Abstract
OBJECTIVE
The plasma adiponectin level, a potential upstream and internal facet of metabolic and cardiovascular diseases, has a reasonably high heritability. Whether other novel genes influence the variation in adiponectin level and the roles of these genetic variants on subsequent clinical outcomes has not been thoroughly investigated. Therefore, we aimed not only to identify genetic variants modulating plasma adiponectin levels but also to investigate whether these variants are associated with adiponectin-related metabolic traits and cardiovascular diseases.
RESEARCH DESIGN AND METHODS
We conducted a genome-wide association study (GWAS) to identify quantitative trait loci (QTL) associated with high molecular weight forms of adiponectin levels by genotyping 382 young-onset hypertensive (YOH) subjects with Illumina HumanHap550 SNP chips. The culpable single nucleotide polymorphism (SNP) variants responsible for lowered adiponectin were then confirmed in another 559 YOH subjects, and the association of these SNP variants with the risk of metabolic syndrome (MS), type 2 diabetes mellitus (T2DM), and ischemic stroke was examined in an independent community–based prospective cohort, the CardioVascular Disease risk FACtors Two-township Study (CVDFACTS, n = 3,350).
RESULTS
The SNP (rs4783244) most significantly associated with adiponectin levels was located in intron 1 of the T-cadherin (CDH13) gene in the first stage (P = 7.57 × 10−9). We replicated and confirmed the association between rs4783244 and plasma adiponectin levels in an additional 559 YOH subjects (P = 5.70 × 10−17). This SNP was further associated with the risk of MS (odds ratio [OR] = 1.42, P = 0.027), T2DM in men (OR = 3.25, P = 0.026), and ischemic stroke (OR = 2.13, P = 0.002) in the CVDFACTS.
CONCLUSIONS
These findings indicated the role of T-cadherin in modulating adiponectin levels and the involvement of CDH13 or adiponectin in the development of cardiometabolic diseases.
Adiponectin plays vital roles in modulating insulin sensitivity, glucose homeostasis, lipid metabolism, and antiatherosclerotic and anti-inflammatory responses in the vascular system (1,2). The concentration of adiponectin, the most abundant adipokine secreted by adipocytes, ranges from 4 to 30 μg/mL in the blood, which is much higher than the concentrations of various other hormones and cytokines (3). Decreased levels of plasma adiponectin are associated with an increased risk of not only obesity (4) and metabolic syndrome (MS) (5) but also type 2 diabetes mellitus (T2DM) (6), hypertension (7), myocardial infarction (8), and ischemic stroke (9). Animal studies and cell culture experiments have shown that direct stimulation of nitric oxide synthesis is responsible for the anti-inflammatory mechanism and antiatherogenic effects of adiponectin (10). These findings give biological plausibility to the phenomenon that the decreased plasma levels of adiponectin may directly lead to the development of insulin resistance, diabetes, and cardiovascular disease (CVD) and not merely be a consequence of the MS. Therefore, understanding the genetic mechanisms involved in the modulation of plasma adiponectin levels in the human body will provide insights into the cause and management of MS.
The plasma adiponectin levels, a potential upstream and internal facet of metabolic disease and CVD (11), have a reasonably high heritability with an estimated range of 40–80% (12,13). Although the ADIPOQ and ARL15 genes identified by genome-wide association studies (GWAS) have been associated with adiponectin levels in white populations (14–16), whether other genes influence the changes in adiponectin level and the roles of these genetic variants on subsequent clinical outcomes, including MS, T2DM, and coronary artery disease, has not been carefully investigated, especially in Asian populations.
We performed a GWAS to identify the quantitative trait loci (QTL) regulating the adiponectin levels by using phenotypic and genotypic information of 941 young-onset hypertensive (YOH) subjects, including the Illumina HumanHap550 SNP data for the initial 382 subjects. Three single nucleotide polymorphism (SNP) variants responsible for lowered adiponectin levels showed genome-wide significance in both the first-stage (with 382 YOH cases) and the second-stage (with 559 YOH cases) studies; subsequently, we determined the association of these SNP variants with the risk of MS, T2DM, and ischemic stroke in an independent large-scale, community-based prospective cohort study, the CardioVascular Disease risk FACtors Two-township Study (CVDFACTS).
RESEARCH DESIGN AND METHODS
GWAS using the data collected for YOH patients to determine QTL influencing the plasma adiponectin levels.
We performed a two-stage GWAS to identify the genes/loci that influence the plasma adiponectin levels. In this study, we included 941 hypertensive subjects recruited by the Academia Sinica Multicentered Young-Onset Hypertension Genetic Study: 382 in the first-stage genome-wide scan and 559 in the second-stage confirmatory study. The inclusion criteria for hypertensive subjects are as follows: 1) systolic blood pressure ≥140 mmHg or diastolic blood pressure ≥90 mmHg over a 2-month period or systolic blood pressure/diastolic blood pressure ≥120/80 mmHg in patients taking antihypertensive medications in two consecutive visits within 2 months; 2) age between 20 and 51 years at the first diagnosis of hypertension; 3) no secondary causes of hypertension, such as chronic renal disease, renal arterial stenosis, primary aldosteronism, coarctation of the aorta, thyroid disorders, Cushing syndrome, and pheochromocytoma confirmed by extensive clinical examinations, including blood chemistry examination, renal function tests, endocrine examination, and abdominal sonogram; 4) fasting glucose (FG) level <126 mg/dL; 5) BMI <35 kg/m2, where BMI was defined as body weight in kilograms divided by height in meters squared; and 6) self-reported Han Chinese ethnicity in more than 2 generations.
