Abstract
Background
Congenital adrenal hyperplasia due to 21-hydroxylase deficiency is caused by deletions, large gene conversions or mutations in CYP21A2 gene. The human gene is located at 6p21.3 within a locus containing the genes for putative serine/threonine Kinase RP, complement C4, steroid 21-hydroxylase CYP21 tenascin TNX, normally, in a duplicated cluster known as RCCX module. The CYP21 extra copy is a pseudogene (CYP21A1P). In Brazil, 30-kb deletion forming monomodular alleles that carry chimeric CYP21A1P/A2 genes corresponds to ~9% of disease-causing alleles. Such alleles are considered to result from unequal crossovers within the bimodular C4/CYP21 locus. Depending on the localization of recombination breakpoint, different alleles can be generated conferring the locus high degree of allelic variability. The purpose of the study was to investigate the variability of deleted alleles in patients with 21-hydroxylase deficiency.
Methods
We used different techniques to investigate the variability of 30-kb deletion alleles in patients with 21-hydroxylase deficiency. Alleles were first selected after Southern blotting. The composition of CYP21A1P/A2 chimeric genes was investigated by ASO-PCR and MLPA analyses followed by sequencing to refine the location of recombination breakpoints. Twenty patients carrying at least one allele with C4/CYP21 30-kb deletion were included in the study.
Results
An allele carrying a CYP21A1P/A2 chimeric gene was found unusually associated to a C4B/C4A Taq I 6.4-kb fragment, generally associated to C4B and CYP21A1P deletions. A novel haplotype bearing both p.P34L and p.H62L, novel and rare mutations, respectively, was identified in exon 1, however p.P30L, the most frequent pseudogene-derived mutation in this exon, was absent. Four unrelated patients showed this haplotype. Absence of p.P34L in CYP21A1P of normal controls indicated that it is not derived from pseudogene. In addition, the combination of different approaches revealed nine haplotypes for deleted 21-hydroxylase deficiency alleles.
Conclusions
This study demonstrated high allelic variability for 30-kb deletion in patients with 21-hydroxylase deficiency indicating that a founder effect might be improbable for most monomodular alleles carrying CYP21A1P/A2 chimeric genes in Brazil.
Background
Congenital adrenal hyperplasia (CAH) is an autosomal recessive inborn error of metabolism due to the deficiency of one of the enzymes involved in the adrenal steroidogenesis [1]. More than 95% of the cases are due to deficiency of 21-hydroxylase enzyme (21-OHD; OMIM +201910). Diminished or absent 21-hydroxylase activity leads to reduction or abolishment of cortisol and aldosterone syntheses, consequently, an over production of androgens occurs [1,2]. Different clinical presentations are classified in classical form that can be salt-wasting (SW) or simple-virilizing (SV), and late-onset non-classical (NC) form [1-3]. Mutations on CYP21A2 gene, which encodes the enzyme, are responsible for the disease [2,3].
The CYP21A2 gene and CYP21A1P, its pseudogene, are located within the class III locus of the human major histocompatibility complex at the short arm of chromosome 6 [4,5]. Both have 10 exons, except that CYP21A1P is inactive due to deleterious mutations [6,7]. Approximately 30 kb separate CYP21A2 from CYP21A1P. The genes of the fourth component of complement C4A and C4B, also map to this locus and are located, respectively, upstream and downstream to the CYP21A1P [4,5].
In addition other genes, such as RP1 and TNXB and their truncated gene fragments or pseudogenes, RP2 and TNXA, are present within this genetic unit [8]. Each RP, C4, CYP21, TNX copy together forms the RCCX module [8] that maps within approximately 30 kb [9-11]. In general, 70% of alleles are bimodular (two RCCX units) in most populations [12]. Nevertheless, monomodular (16%), and trimodular (14%) alleles with long and short C4B gene variants are not uncommon [12].
Deletion or duplication of CYP21A1P or CYP21A2 together with C4A or C4B always occurs [8,13-15]. The deletion and large gene conversion were the first mutations described in 21-hydroxylase deficiency alleles [16-19]. Generally, disease-causing 30-kb deletions extend from 3'end CYP21A1P and include C4B and 5' end CYP21A2 to form inactive CYP21A1P/A2 chimeric genes [20-27]. Whereas monomodular alleles with C4A/C4B chimeric genes may also be formed after unequal crossovers, they are frequently associated to a functional CYP21A2 gene copy [13,22]. Such rearrangements can be recognized in Southern blots using Taq I restriction enzyme where C4 and CYP21 chimeric genes produce 6.4- and 3.2-kb fragments, respectively [22,23]. Deletions, large gene conversions and duplications may be evaluated either by Southern blot [3] or by the recently developed Multiplex Ligation-dependent Probe Amplification (MLPA) technique [24]. As CYP21A1P/A2 chimeric genes may carry different pseudogene-derived mutations, disease-causing deletion alleles may vary according the chimeric gene composition [21,22,25,26]. Novel CYP21A1P/A2 chimeric genes have been continuously described in different studies [27,28].
In Brazil, 30-kb deletion monomodular alleles correspond to ~9% of disease-causing alleles [29,30]. The present study describes the variability of such alleles in Brazilian patients (n = 20) with 21-hydroxylase deficiency using the conventional Southern blot, ASO-PCR and sequencing techniques combined to recently developed MLPA technique. Nine different and two novel monomodular haplotypes are reported. A 6.4-kb Taq I fragment corresponding to a C4B/C4A chimeric gene was unusually associated to CYP21A1P/A2 chimeric gene that also bore novel and rare single nucleotide polymorphisms (SNPs). Another allele was identified with a CYP21A1P/A2 gene carrying both p.P34L and p.H62L mutations in exon 1, however p.P30L, the most frequent pseudogene-derived mutation in this exon, was absent. The study of 59 healthy controls revealed that the novel p.P34L mutation is not pseudogene-derived and occurred only in alleles carrying the 30-kb deletion whereas the rare p.H62L mutation can be pseudogene-derived since it was found in 3.4% pseudogene sequences.
