Abstract
Background
Escherichia coli cell-free expression systems use bacteriophage RNA polymerases, such as T7, to synthesize large amounts of recombinant proteins. These systems are used for many applications in biotechnology, such as proteomics. Recently, informational processes have been reconstituted in vitro with cell-free systems. These synthetic approaches, however, have been seriously limited by a lack of transcription modularity. The current available cell-free systems have been optimized to work with bacteriophage RNA polymerases, which put significant restrictions to engineer processes related to biological information. The development of efficient cell-free systems with broader transcription capabilities is required to study complex informational processes in vitro.
Results
In this work, an efficient cell-free expression system that uses the endogenous E. coli RNA polymerase only and sigma factor 70 for transcription was prepared. Approximately 0.75 mg/ml of Firefly luciferase and enhanced green fluorescent protein were produced in batch mode. A plasmid was optimized with different regulatory parts to increase the expression. In addition, a new eGFP was engineered that is more translatable in cell-free systems than the original eGFP. The protein production was characterized with three different adenosine triphosphate (ATP) regeneration systems: creatine phosphate (CP), phosphoenolpyruvate (PEP), and 3-phosphoglyceric acid (3-PGA). The maximum protein production was obtained with 3-PGA. Preparation of the crude extract was streamlined to a simple routine procedure that takes 12 hours including cell culture.
Conclusions
Although it uses the endogenous E. coli transcription machinery, this cell-free system can produce active proteins in quantities comparable to bacteriophage systems. The E. coli transcription provides much more possibilities to engineer informational processes in vitro. Many E. coli promoters/operators specific to sigma factor 70 are available that form a broad library of regulatory parts. In this work, cell-free expression is developed as a toolbox to design and to study synthetic gene circuits in vitro.
Background
Cell-free systems are used to synthesize large amounts of recombinant proteins in a few hours. Cell-free expression, which has many advantages in comparison to cell-based expression, is employed in an increasing number of biotechnology and proteomic applications [1]. Many efforts are made to increase the protein productivity and the functionality of cell-free systems. The energy regeneration is frequently optimized [2-6], new E. coli strains are engineered to stabilize some amino acids or to express proteins with PCR products [7,8] and preparation of the crude extract is simplified [9]. Current extracts prepared from E. coli cells can deliver between half and one milligram per milliliter of reporter protein after a few hours of incubation in a test tube. To achieve high protein yield, these extracts use bacteriophage RNA polymerases for transcription, such as T7 and T3, with their specific promoters, because they are the most efficient polymerases. Although well established, however, cell-free expression is narrowed to bacteriophage transcription and is not necessarily adapted to all types of applications. Bacteriophage transcription cannot replace endogenous RNA polymerases for the synthesis of particular genes [10]. In cell-free systems, bacteriophage transcription has to be modified to improve expression of specific genes [11]. Reconstitution of cell-free informational processes, and on a broader range the field of in vitro synthetic biology, is limited to bacteriophage RNA polymerases [12-17]. Only a few synthetic bacteriophage promoters regulated by operators have been characterized, such as the T7/lacO system. The reconstitution of informational processes in vitro would greatly benefit from having efficient cell-free systems that offer a greater modularity at the level of transcription. Promoter/operators from E. coli provide much more modularity for transcription [18]. E. coli cell-free extracts working with the endogenous core RNA polymerase and the housekeeping sigma factor 70 were originally prepared to study gene expression mechanisms [19,20]. However, the endogenous transcription of E. coli extract, which is slower and less efficient than bacteriophage transcription, is not believed to allow efficient in vitro synthesis of proteins. For this reason, bacteriophage transcription has been largely favored at the expense of endogenous transcription systems.
In this work, the preparation of an efficient cell-free expression system that uses the endogenous E. coli RNA polymerase only with sigma factor 70 for transcription is described. Using recent improvements in cell-free preparation and a plasmid with optimized regulatory parts, 0.75 mg/ml (0.6 mg/ml active) of Firefly luciferase (Luc) and 0.75 mg/ml (0.65 mg/ml active) of enhanced green fluorescent protein (eGFP) were produced. The best protein production was obtained with the 3-PGA energy regenerating system. The expression was also characterized with buffers containing either CP or PEP for the energy regeneration. The preparation of the crude extract, which takes only half a day without freezing the cells, was reduced to a routine procedure faster than the preparation of bacteriophage cell-free systems. A new eGFP cDNA was engineered that allow much higher production of the reporter in cell-free system when it is transcribed from an E. coli promoter. The expression and the protein production were characterized with this new enhanced green fluorescent protein and with Luc. This cell-free system can deliver large amounts of proteins without addition of exogenous RNA polymerase.
Methods
Extract preparation
The crude extract was prepared with E. coli BL21 Rosetta2 cells (Novagen) grown at 37°C in 2xYT medium up to OD600 = 1.5, according to Kigawa et al [21] and Liu et al [9]. All the following steps were performed either on ice or at 4°C except for the pre-incubation at 37°C. The cells were washed twice and resuspended in S30 buffer A (50 mM Tris, 60 mM potassium glutamate, 14 mM magnesium glutamate, pH 7.7, 2 mM DTT). The cells were broken with a bead-beater (Biospecs Products Inc, mini bead-beater-1) using 0.1 mm glass beads. The extract was clarified by centrifugation at 30000 g for 25 minutes. The clear supernatant was pre-incubated 80 minutes at 37°C followed by a centrifugation at 30000 g for 10 minutes. The clear supernatant was dialyzed against S30 buffer B (5 mM Tris, 60 mM potassium glutamate, 14 mM magnesium glutamate, pH 8.2, 1 mM DTT) for 3 hours with a Slide-A-Lyzer cassette (MWCO 10 kDa, Pierce Biotechnology). The crude extract was stored at -80°C after dialysis. A typical concentration of 27-30 mg/ml of proteins in the crude extract was measured by Bradford assay. The crude extract is stable at least 1 year at -80°C.
Cell-free reaction
The cell-free reactions were composed of 33% crude extract (9-9.5 mg/ml of proteins), the other 66% contained the reaction buffer and the plasmid. The reaction buffers were prepared with slight modifications according to Sitaraman and co-workers for 3-PGA [4], Kim and Swartz for PEP [3] and Kim and co-workers for CP [22]. 3-PGA and PEP are natural E. coli substrates, therefore no enzyme was added to the reaction. Creatine phosphate is not a natural substrate for E. coli. Creatine kinase was added according to Kim and co-workers [22].
