD1. NOD1−/− mice did not display insulin intolerance in response to NOD1 ligand (Fig. 2B) or reduce fasting blood glucose (data not shown), responses that were manifested in matched WT mice (P < 0.05; Fig. 2A). In NOD2−/− mice, NOD1 activation lowered fasting blood glucose by 45% (data not shown) and increased the AUC during an ITT by 35%, indicating that the NOD2 protein is not involved in NOD1 ligand–mediated insulin intolerance (P < 0.05; Fig. 2C). All mouse genotypes responded similarly to a high dose of LPS (2.0 mg/kg), with a significant reduction in fasting blood glucose (not shown) and insulin intolerance during the ITT (P < 0.05; Fig. 2A–C), indicating that acute LPS-induced insulin resistance is independent of NOD proteins.
FIG. 2.
Ligand specificity of NOD activators that induce insulin intolerance. Relative changes in (A) blood glucose and (inset) AUC during an ITT 6 h after injection of FK156 (100 μg i.p.), and high (2.0 mg/kg LPS i.p.) or low (0.05 mg/kg LPS i.p.) doses of endotoxin in WT mice. Relative changes in (B) blood glucose and AUC during an ITT 6 h after injection of each treatment in NOD1−/− mice. Relative changes in (C) blood glucose and AUC during an ITT 6 h after each treatment in NOD2−/− mice. *P < 0.05, compared with saline injection. Data are means ± SE, n ≥3 per group. (A high-quality color representation of this figure is available in the online issue.)
To determine whether NOD1 ligand–mediated insulin resistance involved peripheral (muscle or adipose tissue) or hepatic components, we performed hyperinsulinemic-euglycemic clamps on WT mice after ligand injection. The glucose infusion rate during the last 30 min of the clamp was markedly suppressed (by 71%, P < 0.05) 6 h postinjection of NOD1 ligand, and was more modestly reduced (by 31%, P < 0.05) by the NOD2 ligand (Fig. 3A). NOD1 and NOD2 activation decreased peripheral glucose disposal (Rd), the NOD1 ligand being significantly more effective, without correcting for basal glucose (P < 0.05; Fig. 3B). Insulin suppression of endogenous glucose production (EGP) during the clamp was only impaired upon NOD1 activation (P < 0.05; Fig. 3C). In contrast to the ITT results, the more sensitive clamp revealed that NOD2 ligand reduced peripheral glucose utilization. Given that skeletal muscle is responsible for the majority of Rd, this result is consistent with our recent finding that NOD2 ligand can directly elicit insulin resistance on muscle cells via cell-autonomous innate immune responses (22), but this action alone may not suffice to cause more overt insulin intolerance required for detection by ITT.
Skeletal muscle excised after the hyperinsulinemic-euglycemic clamp of NOD1 ligand-challenged mice showed a marked reduction in insulin-dependent phosphorylation of AS160 (P < 0.05; Fig. 3D), a key determinant of glucose uptake (35). The insulin-dependent phosphorylation of muscle Akt was not altered by injection of either NOD1 or NOD2 ligands (Fig. 3D), consistent with the notion that net phosphorylation of this protein may not necessarily correlate with insulin sensitivity (36) or that prolonged hyperinsulinemia during the clamp may mask such effects. Liver tissue excised from NOD1 ligand–challenged mice after the clamp also showed marked insulin resistance, evinced by the decrease in FOXO1 phosphorylation (P < 0.05; Fig. 3E). Injection of WT mice with NOD2 ligand caused muscle inflammation, demonstrated by an increased iNOS/arginase transcript ratio (P < 0.05; Fig. 3F). This result is consistent with our previous results showing muscle cell–autonomous inflammation and insulin resistance via NOD2 but not NOD1 (22). NOD1 ligand–mediated hepatic insulin resistance corresponded with liver inflammation, demonstrated by an increased iNOS/arginase transcript ratio, which did not occur after injection of NOD2 ligand (P < 0.05; Fig. 3G).
NOD ligands cause low-level elevation in serum cytokines.