Data were collected according to standardized protocols. Blood pressure was measured according to the protocol established for the Nutrition and Health Survey in Taiwan (17). Serum levels of high molecular weight forms of adiponectin for all samples were assayed by Taipei Institute of Pathology (Taipei, Taiwan) using the enzyme immunoassay kit (adiponectin [human] ELISA kit; Phoenix Pharmaceuticals Inc., Belmont, CA). A subsample of 40 samples was analyzed in duplicate. The averaged coefficient of variation was 4.97%. In addition, the data on sociodemographic factors, smoking and drinking habits, medical history, and current medications were obtained by interviewing the subjects. Our multicenter study was approved by the Human Investigation Committee of Academia Sinica. Informed consent was obtained from each participant at his/her first visit to the clinic.
Prospective study to determine the association between QTL of adiponectin and metabolic traits, T2DM, and ischemic stroke.
The study subjects included those who were recruited for the CVDFACTS, which has been described (18,19). In brief, the CVDFACTS is a community-based follow-up cohort study designed to evaluate the risk factors of CVD and metabolic disease in Taiwan. This study was initiated in 1993, and all residents aged more than 3 years in Chu-Dung (northwest Taiwan) and Pu-Tzu (southwest Taiwan) were invited to participate in the baseline examination. The follow-up examinations were performed in 1994–1997, 1997–1999, and 2000–2002. Data on sociodemographic factors, anthropometric parameters, smoking and drinking habits, medical history, and current medications were obtained by interviewing the subjects, and fasting blood was drawn for biochemical examination, including measurement of the levels of serum glucose, triglyceride (TG), insulin, and so forth. Insulin sensitivity was estimated based on the homeostasis model assessment of insulin resistance (HOMA-IR) formula (serum glucose levels × insulin/22.5). Intima–media thickness (IMT) of the carotid artery was assessed for those aged ≥30 years by high-resolution B-mode carotid ultrasonography using a 7.5-MHz transducer (Phillips Medical Systems NA, Bothell, WA). The IMT as the distance between the leading edges of the lumen–intima and media–adventitia interfaces was measured along the near and far wall of the distal 10-mm portion of the common carotid artery and along a 15-mm section from the carotid bifurcation in the plaque-free area. The thickness was measured manually and recorded. The largest value among all the readings was considered as the maximal IMT. Participants in our study were aged ≥20 years with no history of stroke, cancer, or CVD at the time of data entry. Ischemic stroke status was determined based on information in death certificate data, insurance claim records, and hospital record. Detailed rules have been published (19). More detailed information of the baseline characteristics of CVD cohort study is provided in Supplementary Table 1. Our study was approved by the Human Investigation Committee of Academia Sinica.
Genotyping methods.
Genomic DNA was extracted from peripheral blood samples of hypertensive subjects using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN) for the YOH genetic study and the phenol/chloroform method for the CVDFACTS. For the GWAS, genotyping experiments were performed by deCODE genetics (Reykjavik, Iceland) by using the Illumina Infinium II HumanHap550 SNP chips (Illumina, San Diego, CA) to analyze data from 382 leukocyte DNA samples. We followed the Wellcome Trust Case Control Consortium criteria for quality control (20); only those individuals in whom more than 3% of the required data were missing were excluded. The SNPs were excluded if they showed violation of the Hardy–Weinberg equilibrium (P < 1 × 10−7), call rates <95%, or minor allele frequency <1%. One SNP located at intron 1 of CDH13 with –log P ≥7 and two additional SNPs located at intron 1 (P = 4.10 × 10−6) and promoter (P = 4.88 × 10−5) of CDH13 gene in the first stage were genotyped in the second stage for further confirmation. The genotyping of the three SNPs for the samples in the second-stage confirmation study and for the CVDFACTS subjects was performed using the Sequenom MassARRAY System (San Diego, CA) by the Academia Sinica National Genotyping Center (Taipei, Taiwan).
Statistical analysis.
We performed a two-stage genome-wide QTL mapping for adiponectin levels. The general linear model (GLM) was used for associating adiponectin levels with genotype data, making adjustments for sex, age, smoking, and BMI in the first- and second-stage analyses, where the distribution of adiponectin levels was normalized by taking a square root transformation of the original values because of the skewed distributions of the original values. In the first stage, the genome-wide significance level was set to be 1 × 10−7 (≈0.05/509,174) according to the Bonferroni multiple testing correction (20). A multiple regression model was used to estimate the degree of variation in plasma adiponectin levels explained by the selected SNPs using the combined data from the first and second stages. An examination of possible population stratification was carried out using multidimensional scaling analysis of PLINK software (21). Quantile–quantile plot of the genome-wide QTL mapping was also drawn to examine P value distributions based on 382 YOH patients.