(i) 3-PGA buffer
50 mM Hepes pH 8, 1.5 mM ATP and GTP each, 0.9 mM CTP and UTP each, 1 mM spermidine, 0.75 mM cAMP, 0.33 mM NAD, 0.26 mM coenzymeA, 30 mM 3-PGA, 0.068 mM folinic acid, 0.2 mg/ml tRNA, 1 mM IPTG.
(ii) PEP buffer
50 mM Hepes pH 8, 1.5 mM ATP and GTP each, 0.9 mM CTP and UTP each, 1 mM putrescine, 0.75 mM cAMP, 0.33 mM NAD, 0.26 mM coenzymeA, 30 mM PEP, 0.068 mM folinic acid, 0.2 mg/ml tRNA, 1 mM IPTG.
(iii) CP buffer
50 mM Hepes pH 8, 1.5 mM ATP and GTP each, 0.9 mM CTP and UTP each, 0.5 mM spermidine, 0.75 mM cAMP, 0.33 mM NAD, 0.26 mM coenzymeA, 67 mM CP, 0.0032 mg/ml creatine kinase, 0.068 mM folinic acid, 0.17 mg/ml tRNA, 1 mM IPTG.
The concentrations of amino acids (0.5-1.5 mM for eGFP and 1.5 mM for Luc, for each amino acid), PEG 8000 (0.5-2%), additional magnesium glutamate (0-10 mM) and additional potassium glutamate (0-120 mM) were adjusted depending on the reporter and the buffer used. The optimum plasmid concentration is discussed in the results and discussion section. The volume of each reaction was 10 μl. The reactions were incubated at 22°C for Luc and 29°C for eGFP with no shaking. The reagents were purchased from Sigma, USB Corporation (GTP, CTP, UTP) and Roche (tRNA, amino acids). The concentrations of magnesium and potassium shown in the graphs are additional to the magnesium and potassium provided by the crude extract. The crude extract brings an additional 4.5 mM of magnesium glutamate and 20 mM of potassium glutamate.
Plasmids
All the plasmids used in this study originate from the plasmid pBEST-Luc (Promega). The list and sequences of the different regulatory parts are reported in the additional file 1 (List on page 1). Luc refers to Firefly luciferase [GenBank: CAA59281.1], eGFP to the enhanced green fluorescent protein [GenBank: CAD97424.1], eGFP-Del6 to the enhanced green fluorescent protein truncated and modified in N-terminal, eGFP-Del6-229 to the enhanced green fluorescent protein truncated and modified in N- and C-terminal, PtacI to the consensus E. coli sigma factor 70 promoter [23], OR2-OR1-Pr to the lambda repressor Cro promoter [GenBank: J02459.1], UTR1 to the untranslated region containing the T7 g10 leader sequence for highly efficient translation initiation [24] [GenBank: M35614.1] and T500 to the transcription terminator [25]. The plasmids were prepared according to the standard molecular cloning procedures. Picogreen (Invitrogen) was used to measure plasmid concentrations. The E. coli strain KL740 was purchased from the Coli Genetic Stock Center (Yale) [CGSC#: 4382].
Measurements
A Wallac Victor III platereader (PerkinElmer) was used to measure eGFP, eGFP-Del6 and eGFP-Del6-229 expression (kinetics and end-point measurements). Pure recombinant eGFP (Clontech) was used for calibration. Luc expression was measured with a custom-built luminometer described previously [12]. Pure Luc and Luc assay kit (Promega) were used for calibration and measurements. Polyacrylamide gel electrophoresis was carried out according to the standard procedures and labeled with SimplyBlue safestain (Invitrogen).
Results and discussion
Material and cell-free preparation
The crude extract was prepared according to Kigawa et al [21] and Liu et al [9] with some modifications. The usual buffers S30 A and S30 B used for the preparation of cell-free systems were composed of magnesium glutamate and potassium glutamate instead of the acetate form of the ions [26]. DTT only was used for the washing and the resuspension steps (S30 buffer A) at a concentration of 2 mM instead of the usual combination of 2-mercaptoethanol and DTT. The optimum pH for the S30 buffer A and the S30 buffer B was 7.7 and 8.2 respectively. The cells were broken with a mini bead-beater. This lysis method is more reproducible than the other methods [21] and this technique is also more affordable. Typically, 4.5 g of cells were resuspended in 4.05 ml of S30 buffer A before adding 23 g of dry beads. The mixture was homogenized before bead beating twice at 4600 rpm for 30 seconds with a pause of 30 seconds in between. The mixture was centrifuged at 30000 g for 25 minutes followed by a pre-incubation of the clear supernatant at 37°C for 80 minutes. The extract was clarified again at 30000 g for 10 minutes before 3 hours of dialysis. The entire preparation, from cell culture to crude extract storage, takes 12 hours and does not present any particular constraints compared to other procedures. The cells do not need to be frozen at any time during the process. The crude extract can be thawed once without any loss of activity. Two liters of cell culture give approximately 6 ml of crude extract with a final protein concentration of 27-30 mg/ml.