Because NOD1 and NOD2 typically activate inflammatory pathways leading to cytokine production from macrophages, and cytokines/chemokines such as TNFα and chemokine CXCL1 have been causally linked to impaired insulin sensitivity (37,38), we searched for changes in specific circulating cytokines that might precede or accompany NOD ligand–mediated insulin resistance. Although several rose modestly, none of this subset of the 40 cytokines/chemokines assayed in serum of mice challenged with NOD1 or NOD2 ligands reached the levels induced by low-dose LPS injection (Fig. 4A; Supplementary Fig. 5). Of note, IL1α, IL1β, and TNFα were not found to be elevated, and IL-6 was not higher, compared with low-dose LPS (Supplementary Fig. 5). Quantification by ELISA confirmed that NOD1 ligand injection did not alter circulating TNFα levels, which rose in response to low-dose LPS (0.05 mg/kg), despite the latter not causing insulin resistance (P < 0.05; Fig. 4B). NOD1 activation did elevate serum CXCL1 levels in WT (P < 0.05; Fig. 4C), but not NOD1−/−, mice (Fig. 4E). By comparison, low-dose LPS elicited a marked increase in CXCL1 serum levels (P < 0.05; Fig. 4C) independently of NOD1 or NOD2 expression (P < 0.05; Fig. 4E and G).
FIG. 4.
NOD ligand–mediated circulating cytokine responses. A: Simultaneous analysis of the relative levels of 40 serum cytokines in WT mice 2 h after injection with saline, a NOD1 ligand (100 μg FK156 i.p.), a NOD2 ligand (100 μg MDP i.p.), or low dose of endotoxin (0.05 mg/kg of LPS). Quantification of serum levels of (B) TNFα and (C) CXCL1 by ELISA 2 h after injection of a NOD1 or NOD2 ligand or a low dose of LPS in WT mice. Quantification of serum levels of (D) TNFα and (E) CXCL1 in NOD1−/− mice. Quantification of serum levels of (F) TNFα and (G) CXCL1 in NOD2−/− mice. *P < 0.05, compared with saline injection. Data are means ± SE, n = 3 per group. See also Supplementary Fig. 5. A complete map of the cytokines corresponding to immunoreactive spots for the Mouse Cytokine Antibody Array, panel A (catalog no. ARY006) is available from R&D Systems. (A high-quality color representation of this figure is available in the online issue.)
Elevated circulating free fatty acids (FFAs) are a hallmark of many insulin-resistant states, and even brief periods of elevated circulating lipids (i.e., 6–7-h lipid infusion) can result in insulin resistance (39,40). NOD1 or NOD2 ligands did not alter circulating FFA during the 6-h period preceding the assessments of insulin sensitivity (Supplementary Fig. 6), indicating that the insulin resistance cannot be ascribed to the concomitant hypoglycemia that might have potentially acted through a counterregulatory mechanism involving elevation in FFA (40).
NOD1 ligand provokes adipose tissue inflammation and has direct actions on adipocytes.
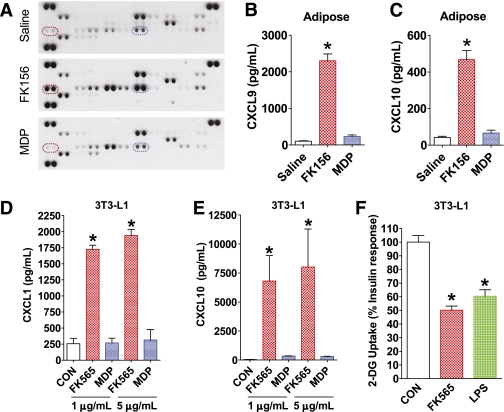
Analysis of cytokine/chemokine levels in adipose tissue after NOD1 or NOD2 activation in vivo revealed that NOD1 activation preferentially increased levels of CXCL1, CXCL9, CXCL10, and CCL5, whereas NOD2 activation elevated TREM-1, and neither ligand increased TNFα (Fig. 5A; Supplementary Fig. 7). By ELISA-based quantification, CXCL9 and CXCL10 rose by >23- and 11-fold, respectively (P < 0.05; Fig. 5B and C), exclusively in response to NOD1 activation.
FIG. 5.