The levels of serum glucose, HOMA-IR, and TG were analyzed after obtaining the square root of the original value because of the skewed distributions of these values in CVDFACTS in which adiponectin QTL was examined against the aforementioned cardiometabolic outcomes. Analysis of GLM was used to compare the mean levels of metabolic parameters among genotype groups with adjustments for age, sex, smoking, and medication for T2DM as covariates. The relationship between the SNP rs4783244 and the presence of MS, T2DM, hypertension, and stroke was examined by using dichotomous logistic regression analyses with adjustments for age, BMI, sex, and smoking. With the exception of the evaluation of population stratification, all other statistical analyses were performed using SAS version 9.2 (SAS Institute Inc., Cary, NC).
RESULTS
Characteristics of subjects in the Academia Sinica Multicentered Young-Onset Hypertension Genetic Study.
No significant differences were observed between subjects in the first and second stages (Table 1) with respect to sex distribution, mean levels of LDL and adiponectin, and proportion of smoking status. Significant differences were noted in the mean age and BMI of the subjects. The subjects in the second stage were 4 years older than those in the first stage, but the magnitude of the difference in BMI was small.
TABLE 1.
Characteristics of the study subjects in the first two stages
| Characteristic | Initial stage | Second stage | P† |
|---|---|---|---|
| n | 382 | 559 | |
| Male (%) | 68 | 69 | 0.67 |
| Age (years) | 38.4 ± 0.4 | 42.9 ± 0.2 | <0.0001 |
| BMI (kg/m2) | 26.2 ± 0.2 | 26.7 ± 0.1 | 0.03 |
| LDL (mg/dL) | 126.2 ± 1.7 | 126.2 ± 1.3 | 0.97 |
| Adiponectin (μg/mL) | 4.1 ± 0.23 | 4.2 ± 0.17 | 0.81 |
| Smoking status (%) | 21.9 | 24.7 | 0.36 |
Data are given in percentage or mean ± SE.
†A t or χ2 test was used to make comparisons between the first- and second-stage samples in the GWAS with a significance level set at P = 0.05.
GWAS findings.
The results of principal component analysis in stage 1 revealed no evidence for population stratification for hypertensive subjects. Multidimensional scaling analysis using PLINK also showed similar results (Supplementary Fig. 1).
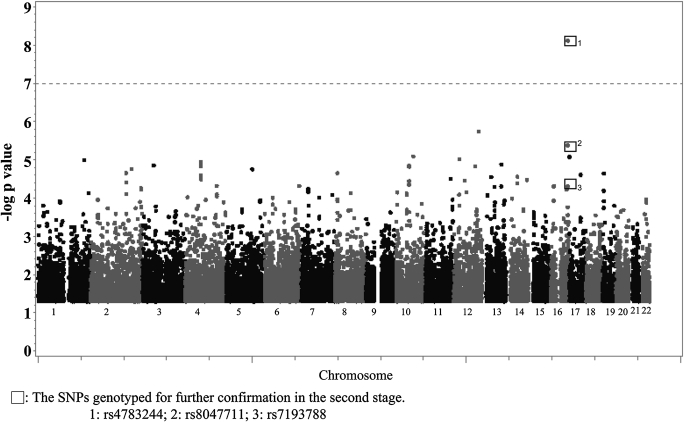
To identify the QTLs influencing the adiponectin levels, we performed GLM with adjustments for age, sex, BMI, and smoking to reduce the potential confounding effects. The major results of the GWAS for adiponectin levels are shown in Fig. 1 and Supplementary Fig. 2. The SNP (rs4783244) most significantly associated with adiponectin levels was located in intron 1 of the CDH13 gene (P = 7.57 × 10−9); this SNP was selected for the second-stage confirmatory study, because no other SNPs had a –log P value greater than 7. In addition to the most significant SNP (rs4783244), two SNPs located at promoter (rs7193788; P = 4.88 × 10−5) and intron 1 of CDH13 locus (rs8047711; P = 4.10 × 10−6) were also selected based on the location of SNPs and P value of the genome-wide scan. We selected these three candidate SNPs for confirmation in the second stage by examining an additional 559 YOH patients. The three SNPs again showed significant associations with adiponectin levels (Table 2): rs4783244 (P = 5.70 × 10−17), rs8047711 (P = 6.32 × 10−8), and rs7193788 (P = 1.36 × 10−9). The SNP rs4783244 that showed the most significant association accounted for 9.28% of the total variance of adiponectin levels in the combined data analysis of 941 hypertensive subjects (Table 2), which was more significant than the other two SNPs.
FIG. 1.
Illumina HumanHap550 SNPs associated with adiponectin levels in the GWAS. The GLM was used to examine associations between SNPs and adiponectin levels after adjusting for age, sex, BMI, and smoking status with the significance level set at –log P = 7 (gray dashed horizontal line).
TABLE 2.