Methods
Subjects
This study was approved by the Ethics Committee from Universidade Estadual de Campinas (São Paulo, Brasil) and an informed consent was obtained from patients and relatives.
Twenty patients (8 males, 12 females) with 21-hydroxylase deficiency were included in this study (table 1). They represent 20 unrelated families. Consanguinity was reported in two families. Fourteen patients presented with the salt-wasting (SW) form, whereas five manifested the simple-virilizing (SV) form and one with the non-classic form (NC). Fifty-two family members were genotyped. Fifty non-related health individuals were screened for novel and rare mutations.
Table 1.
Clinical data of CAH patients
| Case | Sex 1 | Age at diagnosis 2 | Clinical Data | basal 17OHP (nmol/L) | Na + /K + (mmol/L) | Phenotype 3 |
|---|---|---|---|---|---|---|
| 1 | M | 1 m 28 d | vomiting, dehydration | na4 | 129/6.2 | SW |
| 2 | F | 12 d | ambiguous genitalia; Prader III | > 20 ndm5 | 137/6.2 | SW |
| 3 | M | 2 m 24 d | vomiting, dehydration | 49.3 | 116/9.2 | SW |
| 4 | F | 19 d | vomiting, dehydration; registered as male | na | 128/7.8 | SW |
| 5 | M | 31 d | vomiting, dehydration | > 20 ndm | 120/5.5 | SW |
| 6 | M | 2 m 8 d | vomiting, dehydration | > 200 ndm | 121/6.8 | SW |
| 7 | F | 10 m | ambiguous genitalia; Prader III | > 25 ndm | 142/5.0 | SV |
| 8 | F | 6 d | ambiguous genitalia; Prader III/IV | > 200 ndm | 123/7.4 | SW |
| 9 | F | np | ambiguous genitalia; Prader IV | SW | ||
| 10 | M | 1 yr 1 m | vomiting, dehydration | > 25 ndm | 120/6.8 | SW |
| 11 | F | 6 yr 3 m | precocious pubarche since 4 yr 6 m; high stature | > 25 ndm | nt | NC |
| 12 | F | 10 d | ambiguous genitalia; Prader III | > 20 ndm | 121/6.5 | SW |
| 13 | F | np | ambiguous genitalia; Prader III/IV; vomiting, dehydration; registered as male | na | na | SW |
| 14 | F | np | ambiguous genitalia; Prader III | na | na | SW |
| 15 | F | 14 d | ambiguous genitalia; Prader IV | > 6 ndm | 119/5.3 | SW |
| 16 | M | 28 d | vomiting, dehydration, adrenal crysis | > 200 ndm | 119/9.7 | SW |
| 17 | M | 3 yr 5 m | precocious pubarche since 2 yr | > 20 ndm | na | SV |
| 18 | M | 6 yr | precocious pubarche | > 20 ndm | na | SV |
| 19 | F | 5 yr 7 m | precocious pubarche, ambiguous genitalia; Prader IV | 238 | na | SV |
| 20 | F | 37 d | ambiguous genitalia; Prader III | > 20 ndm | 136/5.7 | SV |
1F = Female, M = male; 2yr = years, m = months, d = days, np = neonatal period; 3SW = salt-wasting, SV = simple virilizing, NC = non-classic; 4na = not available; 5ndm = non-diluted measurements (ref value 5.97 nmol/L).
Methods
Southern blot and Allele-specific PCR (ASO-PCR)
Genomic DNA was obtained from peripheral blood by Proteinase K digestion and phenol/chloroform extraction [31]. Southern blots with Taq I digested DNA were performed following standard procedures [31]. Membranes were hybridized with CYP21 (pC21/3c) and C4 (C4B550) probes [29]. The probes were radiolabeled in a random priming reaction (Invitrogen, CA, USA) with α-32P-dCTP (Amersham, Uppsala, Sweden). The blots were washed using conventional conditions [31] and a final wash using 0.1 × SSC/0.1% SDS at 65°C was performed. Autoradiographies on Hyperfilm MP X-ray films (Amersham) were obtained. Blots were analysed following Taq I RFLP in C4 and CYP21 genes (Figure 1a) and relative band intensities.
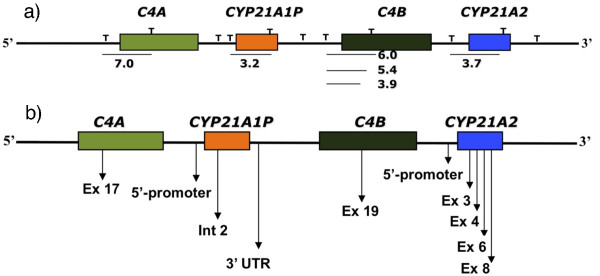
Figure 1.
Schematic representation illustrating the hybridization sites for C4 and CYP21 probes. a) Southern blot: bars indicate hybridization regions for each probe; T denote Taq I restriction sites; numbers indicate sizes in kb of fragments recognized by each probe; b) MLPA experiment: arrows indicate the location where each probe hybridizes. Ex = exon, Int - intron.
ASO-PCR was performed with primers for the eight most common CYP21A1P-derived mutations as described elsewhere [32]. Six single nucleotide polymorphisms (SNPs) were also investigated either using ASO-PCR with primers specifically designed on basis the gene sequences or by digestion with restriction enzymes. Primers for exon 6 normal or mutant sequences (CL6N or Cl6M) were used as selective primers. As pseudogenes are also amplified in the analysis of CYP21A1P/A2 chimeric genes with exon 6 mutant primer, chimeric genes were identified upon analysis of all family members.