NOD protein–induced inflammation in adipose tissue and NOD1-mediated inflammation and insulin resistance directly in adipocytes. A: Simultaneous analysis of 40 cytokines in adipose tissue from WT mice 6 h after injection with saline, a NOD1 ligand (100 μg FK156 i.p.), or a NOD2 ligand (100 μg MDP i.p.). Highlighted by dashed ellipses are duplicate immunoreactive spots for CXCL9 (red) and CXCL10 (blue). Quantification of (B) CXCL9 and (C) CXCL10 in adipose tissue by ELISA, 6 h after injection of WT mice with saline, a NOD1 ligand (100 μg FK156 i.p.), or a NOD2 ligand (100 μg MDP i.p.). Quantification of (D) CXCL1 and (E) CXCL10 secreted from 3T3-L1 adipocytes after exposure to different doses of NOD1 ligand (FK565) or NOD2 ligand (L18-MDP) for 18 h. F: Insulin-mediated glucose uptake in 3T3-L1 adipocytes after direct exposure to a NOD1 ligand (5 μg/mL of FK565) or a high dose of LPS (100 ng/mL) for 3 h. *P < 0.05, compared with saline injection or to vehicle control (CON). Data are means ± SE, n >3 per group. See also Supplementary Fig. 7. A complete map of the cytokines corresponding to immunoreactive spots for the Mouse Cytokine Antibody Array, panel A (catalog no. ARY006) is available from R&D Systems. (A high-quality color representation of this figure is available in the online issue.)
Direct treatment of 3T3-L1 adipocyte cultures with NOD1 ligand elicited inflammation defined by 7- and 150-fold increases in CXCL1 and CXCL10 production, respectively, effects that were not seen with NOD2 ligand (P < 0.05; Fig. 5D and E). Moreover, direct exposure to NOD1 ligand rendered 3T3-L1 adipocytes insulin resistant, provoking a 50% drop in insulin-dependent uptake of 2-DG within 3 h, similar in magnitude to the effect of a high dose of LPS (100 ng/mL) (P < 0.05; Fig. 5F). The responses of cultured adipocytes sharply contrast with our recent observations that NOD2, but not NOD1, ligands elicit inflammation and insulin resistance in a muscle cell line (22). Hence, the NOD1-induced drop in Rd observed in vivo during the clamp might be due to an indirect rather than direct action on skeletal muscle (see DISCUSSION). The inherent limitations of any cultured cell model should be considered, which in the case of 3T3-L1 adipocytes may include expressing relatively low NOD2 transcript. However, our previous results suggest that levels of NOD transcripts do not necessarily predict cell-autonomous responsiveness to ligand-mediated effects (22).
NOD1 ligand directly causes hepatocyte inflammation and insulin resistance.
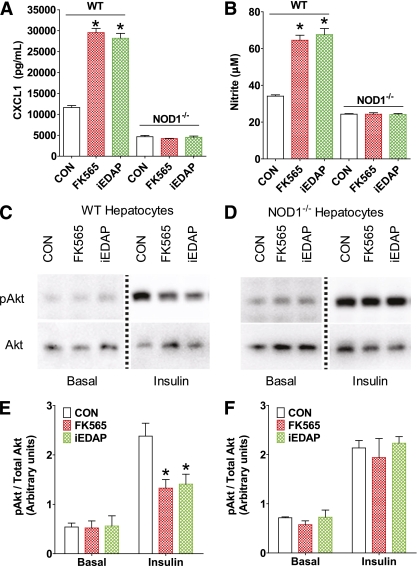
Given the potent suppressive effect of NOD1 ligand on EGP during the clamp in vivo, we explored effects on isolated primary hepatocytes derived from WT and NOD1−/− mice. The NOD1 ligands FK565 and iEDAP elevated the secretion of the inflammatory chemokine CXCL1 by over twofold (P < 0.05) in hepatocytes from WT, but not NOD1−/−, mice (Fig. 6A). NOD1 ligands also increased nitrite production (an index of iNOS activity) in hepatocytes from WT (P < 0.05), but not NOD1−/−, mice, providing further evidence of NOD1-mediated inflammation in these cells (Fig. 6B). These observations are consistent with very recent findings demonstrating that hepatocytes mount an inflammatory response to NOD1, but not NOD2, ligands (41). Importantly, the NOD1 ligand directly suppressed insulin-stimulated Akt phosphorylation by ∼40% in primary hepatocytes of WT mice (P < 0.05), but not from NOD1−/− mice (Fig. 6C–F). These observations indicate that direct NOD1 activation in hepatocytes can evoke insulin resistance.
FIG. 6.