Optimization of the reaction
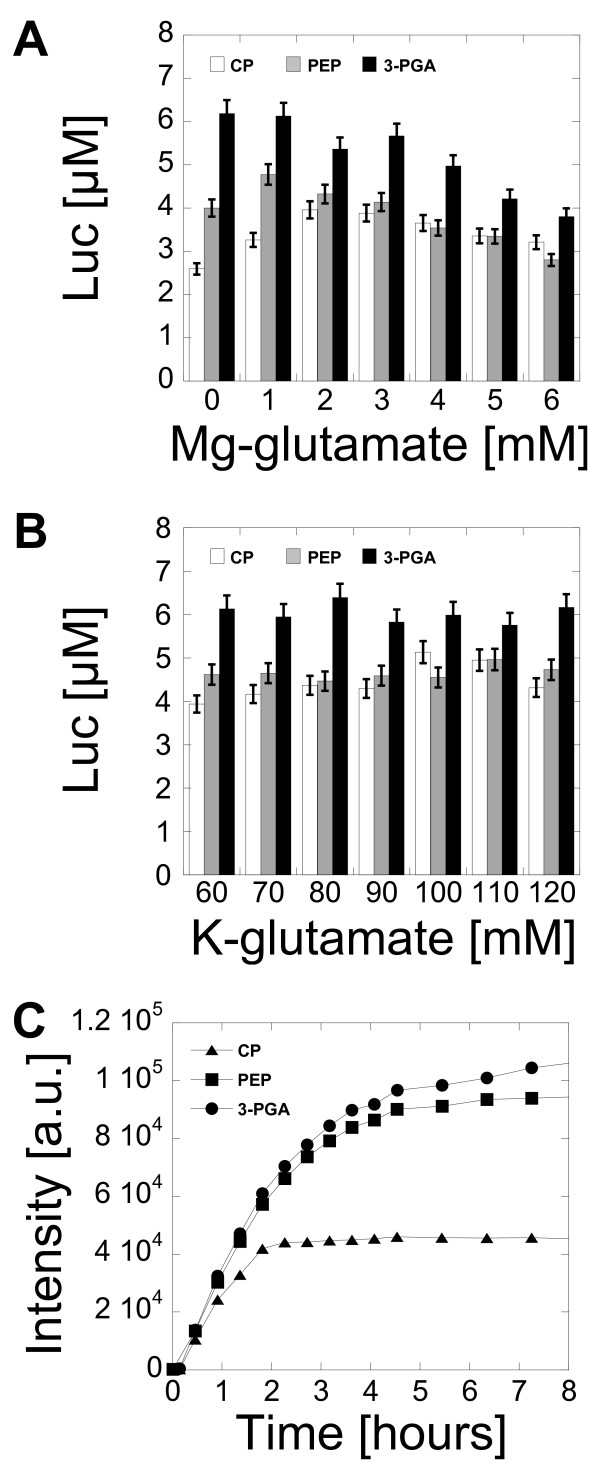
The cell-free reaction contained all the usual components of bacteriophage systems with minor modifications. Almost all the components in the reaction were optimized. The results for DTT, tRNA, and amino acids are reported in the additional file 1 (Figures S1, S2 and S3). The most important parameters for the optimization of the cell-free reactions were the magnesium concentration, the potassium concentration and the plasmid construction. The magnesium and the potassium concentrations were adjusted with the plasmids pBEST-UTR1-Luc and pBEST-UTR1-eGFP. The reporter genes were transcribed with a PtacI promoter [23]. PtacI, a consensus E. coli sigma factor 70 promoter, is one of the strongest E. coli promoters. The untranslated region of the plasmid pBEST-Luc was replaced by the untranslated region called UTR1 in this work. The UTR1 sequence includes the T7 gene 10 leader RNA, which contains a strong ribosome binding site [24]. The lac operator overlapping the -10 part of the promoter was conserved. With this modification, the expression was increased by a factor of five to ten on average in any of the buffers. The protein production was characterized with the three most common ATP regeneration systems used in cell-free studies: creatine phosphate (CP), phosphoenolpyruvate (PEP) and 3-phosphoglyceric acid (3-PGA). In each of these buffers, we found that the concentration of magnesium had to be precisely adjusted in order to maximize protein production (Figure 1A). This result was similar to the results obtained with cell-free systems using bacteriophage RNA polymerases. The best expression was obtained with 3-PGA, followed by PEP and CP. In the three buffers, the protein production was relatively independent of the potassium concentration over a wide range of concentrations (Figure 1B). The best protein production was also obtained with 3-PGA, followed by PEP and CP. For eGFP, the best expression was obtained with the 3-PGA buffer although the results obtained with the PEP buffer were close to 3-PGA (Figure 1C). All the following experiments were carried out with the 3-PGA buffer, which gave a greater protein production than PEP and CP buffers. In the 3-PGA buffer, the protein production was independent of the nucleotide concentrations over a wide range of concentrations (additional file 1, Figure S4.
Figure 1.
Protein synthesis as a function of magnesium glutamate and potassium glutamate in three different buffers CP, PEP and 3-PGA. (A) Luc synthesized as a function of magnesium glutamate with 5 nM plasmid pBEST-UTR1-Luc (CP, PEP and 3-PGA: 90 mM potassium glutamate, 1.5 mM of each amino acid, 2% PEG 8000). (B) Luc synthesized as a function of potassium glutamate with 5 nM plasmid pBEST-UTR1-Luc (CP: 2 mM magnesium glutamate, 1.5 mM of each amino acid, 2% PEG 8000; PEP: 1 mM magnesium glutamate, 1.5 mM of each amino acid, 2% PEG 8000; 3-PGA: 0 mM magnesium glutamate, 1.5 mM of each amino acid, 2% PEG 8000). (C) Kinetics of eGFP expression in the best conditions for the three different buffers with 5 nM plasmid pBEST-UTR1-eGFP (CP: 10 mM magnesium glutamate, 50 mM potassium glutamate, 1 mM of each amino acid, 2% PEG 8000; PEP: 9 mM magnesium glutamate, 30 mM potassium glutamate, 1 mM of each amino acid, 2% PEG 8000; 3-PGA: 6 mM magnesium glutamate, 40 mM potassium glutamate, 1 mM of each amino acid, 2% PEG 8000). Concentrations of magnesium glutamate and potassium glutamate reported here are the concentrations added to the cell-free reaction. The crude extract, dialyzed against the S30 buffer B, brings an additional 4.5 mM magnesium glutamate and 20 mM potassium glutamate to the cell-free reaction.