Association between adiponectin levels and SNPs identified at the initial stage, confirmed at the second stage, and some common SNPs of candidate genes
| SNP | Gene | Position | Allele | MAF | β (SE) | Initial stage (n = 382) |
Second stage (n = 559) |
Two stages combined (n = 941) |
|---|---|---|---|---|---|---|---|---|
| P value§ | P value|| | R2 (%)¶ | ||||||
| rs7193788 | CDH13 | Promoter | A/G* | 0.398 | 0.254 (0.056) | 4.88 × 10−5 | 1.36 × 10−9 | 0.03 |
| rs4783244 | CDH13 | Intron 1 | G*/T | 0.298 | 0.346 (0.057) | 7.57 × 10−9 | 5.70 × 10−17 | 9.28 |
| rs8047711 | CDH13 | Intron 1 | G*/A | 0.204 | 0.323 (0.064) | 4.10 × 10−6 | 6.32 × 10−8 | 0.41 |
| rs4311394† | ARL15 | Intron 4 | A/G* | 0.442 | 0.034 (0.056) | 0.027 | —# | —# |
| rs16861194† | ADIPOQ | Promoter | A/G* | 0.139 | −0.03 (0.08) | 0.648 | —# | —# |
| rs3774262† | ADIPOQ | Intron 2 | A/G* | 0.279 | −0.07 (0.06) | 0.529 | —# | —# |
| rs10753929‡ | ADIPOR1 | Intron 2 | C/T* | 0.096 | 0.118 (0.099) | 0.03 | —# | —# |
| rs4766413‡ | ADIPOR2 | Intron 1 | A/G* | 0.298 | −0.067 (0.063) | 0.128 | —# | —# |
Major allele/minor allele. MAF, minor allele frequency; β, estimated effect size.
*Allele with higher adiponectin value.
†SNPs reported in previous studies.
‡The most significant SNPs in adiponectin receptor found by the authors.
§Statistics corresponding to GLM testing association between genotypes and adiponectin levels after adjustments for age, sex, BMI, and smoking in the first-stage study comprising 382 subjects.
||Statistics corresponding to GLM testing association between genotypes and adiponectin levels after adjustment for age, sex, BMI, and smoking in the second-stage confirmatory study comprising 559 subjects.
¶R2 was obtained by conducting multiple regression analysis for the samples obtained from both the first and second stages.
#SNP was not replicated in the second stage.
We further examined the association for loci previously reported in the GWAS or candidate gene study. We did not observe the associations with SNPs in ADIPOQ (rs16861194 and rs3774262, P = 0.648 and P = 0.529), ARL15 (rs4311394, P = 0.027), ADIPOR1 (rs10753929, P = 0.03), and ADIPOR2 (rs4766413, P = 0.128) after multiple testing corrections (Table 2).
Association between rs4783244 in CDH13 and metabolic traits.
Because of a strong linkage disequilibrium among these three SNPs in the CDH13 gene (D′ = 1 among three SNPs), we selected the most significant SNP, rs4783244, to further determine its association with other metabolic parameters, including hypertension, waist circumference (WC), and the levels of HDL-cholesterol, TG, and FG for the samples in CVDFACTS. The SNP rs4783244 was significantly associated with WC (P = 0.014) and the levels of TG (P = 0.010) and FG (P = 0.024), whereas it was not significantly associated with HDL-cholesterol levels and hypertension in the CVDFACTS samples (Table 3). The GG genotype of rs4783244 was associated with WC, a low adiponectin level, a high level of TG and FG, and an increased risk of MS (odds ratio [OR] = 1.42 [CI 1.04–1.95], P = 0.0273; Table 3).
TABLE 3.
Associations between genotypes of rs4783244 on CDH13 and metabolic parameters
| Trait |
rs4783244 |
|||
|---|---|---|---|---|
| G/G (n = 1,474) |
G/T (n = 1,451) |
T/T (n = 340) |
P value for trend |
|
| Mean ± SD | ||||
| TG (mg/dL) | 110.72 ± 2.3 | 111.90 ± 2.8 | 98.3 ± 3.8 | 0.010 |
| WC (cm) | 81.50 ± 9.6 | 80.59 ± 9.1 | 80.29 ± 9.4 | 0.014 |
| Glucose (mg/dL) | 100.48 ± 22.5 | 99.50 ± 20.6 | 97.78 ± 17.1 | 0.024 |
| HDL-cholesterol (mg/dL) | 42.65 ± 11.8 | 42.60 ± 12.2 | 43.48 ± 11.9 | 0.554 |
| OR (95% CI) | ||||
| Hypertension | 1.28 (0.97–1.69) | 1.18 (0.90–1.55) | 1 | |
| P = 0.07† | P = 0.23† | |||
| MS | 1.42 (1.04–1.95) | 1.30 (0.95–1.79) | 1 | |
| P = 0.03† | P = 0.02† | |||
TG, glucose, and HDL-cholesterol levels and WC were averaged by genotypes of rs4783244.
†P values were obtained from logistic regression analysis with adjustments for age, sex, and smoking.
No significant associations were observed between the genotypes of rs4783244 and HOMA-IR or T2DM without stratification of the sex (data not shown). However, in the male subgroup, the SNP rs4783244 was significantly associated with HOMA-IR (P = 0.033). The HOMA-IR levels increased with the number of the G allele of rs4783244 in a dominant fashion; GG and GT genotypes of rs4783244 were significantly associated with T2DM in a similar manner (OR = 3.25 [CI 1.15–9.19], P = 0.0259 and OR = 3.50 [1.24–9.8], P = 0.0184, respectively; Table 4). However, none of the associations with HOMA-IR and T2DM were detected in the female subgroup. We further conducted a test for the rs4783244* interactions between the sexes. We observed a statistically significant interaction effect between the rs4783244 genotypes and sex (P for interaction = 0.05). Because a higher proportion of smokers are men than women (<1%), we investigated whether it is in smokers that rs4783244 exacts its effect. However, we found that there was no interaction between smoking and rs4783244 in men (P for interaction = 0.159).