NOD1-mediated hepatocyte inflammation and insulin resistance. Quantification of (A) CXCL1 secretion and (B) nitrite from primary hepatocytes of WT and NOD1−/− mice after exposure to NOD1 ligands (5 μg/mL of FK565 or 100 ng/mL of c12-iEDAP) for 18 h. Representative immunoblots of phospho-Akt (pAkt) and total Akt from primary hepatocytes of (C) WT and (D) NOD1−/− mice. The dotted line indicates splicing of the gel to remove unrelated samples. Densitometric quantification of pAkt/Akt in (E) WT and (F) NOD1−/− primary hepatocytes under basal and insulin-stimulated (100 nmol/L for 2 min) conditions. *P < 0.05, compared with vehicle control (CON). Data are means ± SE, n ≥3 per group. (A high-quality color representation of this figure is available in the online issue.)
DISCUSSION
Given the integration of pathogen sensing and metabolic responses and the role of inflammation in insulin resistance, it is paramount to identify the factors and sensors that evoke insulin resistance. We show that NOD1/2−/− mice are more insulin tolerant and have reduced lipid accumulation and lower inflammation in adipose and hepatic tissues after an HFD. Given the complicated phenotype of NOD1/2−/− mice (including altered activity levels and food intake), we utilized PGN derivatives that are specific for NOD1 or NOD2 to determine if acute activation of NODs is sufficient to cause insulin resistance. Meso-DAP–containing derivatives typical of PGN present in most Gram-negative bacteria acutely induce insulin resistance via NOD1. In contrast, the minimum bioactive PGN motif that activates NOD2 (MDP) only modestly reduced insulin-mediated peripheral glucose disposal. These findings are in line with the notion that diet-induced changes in colonizing microbiota, such as a rise in the Gram-negative to Gram-positive ratio, may be an important factor in precipitating adiposity and insulin resistance (10,13). Given that bacterial components are able to penetrate mucosal barriers to promote metabolic aberrations, the results identify a constituent of Gram-negative bacteria additional to LPS as a potential trigger for metabolic disease (12,42). Future investigation will explore the potential that lipid metabolites are sensed by NOD proteins (43).
TLR4 has been proposed as an immune sensor that can alter metabolism, and mice with defective TLR4 signaling are partially protected from insulin resistance induced by HFD and acute lipid infusion (6). We show that ablation of NOD1 and NOD2 also attenuates HFD-induced insulin resistance and hepatic lipid accumulation. Both adipose and liver tissues showed reduced inflammation in NOD1/2−/− mice after the HFD, positioning NOD proteins as a link between the immune and metabolic systems. Our results and recent evidence implicating the inflammasome responses in insulin resistance demonstrate that TLRs are not the only innate immune component that links inflammation and metabolic outcomes (19–21). Focusing on bacterial cues instigating insulin resistance, we show that acute injection of meso-DAP–containing PGN derivatives causes insulin resistance via NOD1. This obviously occurs in the absence of changes in weight or adiposity, indicating that NOD1 activation per se can rapidly lead to insulin resistance.
The results presented suggest that the whole-body insulin resistance is not likely caused by a profound change in the circulating proinflammatory signature, but may be linked to specific effects on metabolically relevant tissues (adipose, muscle, and liver). NOD1 ligand (but not NOD2 ligand) administered in vivo distinctly augmented adipose tissue CXCL1, CXCL9, and CXCL10, showing that this metabolic tissue is a PGN sensor. The rise in adipose CXCL1 in response to NOD1 ligand is interesting because hematopoietic deletion of CXCR2 protected mice from diet-induced obesity (37).
Given that 3T3-L1 adipocytes directly respond to NOD1 ligand by producing significant levels of CXCL1 and CXCL10, our results suggest that macrophages present in the adipose tissue are unlikely to be the sole responders to NOD1 ligand in vivo. This builds upon findings that a crude preparation of Staphylococcus aureus–derived PGN elevated inflammatory signals and suppressed transcript levels of adiponectin receptors in 3T3-L1 adipocytes (44). However, the relative impurity of that PGN preparation (i.e., contaminating lipoproteins) prevents ascribing the response to this innate immune receptor with confidence (16).