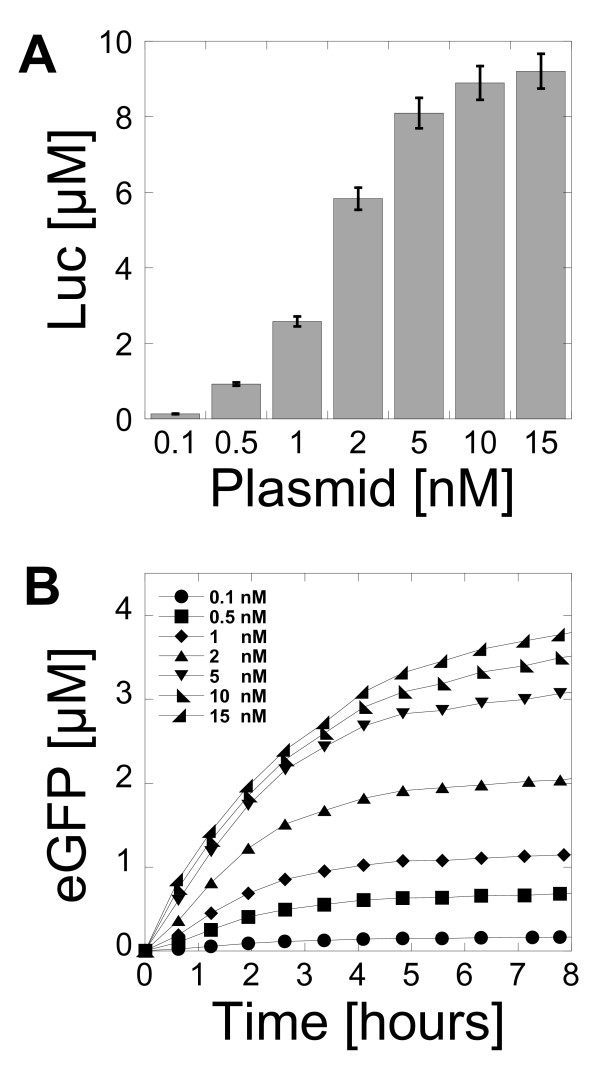
The plasmid concentration is another important parameter in a cell-free reaction. Because the cell-free system described in this study works with the endogenous RNA polymerase and sigma factor 70, it was interesting to determine the quantity of template required to get maximum protein production and to compare it to the amount of plasmid required in bacteriophage-based transcription systems. The best protein production was obtained with a plasmid concentration between 5 nM and 15 nM for Luc (Figure 2A) and eGFP (Figure 2B). Surprisingly, the amount of plasmid required for maximum protein production was comparable to cell-free systems using bacteriophage RNA polymerase for transcription [2-6]. The protein production was linear with respect to the plasmid concentration below 2 nM. At a plasmid concentration of 5 nM, the protein production was 90% of the maximum for Luc and 80% of the maximum for eGFP. In our best extract preparation, a maximum Luc production of 10 μM could be measured, which corresponds to 0.6 mg/ml of active proteins. This protein yield, comparable to bacteriophage cell-free systems, was unexpected because the endogenous E. coli transcription machinery is much less efficient. However, the maximum production of eGFP was much smaller than Luc, with only 0.1 mg/ml produced (4 μM). Addition of pure E. coli RNA polymerase saturated with sigma factor 70 did not increase the protein production (additional file 1, Figure S5). This led us to the conclusion that expression could be optimized with different DNA regulatory parts and that eGFP was poorly translated. Furthermore, the amplification of plasmids with E. coli promoters can be problematic because of potential over-expression of the recombinant proteins. The plasmids pBEST-UTR1-Luc and pBEST-UTR1-eGFP were leaky during plasmid amplification despite the lac operator.
Figure 2.
Protein synthesis as a function of plasmid concentration. (A) Luc synthesized as a function of plasmid pBEST-UTR1-Luc concentration in 3-PGA buffer (conditions: 0 mM magnesium glutamate, 80 mM potassium glutamate, 1.5 mM of each amino acids, 0.5% PEG 8000). (B) Kinetics of eGFP expression as a function of plasmid pBEST-UTR1-eGFP concentration in 3-PGA buffer (conditions: 4 mM magnesium glutamate, 60 mM potassium glutamate, 1 mM of each amino acids, 2.5% PEG 8000). Concentrations of magnesium glutamate and potassium glutamate reported here are the concentrations added to the cell-free reaction. The crude extract, dialyzed against the S30 buffer B, brings an additional 4.5 mM magnesium glutamate and 20 mM potassium glutamate to the cell-free reaction.
Plasmid optimization
The maximum production of Luc was obtained with the plasmid pBEST-UTR1-Luc, which contains the PtacI consensus promoter and UTR1 (Figure 2A). No other modifications increased Luc production. A maximum of 0.6 mg/ml (10 μM) active Luc was measured by luminescence. eGFP production with the plasmid pBEST-UTR1-eGFP was significantly lower with only a few micromolars of eGFP produced (Figure 2B). This was far below the expected amount compared to Luc with the same plasmid. We found that translation initiation of eGFP did not work efficiently in our extract. To resolve this problem, the N-terminal sequence of eGFP was truncated according to the work of Li et al [27] and modified by silent mutations to remove ribosome binding site-like sequences. The new protein was named eGFP-Del6. Compared to pBEST-UTR1-eGFP, the fluorescence intensity at the end of the reaction was approximately 3 times greater with the plasmid pBEST-UTR1-eGFP-Del6 (Table 1). This increase, which is a real increase of protein production as opposed to an increase of the reporter fluorescence, was confirmed later by polyacrylamide gel electrophoresis (Figure 4B). The C-terminal sequence of eGFP was also modified [27]. A 20% increase in protein production was measured with this new reporter, named eGFP-Del6-229 (Table 1).
Table 1.
eGFP and eGFP variants synthesis as a function of DNA regulatory parts
| Plasmid pBEST- | Plasmid concentration [nM] | Protein production [μM] (mg/ml) | |
|---|---|---|---|
| eGFP | 1 | 0.02 | (0.00067) |
| UTR1-eGFP | 1 | 1.05 | (0.0284) |
| UTR1-eGFP-T500 | 1 | 1.10 | (0.0298) |
| UTR1-eGFP-Del6 | 1 | 3.16 | (0.0836) |
| UTR1-eGFP-Del6-229 | 1 | 3.72 | (0.0944) |
| UTR1-eGFP-Del6-229-T500 | 1 | 4.65 | (0.118) |
| OR2-OR1-Pr-UTR1-eGFP | 1 | 1.40 | (0.0375) |
| OR2-OR1-Pr-UTR1-eGFP-Del6 | 1 | 3.30 | (0.0874) |
| OR2-OR1-Pr-UTR1-eGFP-Del6-229 | 1 | 4.24 | (0.108) |
| OR2-OR1-Pr-UTR1-eGFP-Del6-229-T500 | 1 | 5.07 | (0.129) |
| OR2-OR1-Pr-UTR1-eGFP-Del6-229-T500 | 2 | 10.4 | (0.264) |
| OR2-OR1-Pr-UTR1-eGFP-Del6-229-T500 | 5 | 20.2 | (0.513) |
| OR2-OR1-Pr-UTR1-eGFP-Del6-229-T500 | 10 | 25.2 | (0.639) |
| OR2-OR1-Pr-UTR1-eGFP-Del6-229-T500 | 15 | 24.2 | (0.615) |
| OR2-OR1-Pr-UTR1-eGFP-Del6-229-T500 | 20 | 23.4 | (0.593) |
At 1 nM, protein expression with pBEST-OR2-OR1-Pr-UTR1-eGFP-Del6-229-T500 is about 250 times greater than expression with pBEST-eGFP (conditions: 5 mM magnesium glutamate, 50 mM potassium glutamate, 1.5 mM of each amino acid, 2% PEG 8000). Concentrations of magnesium glutamate and potassium glutamate reported here are the concentrations added to the cell-free reaction. The crude extract, dialyzed against the S30 buffer B, brings an additional 4.5 mM magnesium glutamate and 20 mM potassium glutamate to the cell-free reaction.