Multiplex Ligation-dependent Probe Amplification (MLPA)
MLPA was performed exactly as recommended by the manufacturers using SALSA MLPA P050B kit (MRC-Holland, Amsterdam, Netherlands) [33]. Fragment analysis was performed on an ABI 310 Genetic Analyzer (ABI PRISM/PE Biosystems, Foster City, CA, USA) and results were analyzed using Genescan and Genotyper softwares (Applied Biosystems, Foster City, CA, USA). Calculations were performed according to the method described by Taylor et al. [34]. The probemix included in the MLPA kit contains probes for several regions within CYP21 locus (Figure1b). Data were analysed using free Coffalyser MLPA data analysis software [33]. Normalized relative values ranging from 0.8 to 1.2 corresponded to two gene copies in the genotype, whereas values below 0.8 and above 1.2 corresponded to deletion (one gene copy) and duplication (three gene copies), respectively. This confidence interval was established by data obtained with five bimodular controls in each MLPA assay.
Sequencing
A fragment including 600 bp from 5' promoter region to exon 6 (CL6N or CL6M) and a fragment from exon 6 to 3'UTR (CL6N or CL6M) were first amplified. The fragments were directly sequenced with internal primers using Big Dye™ Terminator Cycle Sequencing Kit V3.1 Ready Reaction (ABI PRISM/PE Biosystems). The sequences obtained in an ABI 3700 Automated Sequencer (ABI PRISM/PE Biosystems) were compared to CYP21A2 and CYP21A1P sequences (Ensembl - ENSG00000231852 and ENSG00000204338, respectively).
Results
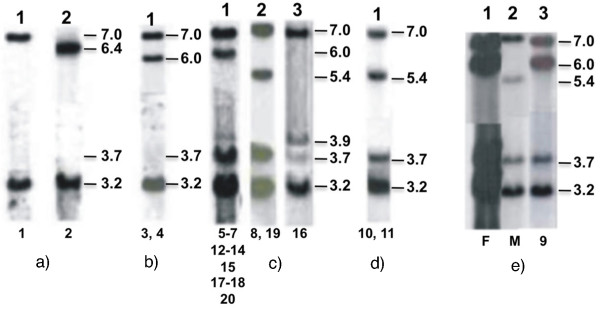
Southern blot analysis with CYP21 and C4 probes was performed to distinguish deletions from large gene conversions. ASO-PCRs using CL6N or CL6M sequences as selective primers were performed to investigate both pseudogene-derived mutation and polymorphisms present in chimeric CYP21A1/A2 and CYP21A2 genes. All family members have been analyzed; therefore, pseudogenes and chimeric genes carrying CL6M sequences could be distinguished. Southern blot revealed 30-kb deletion in 20 patients with different genotypes (table 2; Figure 2).
Table 2.
Genotypes of CAH patients
| Patients | Genotype | |
|---|---|---|
| Paternal Allele | Maternal Allele | |
| 1 | 30-kb del1 | 30-kb del |
| 2 | 30-kb del | 30-kb del |
| 3 | LGC2 | 30-kb del |
| 4 | 30-kb del | LGC |
| 5 | IVS2-2A > G | 30-kb del |
| 6 | 30-kb del | c.920_921insT |
| 7 | p.I172N | 30-kb del |
| 8 | 30-kb del | p.P30L |
| 9 | p.W19X | 30-kb del |
| 10 | p.Q318X | 30-kb del |
| 11 | 30-kb del | p.V281L |
| 12 | 30-kb del | IVS2-13A/C > G |
| 13 | p.R356W | 30-kb del |
| 14 | IVS2-13A/C > G | 30-kb del |
| 15 | c.920_921insT | 30-kb del |
| 16 | 30-kb del | p.R356W |
| 17 | p.I172N | 30-kb del |
| 18 | 30-kb del | p.Q318X |
| 19 | 30-kb del | p.I172N |
| 20 | 30-kb del | IVS2-13A/C > G |
130-kb del = deletion of 30 kb including 3'-end CYP21A1P, C4B and 5'-end CYP21A2,
2LGC = large gene conversion;
Figure 2.
Taq I Southern blots showing patients carrying monomodular alleles. Hybridizations with CYP21 (pC21/3c) and C4 (C4B550) probes are shown. a) Homozygous genotypes for monomodular alleles; b) compound heterozygous genotypes with mono- and bimodular alleles carrying large gene conversion; c) compound heterozygous genotypes with mono- and bimodular alleles carrying pseudogene-derived mutations; d) compound heterozygous genotype with mono- and trimodular alleles carrying pseudogene-derived mutations; e) compound heterozygous genotype with mono- and tetramodular alleles (lane 3); maternal genotype showing compound heterozygosis with mono- and bimodular alleles (lane 2); and paternal genotype with tetramodular and bimodular alleles (lane 1). Numbers below each lane depict patient numbers.
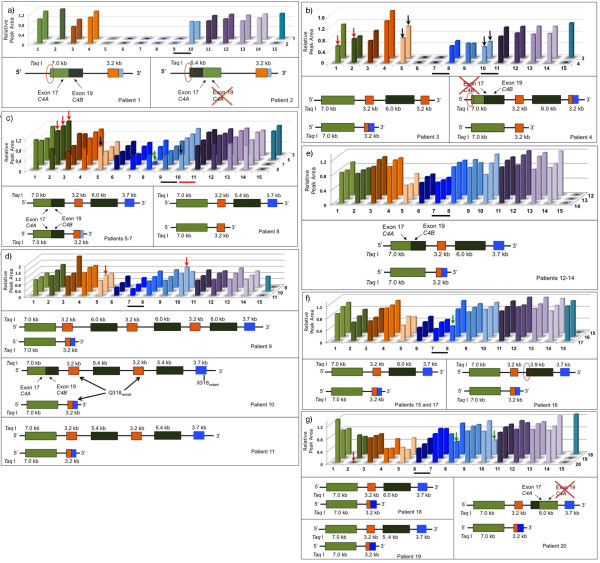
MLPA technique was used to confirm Southern blot results and to obtain data on the composition of chimeric genes since it includes one probe for each C4A and C4B genes, and three and five probes, respectively, for CYP21A1P and CYP21A2 genes (Figure 1b). The MLPA kit available also analyzes TNXB located within the RCCX locus and CREBL1 on chromosome 6p21.3 and nineteen control genes as well. For all patients, normalized data showed two copies for TNXB and CREBL1 genes (Figure 3) on chromosome 6 and also for control genes (data not shown) indicating no copy number variation in other loci.