In contrast to these direct actions of NOD1 ligand on 3T3-L1 adipocytes, where NOD1 activation reduced insulin-stimulated glucose uptake, muscle cells in culture only became insulin resistant when exposed to NOD2 ligand (22). These findings point to important differences in NOD protein activation between muscle and adipose cells. However, skeletal muscle participates in the whole-body insulin resistance provoked by injection of NOD1 ligand, reflected in the reduction in Rd (and AS160 phosphorylation) recorded during euglycemic-hyperinsulinemic clamps. These contrasting observations suggest that, in vivo, skeletal muscle responds to NOD1 activation through additional mechanisms, potentially including crosstalk with adipose tissue (adipocytes and/or macrophages) or liver. While dissecting all of the potential contributions from those tissues to the reduction in Rd is beyond the scope of this study, we did explore whether factors produced by NOD1-activated primary macrophages might render muscle cells resistant to insulin. Although conditioned medium from palmitate-treated macrophages reduced the insulin response of glucose uptake in L6 muscle cells (25), conditioned medium from NOD1 ligand–treated macrophages had no effect despite elevations in proinflammatory factors emanating from treated macrophages (Supplementary Fig. 8). Hence, neither NOD1 ligand itself nor factors secreted by NOD1 ligand–treated macrophages suffices to provoke muscle insulin resistance in cell culture. Future studies will explore the possible participation of changes in adipose or liver-derived factors, including RBP4, myostatin, selenoprotein P, or adiponectin, in the genesis of muscle insulin resistance in vivo.
Anatomically, the liver is likely to monitor inflammatory bacteria or bacterial components that penetrate the gut and enter the portal circulation, in addition to those present in the systemic circulation. Given that NOD1 activation caused hepatocyte-autonomous inflammation and impaired insulin-dependent signaling, we speculate that, in vivo, hepatocytes (in cooperation with input from other resident hepatic cells) act as PGN sensors and responders to both innate immune and metabolic cues and outcomes.
In summary, NOD1/2−/− double knockout mice were protected from HFD-induced insulin intolerance, lipid accumulation, and inflammation in adipose and liver. Strikingly, direct activation of the NOD1 component of the innate immune system precipitated acute and profound insulin resistance in vivo, involving changes in both glucose clearance and glucose production. Adipocytes and hepatocytes appear to be direct targets of NOD1-mediated changes in metabolism, but contribution by myeloid cells is not excluded. Further, a connection between in vivo inflamed adipose tissue and the affected primary determinants of blood glucose (muscle and liver) is a distinct possibility. Overall, the results of this study position the meso-DAP PGN as a potential trigger for metabolic disease and demonstrate that NOD1 is a possible link between innate immunity and metabolism.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant to A.K. from the Canadian Diabetes Association (CDA) and the Canadian Institutes of Health Research (CIHR) (MOP 12601). A.K. holds a Tier I Canada Research Chair. D.J.P., G.R.S., and A.G. were supported by grants from CIHR. D.J.P. is a Howard Hughes Medical Institute International Scholar. J.D.S. was supported by fellowships from CIHR, CDA, and the Degroote Academic Fellowship (McMaster University). A.K.T. was supported by a fellowship from the Better Opportunities for Young Scientists in Chosen Areas of Science and Technology. J.G.M. was supported by fellowships from the Fondation pour la Recherche Medicale (France), the Fundação para a Ciência e Tecnologia (Portugal), and CIHR. S.P. was supported by a Canada Graduate Scholarships Award from CIHR. G.R.S. holds a Tier II Canada Research Chair.
No potential conflicts of interest relevant to this article were reported.
J.D.S. researched data, edited the manuscript, derived the hypothesis, and wrote the manuscript. A.K.T., J.G.M., and S.P. researched data. P.J.B. researched data, derived the hypothesis, and wrote the manuscript. M.D.F. and Z.L. researched data. G.R.S., A.G., and D.J.P. edited the manuscript. A.K. derived the hypothesis and wrote the manuscript.
The authors thank Dr. Dominique Mengin-Lecreulx (University of Paris) for the MDPA-OCH3 and Loretta Lam (University of Toronto) for technical assistance.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-0004/-/DC1.