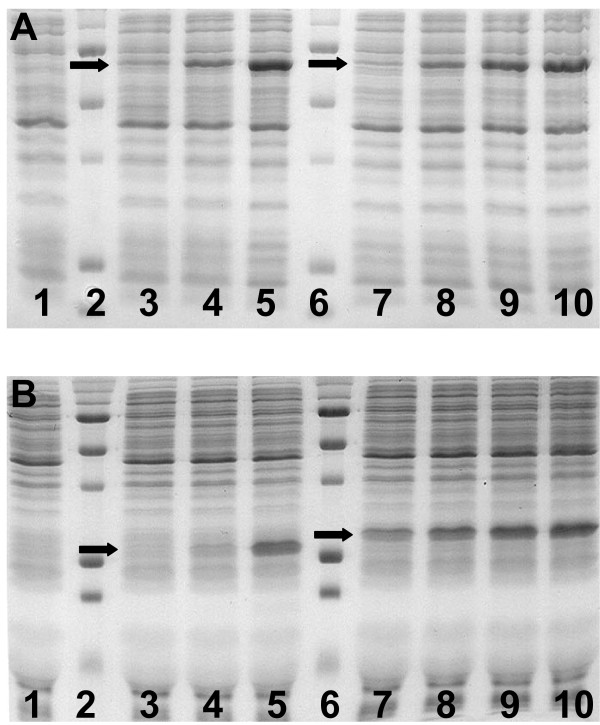
Figure 4.
Analysis of protein production by polyacrylamide gel electrophoresis. For both gel, lane 1 is a negative control (no plasmid added in the reaction), lane 2 and 6 is a protein ladder (5 main bands from top to bottom 75, 50, 37, 25 and 20 kDa). Synthesized proteins are indicated by arrows. (A) Luc synthesized with 10 nM pBEST-Luc plasmid (lane 3), 1.5 nM pBEST-UTR1-Luc plasmid (lane 4), and 15 nM pBEST-UTR1-Luc plasmid (lane 4) (3-PGA buffer, conditions: 1 mM magnesium glutamate, 60 mM potassium glutamate, 1.5 mM of each amino acid, 0.5% PEG 8000). Pure Luc added to the cell-free reaction with no plasmid, 2 μM (lane 7), 6 μM (lane 8), 10 μM (lane 9) and 14 μM (lane 10). (B) eGFP synthesized with 15 nM pBEST-eGFP plasmid (lane 3), 1 nM pBEST-OR2-OR1-Pr-UTR1-eGFP-Del6-229-T500 plasmid (lane 4) and 10 nM pBEST-OR2-OR1-Pr-UTR1-eGFP-Del6-229-T500 plasmid (lane 5), (3-PGA buffer, conditions: 3 mM magnesium glutamate, 30 mM potassium glutamate, 1.5 mM of each amino acid, 2% PEG 8000). Pure eGFP added to the cell-free reaction with no plasmid, 10 μM (lane 7), 15 μM (lane 8), 20 μM (lane 9) and 25 μM (lane 10). Concentrations of magnesium glutamate and potassium glutamate reported here are the concentrations added to the cell-free reaction. The crude extract, dialyzed against the S30 buffer B, brings an additional 4.5 mM magnesium glutamate and 20 mM potassium glutamate to the cell-free reaction.
Cell-free expression with E. coli sigma factor 70 promoters can pose significant problems of toxicity due to a potential over-expression of the recombinant protein during plasmid amplification. Although showing some signs of toxicity, the plasmids pBEST-UTR1-Luc and pBEST-UTR1-eGFP-Del6-229 were stable during amplification. A more general solution to this problem was found by switching to the strong Lambda phage promoter Pr flanked by the two operators OR2 and OR1. The E. coli strain KL740, which over-expresses the temperature sensitive Lambda repressor Cl857, was used to efficiently repress and stabilize the plasmids during cell growth at temperatures below 30°C. Potential problems due to increased background expression during plasmid amplification could also be solved by changing the origin of replication. The plasmid pBEST-Luc has a ColE1 origin of replication (few hundreds copies per cell), which can be replaced by a P15A (10-12 copies per cell) or a PSC101 (2-5 copies per cell) origin of replication. Other tightly regulated transcription systems and strains could be also used [28].
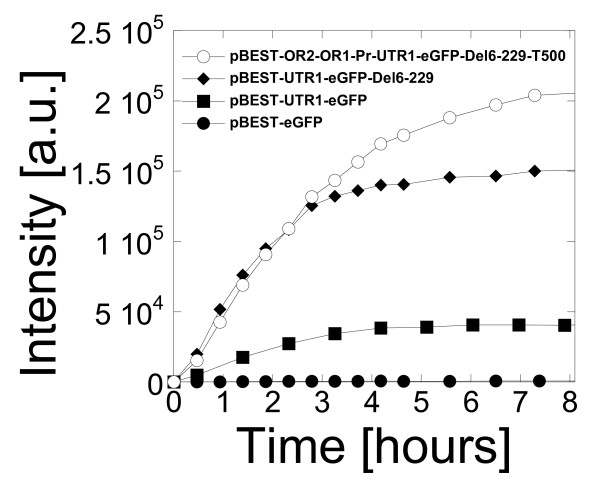
In the case of eGFP-Del6-229, expression with the OR2-OR1-Pr promoter was slightly greater than with the PtacI promoter (Table 1). A 20% increase in eGFP-Del6-229 production was observed with the addition of the strong transcription terminator T500 [25]. For eGFP-Del6-229, the best expression was obtained with the plasmid pBEST-OR2-OR1-Pr-UTR1-eGFP-Del6-229-T500 at a concentration of 10 nM (Table 1). In the best conditions, a concentration of 0.65 mg/ml of active eGFP-Del6-229 was measured. The protein was synthesized at the same rate whether transcribed from a PtacI or a OR2-OR1-Pr promoter (Figure 3). However, the expression lasted a longer time with the lambda promoter. The production of eGFP with the original plasmid pBEST-eGFP was more than 100 times smaller than with the plasmid pBEST-OR2-OR1-Pr-UTR1-eGFP-Del6-229-T500.