TABLE 4.
Associations between genotypes of rs4783244 on CDH13 and HOMA-IR and the risk of diabetes
| Trait |
Sex |
rs4783244 |
P value |
||
|---|---|---|---|---|---|
| G/G |
G/T |
T/T |
|||
| Mean ± SD (n) | |||||
| HOMA-IR | Male | 1.67 ± 0.53 (494) | 1.65 ± 0.52 (468) | 1.54 ± 0.39 (175) | 0.033† |
| HOMA-IR | Female | 1.69 ± 0.52 (793) | 1.69 ± 0.52 (803) | 1.70 ± 0.53 (175) | 0.700† |
| OR (95% CI) | |||||
| DM | Male | 3.25 (1.15–9.19) | 3.50 (1.24–9.8) | 1 | |
| P = 0.026‡ | P = 0.018‡ | ||||
| DM | Female | 1.51 (0.6–2.22) | 0.98 (0.5–1.91) | 1 | |
| P = 0.674‡ | P = 0.960‡ | ||||
HOMA-IR stratified by sex was averaged by genotypes of rs4783244. DM, diabetes mellitus.
†P values were obtained from GLM with adjustments for age and smoking.
‡P values were obtained from logistic regression analysis with adjustments for age and smoking.
Association between rs4783244 in CDH13 and IMT and the risk of ischemic stroke.
Results from the GLM with adjustments for sex, age, and smoking showed that the mean IMT was associated with rs4783244 (P = 0.048). The mean IMT increased with the GG genotype of rs4783244 in a recessive fashion (P = 0.026). We further compared the risk of ischemic stroke between people with GG and people with combined TT and GT genotypes. Similar to the findings for IMT, the risk of ischemic stroke increased with the GG genotype of rs4783244 (OR, 2.13 [CI 1.33–3.39], P = 0.002; Table 5).
TABLE 5.
Associations between rs4783244 on CDH13 and IMT and the risk of ischemic stroke
| Genotype | No. of subjects | IMT (mm) |
Stroke risk |
|||
|---|---|---|---|---|---|---|
| Mean (SD) | P value | No. of events | OR (95% CI) | P value | ||
| rs4783244 | ||||||
| TT | 338 | 0.515 (0.153) | 5 | 1 | ||
| GT | 1,444 | 0.508 (0.159) | 24 | 1.182 (0.445–3.139) | 0.737‡ | |
| GG | 1,469 | 0.524 (0.211) | 0.048† | 50 | 2.436 (0.958–6.192) | 0.061‡ |
| G/T + TT | 1,782 | 0.510 (0.158) | 29 | 1 | ||
| GG | 1,469 | 0.524 (0.211) | 0.026† | 50 | 2.125 (1.332–3.390) | 0.002‡ |
†P values were obtained from the GLM procedure with adjustments for age, sex, and smoking.
‡P values were obtained from logistic regression analysis with adjustments for age, sex, and smoking.
To summarize, the G allele at rs4783244 was consistently associated with deleterious states of the five metabolic traits examined and with an increased risk of MS, T2DM (in men), and ischemic stroke.
DISCUSSION
We performed a GWAS for high molecular weight forms of adiponectin levels and found a QTL on CDH13 that affects adiponectin levels. This adiponectin QTL was recently reported for Filipinos by Wu et al. (22) and Koreans by Jee et al. (23). We are the first to find this adiponectin QTL in Chinese and to show that this QTL was associated with metabolic traits and the risk of MS, T2DM (in men), and stroke. These findings may broaden our understanding of the mechanisms modulating adiponectin levels and the role of adiponectin in the development of metabolic disease and CVD.
Several GWAS have been performed for adiponectin levels. Wu et al. (22) and Jee et al. (23) found that SNP-rs3865188 on the CDH13 locus was associated with high molecular weight forms of adiponectin levels in an Asian population. The rs4783244 (D′ = 0.94; r2 = 0.9) SNP found in our study was in strong linkage disequilibrium with this SNP. Another family study (15) revealed that rs7195409 (which did not reach a genome-wide significance) on intron 7 of the CDH13 locus potentially affects adiponectin levels in subjects of northern and western European origin. We tried to compare allele frequencies of 10 previously reported associated SNPs in the ADIPOQ gene (22) between Asians and Europeans. No polymorphisms were found for two of them in Asians, namely, rs17300539 and rs17366568. Allele frequencies of the other eight SNPs differed from Europeans by 3–16%. Only one of the eight is included in the Illumina Infinium II HumanHap550 SNP chips. We did not have the opportunity to study the association between these seven SNPs and adiponectin levels. In addition to the polymorphisms at the ADIPOQ locus (14–16), a previous study (16) revealed that ARL15, whose functions were unknown, influenced circulating adiponectin levels and moderately increased the risk of coronary heart disease. We showed that a variant of CDH13, but not that of ADIPOQ and ARL15, was associated with adiponectin levels. This discrepancy in results may be explained by population-specific genetic variants, limited sample sizes to detect the modest associations, and varied coverage of different whole-genome SNP–genotyping platforms. Similar to our study, the small scale study conducted by Jee et al. (23) in Korea did not show an association between adiponectin levels and ADIPOQ or ARL15.