Figure 3.
Integrated and normalized MLPA data. C4A, C4B, CYP21A1P, CYP21A2, TNXB, CREB1 are shown. Numbers 1-15 correspond to: (1) C4A exon 17, (2) C4B exon 19, (3-5) CYP21A1P 5' promoter region, intron 2, 3'UTR; (6-10) CYP21A2 5' promoter region, exon 3, exon 4, exon 6, exon 8; (11-13) TNXB exon 32, 15, 1; (14) CREB1 probe. (15) Y-chromosome probe. Horizontal black bars denote recombination breakpoints (RB); red bar, for patient 8. Columns correspond to integrated and normalized electropherogram peak areas, values between 0.8 and 1.2 indicate two copies, below and above correspond to one or more than two copies, respectively. Patients' numbers are on the right. Green arrows indicate heterozygosis for p.I172N and p.Q318X (patient 18). Black arrows indicate 2:1 ratio for two copies of 3'UTR pseudogene (5) and one copy of exon 8 (10). (a-g) Upper panels - MLPA results; lower panels - schematic genotypes. a) Monomodular homozygosis; red circles denote different C4 5'-end; b) Mono- and bimodular alleles carrying chimeric genes; red arrows indicate 1:2 ratio (C4A:C4B); c) heterozygosis for mono- and bimodular alleles; red arrows indicate three copies of C4B; d) heterozygosis for mono- and tri- or tetramodular alleles; red arrows indicate 2:2 ratio for 3'-end CYP21A1P (5) to exon 8 CYP21A2 (10); patient 10 - trimodular allele bearing normal p.Q318 in the two pseudogenes; e) heterozygosis for alleles mono- and bimodular with CYP21A1P/A2 and C4A/C4B, respectively; f-g) heterozygosis for mono- and bimodular alleles; RB between exons 3-4 and 1-3, respectively; red arrow - null C4B hybridization signal (patient 20).
After sequencing CYP21A1P/A2 chimeric genes, novel mutations and novel single nucleotide polymorphisms (SNPs) have been identified. Haplotypes were classified according to C4 gene characteristics and SNPs and mutations present in CYP21A1P/A2 chimeric genes. Table 3 shows nine different haplotypes for monomodular alleles identified in this study.
Table 3.
Haplotypes for monomodular alleles defined on basis of C4 and CYP21A1P/A2 chimeric gene
| Haplotypes | I | II | III | IV | V | VI | VII | VIII | IX | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients | 2 | 1 | 5 | 6 | 7 | 4 | 8 | 3 | 10 | 15 | 16 | 9 | 11 | 12 | 13 | 14 | 17 | 18 | 19 | 20 | ||
| C4 | Sb1 | 6.4 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | |
| MLPA2 | B/A | A/B | A/B | A/B | A/B | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | ||
| CYP21A1P/A2 | 5' | RB3 | 6/8 | 6/8 | 6/8 | 6/8 | 6/8 | 3'end | 3'end | 3/4 | 3/4 | 3/4 | 3/4 | 3/4 | 3/4 | 3/4 | 3/4 | 3/4 | 3/4 | 1/3 | 1/3 | 1/3 |
| -4494 | A | A | A | A | A | A | - | A | A | A | A | A | A | - | G | G | G | A | A | A | ||
| -3085 | C | C | C | C | C | C | - | G | G | G | G | C | C | - | C | C | C | C | C | C | ||
| -2896 | C | T | T | T | T | T | - | T | T | T | T | T | T | - | T | T | T | T | T | T | ||
| -47 | T | C | C | C | C | T | T | C | C | C | C | T | T | T | T | T | T | T | T | T | ||
| E1 | p.P30L8 | T | T | T | T | T | T | T | C | C | C | C | - | T | T | T | - | T | T | T | T | |
| p.P34L9 | C | C | C | C | C | C | C | T | T | T | T | - | C | C | C | - | C | C | C | C | ||
| p.H62L10 | A | A | A | A | A | A | A | T | T | T | T | - | A | A | A | A | A | A | A | A | ||
| I2 | 39511 | T | C | C | C | C | T | T | C | C | C | C | C | C | C | T | T | T | C | C | C | |
| 41912 | - | - | - | - | - | C | C | C | C | C | C | C | C | C | C | C | C | C | C | A | ||
| 54713 | - | - | A | - | - | A | A | A | A | - | A | A | A | A | A | - | A | A | A | C | ||
| 560_566insG14 | - | 6Gs | 6Gs | - | 6Gs | 6Gs | 6Gs | 6Gs | 6Gs | - | 6Gs | 6Gs | 6Gs | 6Gs | 6Gs | - | 6Gs | 6Gs | 6Gs | 7Gs | ||
| 60215 | A | A | A | A | A | A | A | A | A | - | A | A | A | A | A | - | A | C | C | C | ||
| E4 | p.I172N | A | A | A | A | A | A | A | T | T | T | T | T | T | T | T | T | T | T | T | T | |
| I5 | 125316 | A | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | - | - | - | |
| E7 | p.S268T17 | G | C | C | C | C | C | G | G | G | G | G | G | G | G | G | G | G | G | G | G | |
| p.V281L18 | T | T | T | T | T | T | T | G | G | G | G | G | G | G | G | G | G | G | G | G | ||
| E8 | p.Q318X19 | C | C | C | C | C | T | T | C | C | C | C | C | C | C | C | C | C | C | C | C | |
| p.R356W20 | C | C | C | C | C | C | T | C | C | C | C | C | C | C | C | C | C | C | C | C | ||
Numbers are relative to ATG initiation codon in the genomic DNA based on CYP21A2 sequence described by Higashi et al. (1986); mutation caused by nucleotide changes; E, exon; I, intron. 1Sb, size (kb) of C4 Taq I restriction fragments obtained in Southern blots. 2Composition of the C4 gene copy in monomodular alleles indicated by MLPA assays - A, corresponds to C4A and B, to C4B genes. 3RB recombination breakpoint in CYP21A1P/A2 chimeric gene indicated by MLPA and ASO-PCR experiments; 6/8, between exons 6 an 8; 3/4, between exons 3 an 4; 1/3, between exons 1 an 3. CYP21A1P /CYP21A2 consensus nucleotides for polymorphic positions (number at NCBI-SNP database), most frequent nucleotide for each SNP is indicated first: 4A > G (rs28361032)/A (non-polymorphic); 5C (non-polymorphic)/C > G (rs3130676); 6T (non-polymorphic)/T (non-polymorphic); 7T > C (rs6470)/C > T (rs6470); 8T > C (rs9378251)/C > T (rs9378251); 9C (non-polymorphic)/C (non-polymorphic); 10A > T (rs9378252)/A > T (rs9378252); 11C > T (rs28361033)/T > C > G (rs6462); 12A (non-polymorphic)/A > C (rs6448); 13A(non-polymorphic)/C (non-polymorphic); 146Gs (non-polymorphic)/7Gs > 6Gs (not registered); 15A(non-polymorphic)/C > A > G (rs6451); 16G > A (rs28691121)/G (non-polymorphic); 17G (non-polymorphic)/G > C (rs6472); 18T > G (rs41315836)/G > T (rs6471); 19T > C (rs7755898)/C > T (rs7755898); 20T > C (rs7755898)/C > T (rs7769409).