Figure 3.
Cell-free protein synthesis of eGFP variants as a function of DNA regulatory parts. Expression kinetics of four eGFP variants, 1 nM plasmid final concentration (3-PGA buffer, conditions: 5 mM magnesium glutamate, 50 mM potassium glutamate, 1.5 mM of each amino acid, 2% PEG 8000). Concentrations of magnesium glutamate and potassium glutamate reported here are the concentrations added to the cell-free reaction. The crude extract, dialyzed against the S30 buffer B, brings an additional 4.5 mM magnesium glutamate and 20 mM potassium glutamate to the cell-free reaction.
The high protein production for both Luc and eGFP-Del6-229 was confirmed by polyacrylamide gel electrophoresis (Figure 4A and 4B). SDS PAGE analysis showed that eGFP-Del6-229 production was greater than eGFP production. This was essentially due to the modification of the N-terminal of the protein (Table 1). An amount of 0.75 mg/ml for both Luc and eGFP-Del6-229 could be estimated on gel whereas the amount of active reporter proteins measured by luminescence was 0.60 mg/ml (Luc) and by fluorescence 0.65 mg/ml (eGFP-Del6-229). The plasmid maps for pBEST-UTR1-Luc and pBEST-OR2-OR1-Pr-UTR1-eGFP-Del6-229-T500 can be found in the additional file 1 (Figure S6).
The protein productions measured in this work were comparable to the protein productions obtained with cell-free systems using bacteriophage RNA polymerases. Expression of GFP with a T7 transcription, characterized and quantified in two cell-free systems by Iskakova and co-workers [11], was comprised between 0.3 and 0.7 mg/ml. A maximum of 0.15 mg/ml of active Luc was measured in two different cell-free systems [12,14]. Furthermore, the protein production obtained with commercial cell-free systems based on bacteriophage transcription is comprised between 0.5 and 1 mg/ml.
With the CP buffer, the maximum Luc and eGFP-Del6-229 production was only 60% of the amount obtained with 3-PGA. With the PEP buffer, the maximum Luc and eGFP-Del6-229 production was 60% and 100% respectively of the amount obtained with 3-PGA. It is important to note that Luc production was neither greater nor smaller with the different regulatory parts tested with eGFP. The production of Luc with the plasmids pBEST-UTR1-Luc and pBEST-OR2-OR1-Pr-UTR1-Luc-T500 were identical. The plasmid pBEST-OR2-OR1-Pr-UTR1-Luc-T500 was easier to amplify in the E. coli strain KL740.
Expression of reporters as a function of temperature
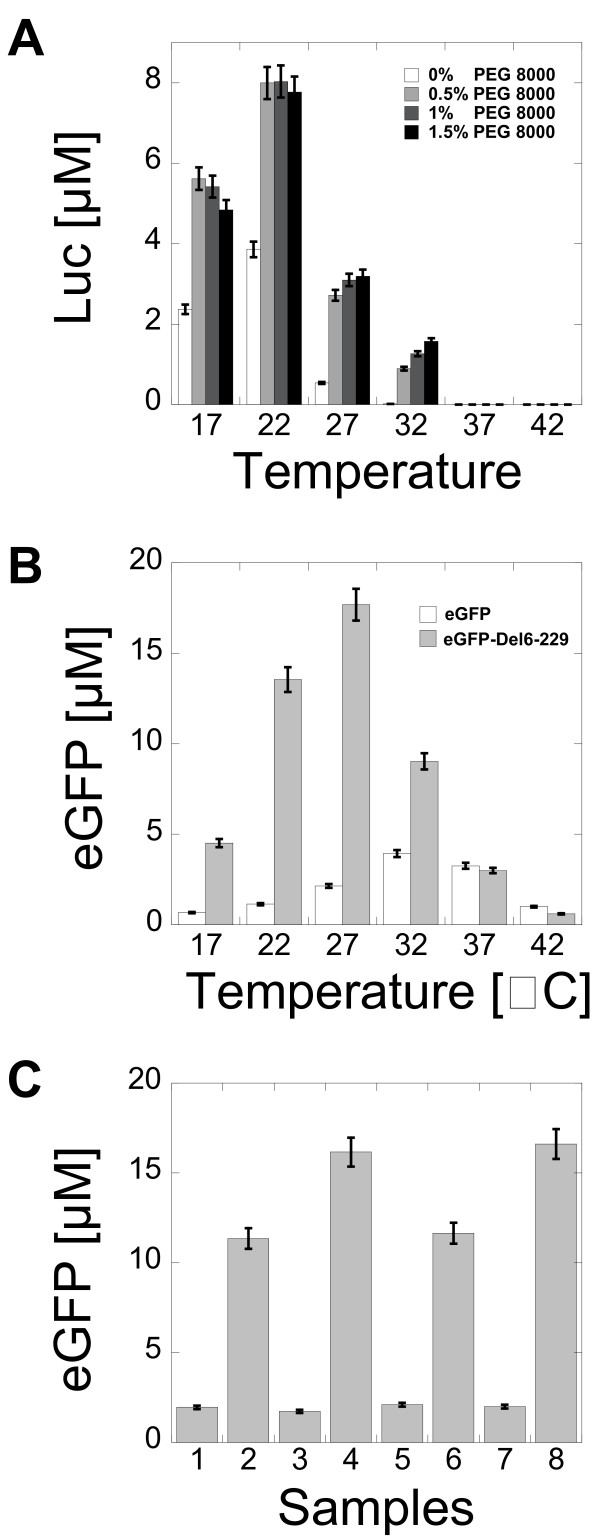
The protein production in cell-free reactions depends on the temperature of incubation. For Luc, the best protein production was obtained in the temperature range 20-25°C (Figure 5A). This result was in agreement with previous studies on the reporter's activity that showed a net decrease of luminescence above room temperature [29-31]. In the case of eGFP, the best protein production was obtained at 32°C (Figure 5B). At this temperature, the expression of eGFP-Del6-229 was twice greater than eGFP. The best production of eGFP-Del6-229 was obtained at 29°C. At 37°C and 42°C, expression of eGFP and eGFP-Del6-229 were comparable. Additional tests were performed to determine if the temperature range 29-32°C was optimum for gene expression or optimum for thermo-stability of the proteins. Cell-free reactions were carried out at 29°C and 32°C for both eGFP and eGFP-Del6-229 during 8 hours followed by 1 hour of incubation at 37°C (Figure 5C). No change in fluorescence was observed. This result confirmed that the production of fluorescent proteins was maximum in the temperature range 29-32°C. Similar results have been obtained for the wild type GFP [11]. In the best conditions, eGFP production never reached more than 25% of the maximum obtained for eGFP-Del6-229. The expression of eGFP and eGFP-Del6-229 was also tested in E. coli using the same plasmid (pBEST-UTR1 with a PtacI promoter). The expression of eGFP-Del6-229 was slightly better than eGFP (additional file 1, Figure S7).