Our study further showed that the CDH13 variant had moderate effects on metabolic traits and on the increased risk of MS, T2DM, and ischemic stroke. CDH13 is located at chromosome 16q24 (24) and encodes the cadherin-related superfamily of transmembrane proteins that mediate calcium-dependent intercellular adhesion. CDH13 is highly expressed in several tissues (heart, aortic wall, neurons of the brain cortex and spinal cord, and small blood vessels) and in a variety of cell types (vascular endothelial cells, smooth muscle cells, pericytes, cardiomyocytes, and cancer cells) (25–28). Studies on cellular signaling have suggested that both LDL and adiponectin were specific ligands for T-cadherin, a product of CDH13 (29–31). Binding of LDL or adiponectin to T-cadherin is capable of activating nuclear factor-κB (NF-κB) signaling pathway (29), which plays a central role in inflammation and serves as a link between obesity and vascular disease (32). T-cadherin expression in the arterial wall after balloon angioplasty is associated with late stages of neointima formation and with the peak of proliferation and differentiation of vascular cells (33). This evidence suggests that T-cadherin, by modulating the levels of adiponectin in the blood and in various tissues, may regulate vascular remodeling, neointima formation, and inflammation-related phenomena and atherosclerosis development. Two additional adiponectin receptors have been identified in the skeletal muscle (AdipoR1) and liver (AdipoR2) by expression cloning (34). Both receptors activate adenosine monophosphate–activated protein kinase and peroxisome proliferator–activated receptor α metabolic pathways that mediate the major metabolic effects of adiponectin, such as those on glucose uptake and fatty-acid oxidation, which are critical components in the development of obesity, T2DM, and CVD (35,36). T-cadherin not only competes with adiponectin receptors R1 and R2 for adiponectin binding but also interferes with the coupling of both receptors to their downstream intracellular targets (37). All of these data provide evidence of the role of CDH13 and of the complexity in the development of cardiometabolic diseases.
In an attempt to dissect the interrelationship among CDH13, obesity, and cardiometabolic diseases, we included BMI as a covariate. Our data show that the direction of association between all components of MS and rs4783244 remained the same, but they were no longer statistically significant, indicating that the genetic effects of CDH13-rs4783244 on these metabolic traits may be mediated in part by BMI. However, the P value for the association with stroke, MS, and diabetes remained significant at 0.0035, 0.026, and 0.03, respectively. These findings indicate that rs4783244 has its independent effects on MS comorbidity, stroke, and diabetes.
An association between rs4783244 and HOMA-IR/T2DM was found in men but not in women. A question arises whether smoking plays any role to augment the effect of this polymorphism in men. Smoking has been known to be associated with increased oxidative stress that induces production of inflammatory cytokines, resulting in endothelial dysfunction and increasing the risk for atherosclerosis (38). The interaction between adiponectin and its receptor CDH13 may trigger the NF-κB signaling pathway and in turn affect the development of cardiometabolic diseases, also via the inflammatory pathway. Smoking may interact with CDH13-rs4783244 and affect the development of cardiometabolic diseases. However, no significant interaction between rs4783244 and smoking was found for T2DM and stroke in men in our study. Among alternative reasons for the difference between the sexes is that plasma adiponectin levels generally are higher in women than in men (39), which may protect women more against T2DM. Because we did not measure adiponectin levels in the CVDFACTS, we could not examine how the sex interacts with adiponectin level to affect the risk of T2DM.
Our study has some other limitations. First, this GWAS was performed in hypertensive subjects. Further studies should be performed in the general population. Second, the number of subjects in our study is generally regarded as small for a GWAS. However, the power for identifying QTLs of adiponectin levels is in general greater than that in complex diseases.
A moderate association was also observed between SNP rs11068544 on chromosome 12 and adiponectin levels (P value = 1.78 × 10−6). This SNP lies in intron 5 of kinase suppressor of ras 2 (KSR2), which acts as a negative regulator of mitogen-activated protein kinase kinase kinase 3 (MAP3K3)-mediated activation of extracellular signal–related kinase (ERK), Jun NH2-terminal kinase, and NF-κB pathways, inhibiting the MAP3K3-mediated proinflammatory pathway and downregulated mitogen-activated protein kinase/ERK kinase kinase 3-induced interleukin-8 production in response to interleukin-1B stimulation (40). Although the magnitude of association did not reach genome-wide threshold for statistical significance, KSR2 also stimulates fatty acids oxidation via the adenosine monophosphate–activated protein kinase pathway. These features may make the KSR2 gene a promising candidate for adiponectin levels regulation. Further large-scale confirmatory studies will be required to clarify this association.
In conclusion, our study provides evidence that a GWAS has the potential to reveal genetic influences on a clear biochemical trait such as adiponectin levels. We have implicated that CDH13 has an influence not only on adiponectin levels in the blood but also on the risk of MS, T2DM, and ischemic stroke. These findings support the pathogenic role of CDH13 in metabolic diseases and in atherothrombotic stroke and suggest the potential application of CDH13 as a target for disease prevention and management.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Department of Health (DOH90-TD-1037), National Science Council (NSC91-3112-P-001-025-YGPCP91-25), Academia Sinica (AS91IBMS2PP-A), and Academia Sinica Genomics and Proteomics Program (2003–2006) in Taiwan.