REFERENCES
- 1.Pratley RE, Weyer C. Progression from IGT to type 2 diabetes mellitus: the central role of impaired early insulin secretion. Curr Diab Rep 2002;2:242–248 [DOI] [PubMed] [Google Scholar]
- 2.Kolb H, Mandrup-Poulsen T. The global diabetes epidemic as a consequence of lifestyle-induced low-grade inflammation. Diabetologia 2010;53:10–20 [DOI] [PubMed] [Google Scholar]
- 3.Hotamisligil GS, Erbay E. Nutrient sensing and inflammation in metabolic diseases. Nat Rev Immunol 2008;8:923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinberg GR. Inflammation in obesity is the common link between defects in fatty acid metabolism and insulin resistance. Cell Cycle 2007;6:888–894 [DOI] [PubMed] [Google Scholar]
- 5.Lumeng CN, Maillard I, Saltiel AR. T-ing up inflammation in fat. Nat Med 2009;15:846–847 [DOI] [PubMed] [Google Scholar]
- 6.Shi H, Kokoeva MV, Inouye K, Tzameli I, Yin H, Flier JS. TLR4 links innate immunity and fatty acid-induced insulin resistance. J Clin Invest 2006;116:3015–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hosoi T, Yokoyama S, Matsuo S, Akira S, Ozawa K. Myeloid differentiation factor 88 (MyD88)-deficiency increases risk of diabetes in mice. PLoS ONE 2010;5:e12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamura T, Furuhashi M, Li P, et al. Double-stranded RNA-dependent protein kinase links pathogen sensing with stress and metabolic homeostasis. Cell 2010;140:338–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vijay-Kumar M, Aitken JD, Carvalho FA, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 2010;328:228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cani PD, Amar J, Iglesias MA, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007;56:1761–1772 [DOI] [PubMed] [Google Scholar]
- 11.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006;444:1027–1031 [DOI] [PubMed] [Google Scholar]
- 12.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med 2010;16:228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cani PD, Bibiloni R, Knauf C, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 2008;57:1470–1481 [DOI] [PubMed] [Google Scholar]
- 14.Carneiro LA, Magalhaes JG, Tattoli I, Philpott DJ, Travassos LH. Nod-like proteins in inflammation and disease. J Pathol 2008;214:136–148 [DOI] [PubMed] [Google Scholar]
- 15.Müller-Anstett MA, Müller P, Albrecht T, et al. Staphylococcal peptidoglycan co-localizes with Nod2 and TLR2 and activates innate immune response via both receptors in primary murine keratinocytes. PLoS ONE 2010;5:e13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Travassos LH, Girardin SE, Philpott DJ, et al. Toll-like receptor 2-dependent bacterial sensing does not occur via peptidoglycan recognition. EMBO Rep 2004;5:1000–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carneiro LAM, Travassos LH, Philpott DJ. Innate immune recognition of microbes through Nod1 and Nod2: implications for disease. Microbes Infect 2004;6:609–616 [DOI] [PubMed] [Google Scholar]
- 18.Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol 2010;11:136–140 [DOI] [PubMed] [Google Scholar]
- 19.Vandanmagsar B, Youm Y-H, Ravussin A, et al. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat Med 2011;17:179–188 [DOI] [PMC free article] [PubMed]
- 20.Stienstra R, Joosten LAB, Koenen T, et al. The inflammasome-mediated caspase-1 activation controls adipocyte differentiation and insulin sensitivity. Cell Metab 2010;12:593–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wen H, Gris D, Lei Y, et al. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat Immunol 2011;12:408–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamrakar AK, Schertzer JD, Chiu TT, et al. NOD2 activation induces muscle cell-autonomous innate immune responses and insulin resistance. Endocrinology 2010;151:5624–5637 [DOI] [PubMed] [Google Scholar]
- 23.Magalhaes JG, Philpott DJ, Nahori MA, et al. Murine Nod1 but not its human orthologue mediates innate immune detection of tracheal cytotoxin. EMBO Rep 2005;6:1201–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dzamko N, van Denderen BJW, Hevener AL, et al. AMPK β1 deletion reduces appetite, preventing obesity and hepatic insulin resistance. J Biol Chem 2010;285:115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samokhvalov V, Bilan PJ, Schertzer JD, Antonescu CN, Klip A. Palmitate- and lipopolysaccharide-activated macrophages evoke contrasting insulin responses in muscle cells. Am J Physiol Endocrinol Metab 2009;296:E37–E46 [DOI] [PubMed] [Google Scholar]