Patients 1 and 2, who were children of consanguineous marriages, were homozygous for a monomodular allele. However, hybridization results with C4 probe indicated a 6.4-kb Taq I fragment in patient 2, whereas patient 1 presented the usual 7.0-kb C4A fragment (Figure 2a). Molecular investigation on patient's 2 family demonstrated that the novel allele has been transmitted through three generations (data not shown). MLPA indicated similar composition of CYP21A1P/A2 chimeric genes in both cases (Figure 3a, upper panel), but once again the two patients differed in the C4 gene composition (Figure 3a, lower panels). Patient 2, who carried the 6.4-kb Taq I fragment, presented MLPA signal for two copies only for C4A probe, whereas patient 1, who carried the 7.0-kb C4A Taq I fragment, showed signals for two copies of C4A and C4B probes, suggesting that both C4 in the genotype have exon 19 sequences corresponding to C4B gene. Therefore, the two different C4 sequence compositions indicate distinct 30-kb deletion alleles. Patient 1 showed haplotype II (table 3) that, besides the C4A/B chimeric copy, also presented the C allelic variant at both -4 (SNP-rs6470) and g.395 (SNP-rs6462) nucleotides. Haplotype I is unique for patient 2 (table 3). In addition to the C4B/A[6.4] gene, it carries the novel -289T > C SNP in CYP21A1P/A2 promoter region. The T variant at g.395 (SNP-rs6462) nucleotide and the rare pseudogene-derived g.1253A variant (SNP-rs28691121) were also detected (table 3).
Patients 3 and 4 presented Southern blot results compatible with compound heterozygosis for 30-kb deletion and large gene conversion in which no signal for 3.7-kb CYP21A2 fragment was observed, whereas C4A[7.0] and C4B[6.0] fragments were present with intensities of 2:1 (Figure 2b). In MLPA experiments C4A:C4B ratios for patients 3 and 4 were 2:1 and 1:2, respectively (Figure 3b, upper panel). Both showed MLPA profiles compatible with mono-/bimodular compound heterozygosis with two different CYP21A1P/A2 chimeric genes (Figure 3b). However, the composition of monomodular allele in each case could only be defined after MLPA analysis of their parents (data not shown). For patient 3, the breakpoint between exon 3 and 4 in the chimeric CYP21A1P/A2 gene was defined for the paternal inherited monomodular allele. The maternal inherited bimodular allele carrying large gene conversion presented a complete pseudogene sequence indicated by the 2:1 ratio for probes CYP21A1P-3'end and CYP21A2-exon 8 in MLPA (Figure 3b, left lower panel) instead of the 1:2 expected if it was a chimeric gene with CYP21A2-3'end. Similar results were obtained for patient 4 except that in this case the complete pseudogene was concluded to be associated to the monomodular allele, whereas the chimeric CYP21A1P/A2 gene with recombination breakpoint between exon 3 and 4 was associated to the bimodular allele with large gene conversion (Figure 3b, right lower panel). In addition, based on MLPA analysis of her parents (data not shown), the maternal bimodular allele in patient 4 demonstrated to carry one of C4 copies in which C4A probe could not hybridize; therefore the 1:3 ratio observed in her mother indicated absence of C4A exon 17 sequences and presence of C4B exon 19 sequences which is suggestive of a chimeric C4A/B gene or a gene conversion at exon 17 (Figure 3b, right lower panel). Considering C4 composition and CYP21A1P/A2 SNPs, the 30-kb deleted alleles in patients 3 and 4 correspond to haplotypes V and III, respectively (table 3). Haplotype V is defined by the presence of the novel p.P34L and the rare p.H62L mutations, but absence of the pseudogene-derived p.P30L, in exon 1. The variant -308G and the variant C at -4 and g.395 nucleotides characterized this haplotype in which the recombination breakpoint was located between exon 3 and 4.
For other sixteen patients (5-20) who were compound heterozygous with monomodular and bi-, tri- or a probable tetramodular alleles bearing pseudogene-derived mutations in the CYP21A2 gene (Figure 2c-e; table 2), different C4A to C4B and CYP21A1P to CYP21A2 probe ratios in MLPA were observed (Figure 3c-g). These patients were divided in two major groups according to the status of exon 6 in the CYP21A1P/A2 chimeric genes: patients 5-8 with mutant exon 6 are shown in Figure 3c, and patients 9-20 with normal exon 6 in Figure 3d-g.