Figure 5.
Luc, eGFP and eGFP-Del6-229 production as a function of temperature. (A) End-point measurement of Luc production as a function of PEG 8000 concentration and as a function of the reaction incubation temperature (3-PGA buffer, conditions: 0 mM magnesium glutamate, 80 mM potassium glutamate, 1.5 mM of each amino acid). (B) End-point measurement of eGFP and eGFP-Del6-229 production as a function of the reaction incubation temperature (3-PGA buffer, conditions: 4 mM magnesium glutamate, 60 mM potassium glutamate, 1.5 mM of each amino acid, 2.5% PEG 8000). (C) Temperature stability of eGFP and eGFP-Del6-229 protein expressed with 2 nM plasmids (pBEST-UTR1-eGFP-T500 and pBEST-UTR1-eGFP-Del6-229-T500). 1) 8 hours expression of eGFP at 32°C, 2) 8 hours expression of eGFP-Del6-229 at 32°C, 3) 8 hours expression of eGFP at 29°C, 4) 8 hours expression of eGFP-Del6-229 at 29°C, 5) 8 hours expression of eGFP at 32°C followed by 1 hour of incubation at 37°C, 6) 8 hours expression of eGFP-Del6-229 at 32°C followed by 1 hour of incubation at 37°C, 7) 8 hours expression of eGFP at 29°C followed by 1 hour of incubation at 37°C, 8) 8 hours expression of eGFP-Del6-229 at 29°C followed by 1 hour of incubation at 37°C. Concentrations of magnesium glutamate and potassium glutamate reported here are the concentrations added to the cell-free reaction. The crude extract, dialyzed against the S30 buffer B, brings an additional 4.5 mM magnesium glutamate and 20 mM potassium glutamate to the cell-free reaction.
Conclusions
In this work, an efficient cell-free expression system driven only by the endogenous E. coli RNA polymerase was prepared. Surprisingly, the protein production with this extract is comparable to bacteriophage systems. This new system is as workable as bacteriophage systems and does present any particular constraints. In order to obtain efficient protein production, the reaction conditions and its components were optimized as well as the regulatory parts of the plasmid carrying the recombinant gene. Our work, however, does not exclude further improvements. Other promoters, untranslated regions and terminators could be tested to increase the protein production. We used the three most common ATP regeneration systems, 3-PGA, PEP and CP. Cell-free protein synthesis with this system, however, could also benefit from recent results at the energy regeneration level [6]. In the course of this study, we found that eGFP was not translated efficiently when it was expressed in vitro from an E. coli promoter. The gene encoding the reporter was modified to get highly translatable reporter messenger RNAs. This cell-free system opens new possibilities to synthesize proteins and it broadens the range of potential applications of cell-free systems.
Abbreviations
ATP: Adenosine triphosphate; 3-PGA: 3-phosphoglyceric acid; PEP: phosphoenolpyruvate; CP: creatine phosphate; Luc: Firefly luciferase; eGTP: enhanced green fluorescent protein; SDS PAGE: polyacrylamide gel electrophoresis.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JS carried out all the experiments. VN carried out the background work of extract preparation and optimization. VN prepared the draft of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Supplementary information for Shin and Noireaux "Efficient cell-free expression with the endogenous E. Coli RNA polymerase and sigma factor 70". supplementary information includes a list of the sequences, data on expression optimization (DTT, tRNA, amino acids, nucleotides), expression using pure E. coli RNAP for transcription, plasmid maps, expression of eGFP and eGFP-Del6-229 in E. coli.
Contributor Information
Jonghyeon Shin, Email: shin@physics.umn.edu.
Vincent Noireaux, Email: noireaux@umn.edu.
Acknowledgements
The authors thank Nadezda Monina for technical assistance in the extract preparation, Jonathan Gapp and Catherine Raach for reading and correcting the manuscript. This work was supported by UMN startup funds, BSF Binational Science Foundation grant 2006398, National Science Foundation grant PHY-0750133.