C.-M.C., J.-W.C., Y.-T.C., and W.-H.P. conceived and designed the experiments. T.-H.L., J.-W.C., H.-B.L., H.-Y.H., C.-T.T., S.-H.S., W.-C.T., J.-H.C., and S.-J.L. performed the experiments. C.-M.C. analyzed the data. T.-H.L., J.-W.C., H.-C.Y., and W.-H.P. contributed reagents, materials, and analysis tools. C.-M.C., H.-C.Y., and W.-H.P. wrote the manuscript.
No potential conflicts of interest relevant to this article were reported.
The authors thank Academia Sinica National Genotyping Center (Taipei, Taiwan) for performing genotyping in the second-stage study and for the prospective study using the CVDFACTS materials.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db10-1321/-/DC1.
REFERENCES
- 1.Ouchi N, Kihara S, Funahashi T, Matsuzawa Y, Walsh K. Obesity, adiponectin and vascular inflammatory disease. Curr Opin Lipidol 2003;14:561–566 [DOI] [PubMed] [Google Scholar]
- 2.Kadowaki T, Yamauchi T, Kubota N, Hara K, Ueki K, Tobe K. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J Clin Invest 2006;116:1784–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Funahashi T, Matsuzawa Y. Hypoadiponectinemia: a common basis for diseases associated with overnutrition. Curr Atheroscler Rep 2006;8:433–438 [DOI] [PubMed] [Google Scholar]
- 4.Weyer C, Funahashi T, Tanaka S, et al. Hypoadiponectinemia in obesity and type 2 diabetes: close association with insulin resistance and hyperinsulinemia. J Clin Endocrinol Metab 2001;86:1930–1935 [DOI] [PubMed] [Google Scholar]
- 5.Kondo H, Shimomura I, Matsukawa Y, et al. Association of adiponectin mutation with type 2 diabetes: a candidate gene for the insulin resistance syndrome. Diabetes 2002;51:2325–2328 [DOI] [PubMed] [Google Scholar]
- 6.Li S, Shin HJ, Ding EL, van Dam RM. Adiponectin levels and risk of type 2 diabetes: a systematic review and meta-analysis. JAMA 2009;302:179–188 [DOI] [PubMed] [Google Scholar]
- 7.Iwashima Y, Katsuya T, Ishikawa K, et al. Hypoadiponectinemia is an independent risk factor for hypertension. Hypertension 2004;43:1318–1323 [DOI] [PubMed] [Google Scholar]
- 8.Pischon T, Girman CJ, Hotamisligil GS, Rifai N, Hu FB, Rimm EB. Plasma adiponectin levels and risk of myocardial infarction in men. JAMA 2004;291:1730–1737 [DOI] [PubMed] [Google Scholar]
- 9.Chen MP, Tsai JC, Chung FM, et al. Hypoadiponectinemia is associated with ischemic cerebrovascular disease. Arterioscler Thromb Vasc Biol 2005;25:821–826 [DOI] [PubMed] [Google Scholar]
- 10.Antoniades C, Antonopoulos AS, Tousoulis D, Stefanadis C. Adiponectin: from obesity to cardiovascular disease. Obes Rev 2009;10:269–279 [DOI] [PubMed] [Google Scholar]
- 11.Pan WH, Lynn KS, Chen CH, Wu YL, Lin CY, Chang HY. Using endophenotypes for pathway clusters to map complex disease genes. Genet Epidemiol 2006;30:143–154 [DOI] [PubMed] [Google Scholar]
- 12.Cesari M, Narkiewicz K, De Toni R, Aldighieri E, Williams CJ, Rossi GP. Heritability of plasma adiponectin levels and body mass index in twins. J Clin Endocrinol Metab 2007;92:3082–3088 [DOI] [PubMed] [Google Scholar]
- 13.Liu PH, Jiang YD, Chen WJ, et al. Genetic and environmental influences on adiponectin, leptin, and BMI among adolescents in Taiwan: a multivariate twin/sibling analysis. Twin Res Hum Genet 2008;11:495–504 [DOI] [PubMed] [Google Scholar]
- 14.Heid IM, Henneman P, Hicks A, et al. Clear detection of ADIPOQ locus as the major gene for plasma adiponectin: results of genome-wide association analyses including 4659 European individuals. Atherosclerosis 2010;208:412–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ling H, Waterworth DM, Stirnadel HA, et al. Genome-wide linkage and association analyses to identify genes influencing adiponectin levels: the GEMS Study. Obesity (Silver Spring) 2009;17:737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards JB, Waterworth D, O’Rahilly S, et al. ; GIANT Consortium A genome-wide association study reveals variants in ARL15 that influence adiponectin levels. PLoS Genet 2009;5:e1000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan WH, Hung YT, Shaw NS, et al. Elderly Nutrition and Health Survey in Taiwan (1999-2000): research design, methodology and content. Asia Pac J Clin Nutr 2005;14:203–210 [PubMed] [Google Scholar]
- 18.Chen HJ, Bai CH, Yeh WT, Chiu HC, Pan WH. Influence of metabolic syndrome and general obesity on the risk of ischemic stroke. Stroke 2006;37:1060–1064 [DOI] [PubMed] [Google Scholar]