Chimeric CYP21A1P/A2 gene composition for alleles in patients 5-7 seemed to be similar to that in patients 1 and 2 with mutant exon 6 and normal exon 8 (Figure 3a-c, upper panels). However, C4A:C4B ratios were 2:3, suggesting that both C4A copies may carry exon 19 corresponding to C4B gene similar to that observed for patient 1. MLPA results for each family (data not shown) indicated the segregation of both C4 and CYP21 chimeric genes in the same allele (Figure 3c, left lower panel). Patient 8 presented 2:1 MLPA ratio for C4A:C4B sequences in agreement with Southern blotting. MLPA showed that 21-hydroxylase gene copy in this case corresponded to a complete pseudogene as the ratio for probes CYP21A1P 3'-end and CYP21A2-exon 8 was 2:1 as described above for patients 3 and 4 (Figure 3c, right lower panel). SNPs in chimeric genes classified the monomodular alleles in patients 5-7 within haplotype II (table 3). Besides deletion, duplication and chimeric gene composition, MLPA analyzes could identify compound heterozygosis for p.I172N in patient 7 (table 2; Figure 3c), because the probe used for CYP21A2 exon 4 anneals to the normal sequence at codon 172. Patient 8 showed haplotype IV, which can be differentiated from III at the SNPs g.1645G > C (SNP-rs6472; p.S268T) and at the g.2110C>T (p.R356W) mutation (table 3).
MLPA results for patients 9-20 were shown in Figure 3d-g. Additionally, the probable localization of recombination breakpoint within CYP21A1P/A2 chimeric genes divided them in two groups: patients 9-17 with breakpoint between exon 3 and 4 (Figure 3d-f) and patients 18-20 between exon 1 and intron 2 (Figure 3g). A combination of mono- and a probable tetramodular allele was considered for patient 9 after analysis of allelic segregation in the family (Figure 2e). This was the most probable combination to explain the relative intensity for C4A[7.0]:C4B[6.0] (2:3) and for CYP21A2[3.7]:CYP21A1P[3.2] (1:4) fragments in the patient. The mother presented ratios of 2:1 for C4A[7.0]:C4B[5.4] and 1:3 for CYP21A2[3.7]:CYP21A1P[3.2] and was positive for p.P30L, IVS2-13A/C > G and Δ8 mutations in ASO-PCR experiments indicating a deleted allele with CL6N sequence. The daughter inherited the 30-kb deleted allele from the mother, therefore she should carry an allele with three copies of each C4B and CYP21A1P to produce the relative intensities observed in the Southern blot (Figure 2e, lanes 2 and 3). Patients 10 and 11 were compound heterozygotes for tri- and monomodular alleles. These genotypes may explain the 2:2 ratio for C4A and C4B genes observed in Southern blot (Figure 2d). However, a 2:2 ratio for C4A:C4B in MLPA experiments has been observed only for patient 11. Patient 10 showed a 2:3 gene ratio, which is probably due to a positive hybridization signal for exon 19 C4B probe within the C4A gene copy in the paternal trimodular allele, deduced from the family analysis, therefore his most probable genotype is shown in Figure 3d (middle panel). An additional difficulty in interpreting MLPA results for patient 10 was that he also carried p.Q318X in CYP21A2 gene copy of the paternal inherited trimodular allele indicated in ASO-PCR experiments (table 2). As CYP21A2 exon 8 MLPA probe anneals to the normal sequence at codon 318, the result expected in this case was a ratio of 2:1 for probes CYP21A1P 3'UTR to CYP21A2 exon 8, if both pseudogenes in the trimodular allele were mutant in codon 318. However a ratio 2:3 was obtained. Analysis of his parents (data not shown) indicated that the two pseudogenes in the paternal trimodular allele might carry the normal sequence in codon 318 (Figure 3d, middle panel). The monomodular alleles carried by patients 10 and 11 were classified, respectively, within haplotype V and VI (table 3).
Patients 12-14 presented 2:2 MLPA ratios for C4A:C4B contrasting with the 2:1 obtained on Southern blot (Figure 2c). In these cases, familial analysis demonstrated that the bimodular alleles they carry in compound heterozygosis with monomodular alleles, probably, contain C4A genes formed by 5'-end C4A[7.0], C4A exon 17, but exon 19 corresponds to C4B specific sequences (Figure 3e). The monomodular allele in patient 12 corresponds to haplotype VI, whereas patients 13 and 14 carry haplotype VII (table 3). Haplotype VI differs from VII by the -449A>G (SNP-rs28361032) and g.395T > C (SNP-rs6462).
Patients 15-19 demonstrated C4A:C4B ratios of 2:1 in both Southern blotting and MLPA (Figure 3f-g). A 3:0 ratio in MLPA for C4A:C4B was observed for patient 20, whereas Southern blotting showed a ratio of 2:1. This result might indicate a C4A gene copy with a 6.0-kb Taq I fragment in which exon 17 sequences correspond to C4A and C4B specific sequences at exon 19 are absent, therefore the most probable genotype in this case is shown in Figure 3g (right lower panel). MLPA indicated compound heterozygosis for p.I172N in patients 17 and 19 (table 2; Figure 3f,g). Likewise, it also detected compound heterozygosis with p.Q318X mutation in patient 18 (table 2; Figure 3g). The recombination breakpoint indicated in MLPA for patients 15-17 was between exon 3 - 4 (Figure 3f). Sequencing data indicated haplotype V for alleles in patients 15 and 16 and haplotype VII for patient 17 (table 3). The recombination breakpoint indicated in MLPA for patients 18-20 was between exon 1 and intron 2 (Figure 3f-g). However, sequencing results suggested distinct alleles within this group. In haplotype VIII, which is shared by patients 18 and 19, the recombination breakpoint was probably located between the SNP 560_566insG and 602A > C (SNP-rs6451) as shown in table 3. Whereas, the recombination breakpoint for the allele carried by patient 20 (table 3, haplotype IX) was probably located at the beginning of intron 2 since it carried the variant A at the g.419 position (SNP - rs6448), which is more frequent in CYP21A2 gene and the nucleotide C at 547 position that is characteristic of the CYP21A2 gene.