References
- He M. Cell-free protein synthesis: applications in proteomics and biotechnology. N Biotechnol. 2008;25:126–32. doi: 10.1016/j.nbt.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Kim DM, Kigawa T, Choi CY, Yokoyama S. A highly efficient cell-free protein synthesis system from Escherichia coli. Eur J Biochem. 1996;239:881–6. doi: 10.1111/j.1432-1033.1996.0881u.x. [DOI] [PubMed] [Google Scholar]
- Kim DM, Swartz JR. Regeneration of adenosine triphosphate from glycolytic intermediates for cell-free protein synthesis. Biotechnol Bioeng. 2001;74:309–16. doi: 10.1002/bit.1121. [DOI] [PubMed] [Google Scholar]
- Sitaraman K, Esposito D, Klarmann G, Le Grice SF, Hartley JL, Chatterjee DK. A novel cell-free protein synthesis system. J Biotechnol. 2004;110:257–63. doi: 10.1016/j.jbiotec.2004.02.014. [DOI] [PubMed] [Google Scholar]
- Kim TW, Oh IS, Keum JW, Kwon YC, Byun JY, Lee KH, Choi CY, Kim DM. Prolonged cell-free protein synthesis using dual energy sources: combined use of creatine phosphate and glucose for the efficient supply of ATP and retarded accumulation of phosphate. Biotechnol Bioeng. 2007;97:1510–5. doi: 10.1002/bit.21337. [DOI] [PubMed] [Google Scholar]
- Kim HC, Kim DM. Methods for energizing cell-free protein synthesis. J Biosci Bioeng. 2009;108:1–4. doi: 10.1016/j.jbiosc.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Michel-Reydellet N, Calhoun K, Swartz J. Amino acid stabilization for cell-free protein synthesis by modification of the Escherichia coli genome. Metab Eng. 2004;6:197–203. doi: 10.1016/j.ymben.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Michel-Reydellet N, Woodrow K, Swartz J. Increasing PCR fragment stability and protein yields in a cell-free system with genetically modified Escherichia coli extracts. J Mol Microbiol Biotechnol. 2005;9:26–34. doi: 10.1159/000088143. [DOI] [PubMed] [Google Scholar]
- Liu DV, Zawada JF, Swartz JR. Streamlining Escherichia coli S30 extract preparation for economical cell-free protein synthesis. Biotechnol Prog. 2005;21:460–5. doi: 10.1021/bp049789y. [DOI] [PubMed] [Google Scholar]
- Lewicki BT, Margus T, Remme J, Nierhaus KH. Coupling of rRNA transcription and ribosomal assembly in vivo. Formation of active ribosomal subunits in Escherichia coli requires transcription of rRNA genes by host RNA polymerase which cannot be replaced by bacteriophage T7 RNA polymerase. J Mol Biol. 1993;231:581–93. doi: 10.1006/jmbi.1993.1311. [DOI] [PubMed] [Google Scholar]
- Iskakova MB, Szaflarski W, Dreyfus M, Remme J, Nierhaus KH. Troubleshooting coupled in vitro transcription-translation system derived from Escherichia coli cells: synthesis of high-yield fully active proteins. Nucleic Acids Res. 2006;34:e135. doi: 10.1093/nar/gkl462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noireaux V, Bar-Ziv R, Libchaber A. Principles of cell-free genetic circuit assembly. Proc Natl Acad Sci USA. 2003;100:12672–7. doi: 10.1073/pnas.2135496100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isalan M, Lemerle C, Serrano L. Engineering gene networks to emulate Drosophila embryonic pattern formation. PLoS Biol. 2005;3:e64. doi: 10.1371/journal.pbio.0030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noireaux V, Libchaber A. A vesicle bioreactor as a step toward an artificial cell assembly. Proc Natl Acad Sci USA. 2004;101:17669–74. doi: 10.1073/pnas.0408236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, White KS, Winfree E. Construction of an in vitro bistable switch from synthetic transcriptional switches. Mol Syst Biol. 2006;2:68. doi: 10.1038/msb4100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takinoue M, Kiga D, Shohda K, Suyama A. Experiments and simulation models of a basic computation element of an autonomous molecular computing system. Phys Rev E. 2008;78:041921. doi: 10.1103/PhysRevE.78.041921. [DOI] [PubMed] [Google Scholar]
- Noireaux V, Bar-Ziv R, Godefroy J, Salman S, Libchaber A. Toward an artificial cell based on gene expression in vesicles. Phys Biol. 2005;2:1–8. doi: 10.1088/1478-3975/2/3/P01. [DOI] [PubMed] [Google Scholar]
- Cox RS, Surette MG, Elowitz MB. Programming gene expression with combinatorial promoters. Mol Syst Bio. 2007;3:145. doi: 10.1038/msb4100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubay G. In vitro synthesis of protein in microbial systems. Annu Rev Genet. 1973;7:267–87. doi: 10.1146/annurev.ge.07.120173.001411. [DOI] [PubMed] [Google Scholar]
- Pratt JM. In: Transcription and Translation: A Practical Approach. Hames BD, Higgins SJ, editor. IRL Press:Oxford U.K; 1984. Coupled transcription-translation in prokaryotic cell-free systems; pp. 179–209. [Google Scholar]
- Kigawa T, Yabuki T, Matsuda N, Matsuda T, Nakajima R, Tanaka A, Yokoyama S. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J Struct Funct Genomics. 2004;5:63–8. doi: 10.1023/B:JSFG.0000029204.57846.7d. [DOI] [PubMed] [Google Scholar]
- Kim TW, Keum JW, Oh IS, Choi CY, Park CG, Kim DM. Simple procedures for the construction of a robust and cost-effective cell-free protein system. J Biotechnol. 2006;126:554–561. doi: 10.1016/j.jbiotec.2006.05.014. [DOI] [PubMed] [Google Scholar]
- de Boer HA, Comstock LJ, Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc Natl Acad Sci USA. 1983;80:21–5. doi: 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins PO, Devine CS, Rangwala SH, Kavka KS. The T7 phage gene 10 leader RNA, a ribosome-binding site that dramatically enhances the expression of foreign genes in Escherichia coli. Gene. 1988;73:227–35. doi: 10.1016/0378-1119(88)90329-0. [DOI] [PubMed] [Google Scholar]
- Larson MH, Greenleaf WJ, Landick R, Block SM. Applied force reveals mechanistic and energetic details of transcription termination. Cell. 2008;132:971–82. doi: 10.1016/j.cell.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Koshiba S, Tochio N, Seki E, Iwasaki N, Yabuki T, Inoue M, Yokoyama S, Kigawa T. Improving cell-free protein synthesis for stable-isotope labeling. J Biomol NMR. 2007;37:225–9. doi: 10.1007/s10858-006-9127-5. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang G, Ngo N, Zhao X, Kain SR, Huang CC. Deletions of the Aequorea victoria green fluorescent protein define the minimal domain required for fluorescence. J Biol Chem. 1997;272:28545–9. doi: 10.1074/jbc.272.45.28545. [DOI] [PubMed] [Google Scholar]
- Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O, and AraC/I1-I2 regulatory elements. Nucl Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundovskikh I, Dementieva E, Ugarova N. Recombinant Firefly Luciferase in Escherichia coli Properties and Immobilization. Appl Biochem Biotech. 2000;88:127–136. doi: 10.1385/ABAB:88:1-3:127. [DOI] [Google Scholar]
- Moradi A, Hosseinkhani S, Naderi-Manesh H, Sadeghizadeh M, Alipour BS. Effect of charge distribution in a flexible loop on the bioluminescence color of firefly luciferases. Biochemistry. 2009;48:575–82. doi: 10.1021/bi802057w. [DOI] [PubMed] [Google Scholar]
- Ataei F, Hosseinkhani S, Khajeh K. Limited proteolysis of luciferase as a reporter in nanosystem biology: a comparative study. Photochem Photobiol. 2009;85:1162–7. doi: 10.1111/j.1751-1097.2009.00583.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information for Shin and Noireaux "Efficient cell-free expression with the endogenous E. Coli RNA polymerase and sigma factor 70". supplementary information includes a list of the sequences, data on expression optimization (DTT, tRNA, amino acids, nucleotides), expression using pure E. coli RNAP for transcription, plasmid maps, expression of eGFP and eGFP-Del6-229 in E. coli.