- 19.Chuang SY, Bai CH, Chen WH, Lien LM, Pan WH. Fibrinogen independently predicts the development of ischemic stroke in a Taiwanese population: CVDFACTS study. Stroke 2009;40:1578–1584 [DOI] [PubMed] [Google Scholar]
- 20.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Y, Li Y, Lange EM, et al. Genome-wide association study for adiponectin levels in Filipino women identifies CDH13 and a novel uncommon haplotype at KNG1-ADIPOQ. Hum Mol Genet 2010;19:4955–4964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jee SH, Sull JW, Lee JE, et al. Adiponectin concentrations: a genome-wide association study. Am J Hum Genet 2010;87:545–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SW. H-cadherin, a novel cadherin with growth inhibitory functions and diminished expression in human breast cancer. Nat Med 1996;2:776–782 [DOI] [PubMed] [Google Scholar]
- 25.Carter BS, Ewing CM, Ward WS, et al. Allelic loss of chromosomes 16q and 10q in human prostate cancer. Proc Natl Acad Sci U S A 1990;87:8751–8755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kremmidiotis G, Baker E, Crawford J, Eyre HJ, Nahmias J, Callen DF. Localization of human cadherin genes to chromosome regions exhibiting cancer-related loss of heterozygosity. Genomics 1998;49:467–471 [DOI] [PubMed] [Google Scholar]
- 27.Tsuda H, Oda T, Sakamoto M, Hirohashi S. Different pattern of chromosomal allele loss in multiple hepatocellular carcinomas as evidence of their multifocal origin. Cancer Res 1992;52:1504–1509 [PubMed] [Google Scholar]
- 28.Ivanov D, Philippova M, Antropova J, et al. Expression of cell adhesion molecule T-cadherin in the human vasculature. Histochem Cell Biol 2001;115:231–242 [DOI] [PubMed] [Google Scholar]
- 29.Hug C, Wang J, Ahmad NS, Bogan JS, Tsao TS, Lodish HF. T-cadherin is a receptor for hexameric and high-molecular-weight forms of Acrp30/adiponectin. Proc Natl Acad Sci U S A 2004;101:10308–10313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Resink TJ, Kuzmenko YS, Kern F, et al. LDL binds to surface-expressed human T-cadherin in transfected HEK293 cells and influences homophilic adhesive interactions. FEBS Lett 1999;463:29–34 [DOI] [PubMed] [Google Scholar]
- 31.Kipmen-Korgun D, Osibow K, Zoratti C, et al. T-cadherin mediates low-density lipoprotein-initiated cell proliferation via the Ca(2+)-tyrosine kinase-Erk1/2 pathway. J Cardiovasc Pharmacol 2005;45:418–430 [DOI] [PubMed] [Google Scholar]
- 32.Ouchi N, Kihara S, Arita Y, et al. Adiponectin, an adipocyte-derived plasma protein, inhibits endothelial NF-kappaB signaling through a cAMP-dependent pathway. Circulation 2000;102:1296–1301 [DOI] [PubMed] [Google Scholar]
- 33.Kudrjashova E, Bashtrikov P, Bochkov V, et al. Expression of adhesion molecule T-cadherin is increased during neointima formation in experimental restenosis. Histochem Cell Biol 2002;118:281–290 [DOI] [PubMed] [Google Scholar]
- 34.Yamauchi T, Kamon J, Ito Y, et al. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature 2003;423:762–769 [DOI] [PubMed] [Google Scholar]
- 35.Halvatsiotis I, Tsiotra PC, Ikonomidis I, et al. Genetic variation in the adiponectin receptor 2 (ADIPOR2) gene is associated with coronary artery disease and increased ADIPOR2 expression in peripheral monocytes. Cardiovasc Diabetol 2010;9:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaxillaire M, Dechaume A, Vasseur-Delannoy V, et al. Genetic analysis of ADIPOR1 and ADIPOR2 candidate polymorphisms for type 2 diabetes in the Caucasian population. Diabetes 2006;55:856–861 [DOI] [PubMed] [Google Scholar]
- 37.Lee MH, Klein RL, El-Shewy HM, Luttrell DK, Luttrell LM. The adiponectin receptors AdipoR1 and AdipoR2 activate ERK1/2 through a Src/Ras-dependent pathway and stimulate cell growth. Biochemistry 2008;47:11682–11692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fröhlich M, Sund M, Löwel H, Imhof A, Hoffmeister A, Koenig W. Independent association of various smoking characteristics with markers of systemic inflammation in men. Results from a representative sample of the general population (MONICA Augsburg Survey 1994/95). Eur Heart J 2003;24:1365–1372 [DOI] [PubMed] [Google Scholar]
- 39.Chandran M, Phillips SA, Ciaraldi T, Henry RR. Adiponectin: more than just another fat cell hormone? Diabetes Care 2003;26:2442–2450 [DOI] [PubMed] [Google Scholar]
- 40.Costanzo-Garvey DL, Pfluger PT, Dougherty MK, et al. KSR2 is an essential regulator of AMP kinase, energy expenditure, and insulin sensitivity. Cell Metab 2009;10:366–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.