Sequencing 170 CYP21A2 genes from obligate heterozygote individuals and CYP21A1P sequences from 59 control individuals indicated that p.P34L was not present in any gene or pseudogene sequences, whereas heterozygosis for p.H62L was observed in the pseudogene of four individuals (data not shown). Therefore, p.P34L seems to be a novel mutation not derived from pseudogene, whereas p.H62L showed a frequency of 3.4% in pseudogenes.
Discussion
This study reports two novel deletion alleles in Brazilian patients with 21-hydroxylase deficiency and describes the variability of C4/CYP21 monomodular alleles evaluated by combining Southern blot, ASO-PCR, MLPA and sequencing techniques. The occurrence of different chimeric genes in Brazilian patients had not been reported before. Although Real-Time PCR and HLA-haplotypes would be alternative techniques to estimate, respectively, C4 and CYP21A2 copy number [35-37] and founder effect they were not available for this work.
By combining different techniques we characterized monomodular alleles carrying CYP21A1P/A2 chimeric genes in genotypes that were compound heterozygous with bi - or trimodular alleles bearing pseudogene-derived mutations, including a bimodular allele carrying p.R356W mutation associated to the rare 3.9-kb C4B fragment (table 2, patient 16; Figure 2c), described before in a non-disease causing allele [38]. Patient 9 who inherited from the mother the monomodular allele might also carry a tetramodular allele inherited from the father, however a more detailed study is required to confirm the tetramodular configuration for this allele that also carry a novel g.60G > A (TGG > TGA) nucleotide change causing the p.W19X mutation. The same mutation had been described before as a result of the TGG > TAG nucleotide change [39].
In general, data were convergent independently on the technique used. However, patients with alleles carrying a C4B/A or C4A/B gene and those in compound heterozygosis with trimodular alleles could be misinterpreted as carrying large gene conversions if only MLPA had been used, because C4 probes resulted in signals indicative of alleles without C4B deletion. Patients 1 and 2, are typical examples. The 6.4-kb Taq I fragment identified in patient 2 is generally a marker for a deletion that results from unequal crossover between a C4A[7.0] and a short C4B[5.4] gene [26,40]. Such fragment is indicative of CYP21A1P deletion in monomodular alleles. It is found with a frequency of 11% in the general population and do not cause 21-hydroxylase deficiency [12] and would not be recognized in MLPA assay. The 6.4-kb gene variant is here described for the first time in association with a CYP21A1P/A2 gene in a case of 21-hydroxylase deficiency. Alleles bearing CYP21A1P deletions have been proposed as premutation for generating CYP21A2 deleted alleles [41]. It can be speculated that an allele bearing C4B/A[6.4] and CYP21A1P deletion might have influenced the formation of the monomodular allele in which the deleted allele seems to have been generated in at least two steps of unequal crossovers. However, those events must have happened several generations ago because the mutated allele had been segregating in this family for three generations. Additionally, those recombination events might have introduced novel nucleotide variants such as the -289T > C SNP in the 5'-promoter region of CYP21A1P/A2. The T variant at nucleotide 395, which is most frequent in CYP21A2 gene and a rare 1253G > A SNP within intron 5, which is described only in the CYP21A1P as a rare polymorphic nucleotide position, were also identified in this CYP21A1P/A2 chimeric gene. Therefore haplotype I was uniquely found in patient 2. In a paper published before [42], data obtained with ELISA assays also showed differences in C4 protein levels between patients 1 and 2. Low level of C4A protein was observed for patient 1 whereas patient 2 showed low level of C4B protein. Although C4A and C4B genes share 99% sequence identities, the proteins they encode have different hemolytic activities, covalent affinities to antigens and immune complexes, and serological reactivities [12,43]. C4A gene copies producing proteins with electrophoretical characteristics of C4A but acting antigenically as C4B have been described in association to 30-kb deletion haplotypes [20,44]. Differences conferring specific binding affinities are located within exon 26 [45], therefore monomodular allele in patient 1 may have 5'-end of C4A as indicated by 7.0-kb fragment and C4B sequences from exon 19 to 3'-end as indicated by MLPA and ELISA assays. Similarly, patients 5, 6 and 7 also carried a C4A[7.0] fragment with 3'-end C4B sequences as denoted by Southern blot and MLPA results. These patients together with patient 1 grouped within haplotype II, which is similar to those described in several populations [15,20,46,47]. In addition, patient 20 showed positive signal for C4A gene but no signal for C4B probe (3:0) in MLPA. This result indicate that exon 17 in the C4B[6.0] gene probably corresponded to C4A sequences and the patient had been described as having very low immune response toward C4B antiserum [42]. Although this allele was not associated to 30-kb deletion, it illustrates C4 gene variability that influences MLPA results.
Both haplotypes III and IV carry a complete CYP21A1P pseudogene copy indicating that the recombination breakpoint should map within the 5 kb that separate 3'-end CYP21A2 and the exon 32 of TNXB gene. They differed because haplotype III showed the C allelic variants in both g.1645 and g.2110 (p.R356W) nucleotides whereas G and T, respectively, were observed in haplotype IV (table 3). Polymorphisms in CYP21A1P pseudogene are known to be common [40], therefore those haplotypes could be originated from unequal crossovers involving bimodular alleles carrying different pseudogene variants. Recombination within 3'-end CYP21A2 seems to be frequent since it has been reported by different research groups [15,41] including as a de novo event in which deleted alleles were generated [48,49].