- 26.Sachithanandan N, Fam BC, Fynch S, et al. Liver-specific suppressor of cytokine signaling-3 deletion in mice enhances hepatic insulin sensitivity and lipogenesis resulting in fatty liver and obesity. Hepatology 2010;52:1632–1642 [DOI] [PubMed] [Google Scholar]
- 27.Park E, Wong V, Guan X, Oprescu AI, Giacca A. Salicylate prevents hepatic insulin resistance caused by short-term elevation of free fatty acids in vivo. J Endocrinol 2007;195:323–331 [DOI] [PubMed] [Google Scholar]
- 28.Finegood DT, Bergman RN, Vranic M. Estimation of endogenous glucose production during hyperinsulinemic-euglycemic glucose clamps. Comparison of unlabeled and labeled exogenous glucose infusates. Diabetes 1987;36:914–924 [DOI] [PubMed] [Google Scholar]
- 29.Fullerton MD, Hakimuddin F, Bonen A, Bakovic M. The development of a metabolic disease phenotype in CTP:phosphoethanolamine cytidylyltransferase-deficient mice. J Biol Chem 2009;284:25704–25713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sweeney G, Garg RR, Ceddia RB, et al. Intracellular delivery of phosphatidylinositol (3,4,5)-trisphosphate causes incorporation of glucose transporter 4 into the plasma membrane of muscle and fat cells without increasing glucose uptake. J Biol Chem 2004;279:32233–32242 [DOI] [PubMed] [Google Scholar]
- 31.Steinberg GR, O'Neill HM, Dzamko NL, et al. Whole-body deletion of AMPK β2 reduces muscle AMPK and exercise capacity. J Biol Chem 2010;285:37198–37209 [DOI] [PMC free article] [PubMed]
- 32.Girardin SE, Boneca IG, Carneiro LAM, et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science 2003;300:1584–1587 [DOI] [PubMed] [Google Scholar]
- 33.Magalhaes JG, Fritz JH, Le Bourhis L, et al. Nod2-dependent Th2 polarization of antigen-specific immunity. J Immunol 2008;181:7925–7935 [DOI] [PubMed] [Google Scholar]
- 34.McGuinness OP, Ayala JE, Laughlin MR, Wasserman DH. NIH experiment in centralized mouse phenotyping: the Vanderbilt experience and recommendations for evaluating glucose homeostasis in the mouse. Am J Physiol Endocrinol Metab 2009;297:E849–E855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kramer HF, Witczak CA, Taylor EB, Fujii N, Hirshman MF, Goodyear LJ. AS160 regulates insulin- and contraction-stimulated glucose uptake in mouse skeletal muscle. J Biol Chem 2006;281:31478–31485 [DOI] [PubMed] [Google Scholar]
- 36.Ng Y, Ramm G, Burchfield JG, Coster ACF, Stöckli J, James DE. Cluster analysis of insulin action in adipocytes reveals a key role for Akt at the plasma membrane. J Biol Chem 2010;285:2245–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neels JG, Badeanlou L, Hester KD, Samad F. Keratinocyte-derived chemokine in obesity: expression, regulation, and role in adipose macrophage infiltration and glucose homeostasis. J Biol Chem 2009;284:20692–20698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uysal KT, Wiesbrock SM, Marino MW, Hotamisligil GS. Protection from obesity-induced insulin resistance in mice lacking TNF-alpha function. Nature 1997;389:610–614 [DOI] [PubMed] [Google Scholar]
- 39.Lewis GF, Carpentier A, Adeli K, Giacca A. Disordered fat storage and mobilization in the pathogenesis of insulin resistance and type 2 diabetes. Endocr Rev 2002;23:201–229 [DOI] [PubMed] [Google Scholar]
- 40.Lucidi P, Rossetti P, Porcellati F, et al. Mechanisms of insulin resistance after insulin-induced hypoglycemia in humans: the role of lipolysis. Diabetes 2010;59:1349–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott MJ, Chen C, Sun Q, Billiar TR. Hepatocytes express functional NOD1 and NOD2 receptors: a role for NOD1 in hepatocyte CC and CXC chemokine production. J Hepatol 2010;53:693–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amar J, Burcelin R, Ruidavets JB, et al. Energy intake is associated with endotoxemia in apparently healthy men. Am J Clin Nutr 2008;87:1219–1223 [DOI] [PubMed] [Google Scholar]
- 43.Zhao L, Kwon M-J, Huang S, et al. Differential modulation of Nods signaling pathways by fatty acids in human colonic epithelial HCT116 cells. J Biol Chem 2007;282:11618–11628 [DOI] [PubMed] [Google Scholar]
- 44.Ajuwon KM, Banz W, Winters TA. Stimulation with peptidoglycan induces interleukin 6 and TLR2 expression and a concomitant downregulation of expression of adiponectin receptors 1 and 2 in 3T3-L1 adipocytes. J Inflamm (Lond) 2009;6:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.