A group of ten genes had recombination breakpoints identified between exon 3 and 4. Haplotype V diverged from the VI and VII because the alleles did not carry the pseudogene-derived p.P30L, but showed p.P34L and p.H62L mutations in exon 1. p.H62L has been recently described in association with p.P453S in patients with the NC form of 21-hydroxylase deficiency [50,51]. In order to investigate whether p.P34L and p.H62L were rare pseudogene-derived mutations that have arisen in this chimeric gene through unequal crossover between gene and pseudogene, since they are flanked by CYP21A1P-like sequences, we tested 170 CYP21A2 genes from obligate heterozygous individuals and CYP21A1P sequences from 59 control individuals. Here we demonstrated that p.H62L might be originated in CYP21A1P since it was found in 3.4% pseudogene sequences. However, p.P34L was not found in any pseudogene sequences, therefore its origin remains unclear. Also, this haplotype showed the G variant at nucleotide -308 in the CYP21A1P/A2 5'-promoter region. Interestingly, this position is considered polymorphic only in CYP21A1P with frequencies varying from 0.1% to 0.3% in Chineses and Africans from Nigeria (NCBI-SNP database). Different mutations in exon 1 together with rare 5'-end SNP characterize a novel haplotype that seems to be frequent in Brazilian patients carrying monomodular alleles since it corresponded to 20% (4/20) of total 30-kb deleted alleles reported here as causing 21-hydroxylase deficiency. Probably, haplotype V have arisen in Brazil from an African ancestral since Brazilian population is highly miscigenated with populations from different origins including Africans of Benin Gulf region [52]. This haplotype might have segregated with 21-hydroxylase deficiency through an isolated founder effect for 30-kb deletion alleles. Main differences between haplotype VI and VII was found in -449 and g.395 nucleotides (table 3). The -449 position is considered to be ambiguous only in the pseudogene (SNP # rs28361032) whereas 395 is polymorphic in both gene and pseudogene sequences (SNP # rs6462) but there are no frequency data for the any of their allelic variants.
Haplotypes VIII and IX were separated because they showed different recombination breakpoints within intron 2. Those two haplotypes presented the CYP21A1P promoter region and p.P30L mutation as pseudogene-derived sequences and they are not very frequent in other populations [15,53].
CYP21A1P/A2 chimeric genes can be correlated with SW or SV forms of 21-hydroxylase deficiency depending on the mutations they carry [15,20,46,47]. The variability verified among 30-kb alleles with CYP21A1P/A2 chimeric genes present in different genotypes did not influence the phenotypes observed (tables 1, 2). Therefore, patients 1-6, 9-10 and 12-16 presented SW form of 21-hydroxylase deficiency as expected for homozygosis or compound heterozygosis for haplotypes I-VII and mutations that severely affect the enzyme activity (tables 1, 2). Similarly, compound heterozygote patients for haplotypes I-VII and p.I172N or p.V281L mutations presented, respectively, SV or NC forms of the disease (tables 1, 2, patients 7, 11, 17). Patients carrying haplotypes VIII and IX presented SV phenotype independently if they were compound heterozygous with a SV (tables 1, 2; p.I172N, patient 19) or SW mutations (tables 1, 2; p.Q318X and IVS2-13A/C > G, patients 18 and 20, respectively) indicating a synergistic effect of CYP21A1P promoter region and p.P30L upon the enzyme activity as proposed before [54].
Our study showed that the combination of Southern blot and ASO-PCR/direct sequencing with MLPA tests may constitute an option for mapping and better characterize chimeric genes on RCCX monomodular alleles, especially in populations with high allelic diversity such as that in Brazil. MLPA has been proposed as a candidate with good potential to be used in neo-natal screening and in pre-natal diagnosis because it can be performed with very low amount of DNA [55]. Eventually, MLPA may substitute time-consuming Southern blot in cases were HCA diagnosis is urgent as it managed to estimate the CYP21A1P/A2 borders in almost all cases. However, to distinguish between deletions and large gene conversions in genetic studies searching for detailed allelic information, it would be more informative if more C4 and CYP21 probes were included in the analysis.
Conclusions
Our study showed that the combination of Southern blot and ASO-PCR/direct sequencing with MLPA tests may constitute an option for mapping and better characterize chimeric genes on RCCX monomodular alleles, especially in populations with high allelic diversity such as that in Brazil. MLPA has been proposed as a candidate with good potential to be used in neo-natal screening and in pre-natal diagnosis because it can be performed with very low amount of DNA [55]. Eventually, MLPA may substitute time-consuming Southern blot in cases were HCA diagnosis is urgent as it managed to estimate the CYP21A1P/A2 borders in almost all cases. However, to distinguish between deletions and large gene conversions in genetic studies searching for detailed allelic information, it would be more informative if more C4 and CYP21 probes were included in the analysis.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
FBC carried out part of Southern blot, the MLPA and sequencing experiments and sequence alignment; FCS, RDB and RJP participated in ASO-PCR experiments for pseudogene-derived mutations and SNPs; MA, LCP and IFL participated in Southern blot and AS0-PCR studies of seven families; SHVLM, MTMB and GGJ were responsible for diagnosis and management of patients and participated in the design of the study; MPM conceived the study, and participated in its design and coordination and also drafted the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Fernanda B Coeli, Email: fbcoeli@bol.com.br.
Fernanda C Soardi, Email: soardi@unicamp.br.
Renan D Bernardi, Email: renandarin@gmail.com.
Marcela de Araújo, Email: maraujo@boldrini.org.br.
Luciana C Paulino, Email: luciana.paulino@ufabc.edu.br.
Ivy F Lau, Email: mpmello276@hotmail.com.
Reginaldo J Petroli, Email: rpetroli@unicamp.br.
Sofia HV de Lemos-Marini, Email: sofia.lemos@terra.com.br.
Maria TM Baptista, Email: terma@uol.com.br.
Gil Guerra-Júnior, Email: gileandrea@uol.com.br.
Maricilda P de-Mello, Email: mmello@unicamp.br.
Acknowledgements
Authors would like to thank Maria Madalena Vasconcelos Rosa for technical support and Dr. Márcio José da Silva from sequencing facility. This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP - grants # 92/03332-6, 97/07622-2, 01/08150-4, 05/00981-5 and FCS received scholarship # 03/01785-0), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (FBC, MA, IFL), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq - Brasil).
References
- New MI. Inborn errors of adrenal steroidogenesis. Mol Cel Endocrinol. 2003;211:75–83. doi: 10.1016/j.mce.2003.09.013. [DOI] [PubMed] [Google Scholar]
- White PC, Speiser PW. Congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Endocrine Reviews. 2000;21:245–291. doi: 10.1210/er.21.3.245. [DOI] [PubMed] [Google Scholar]
- Keegan CE, Killeen AA. An overview of molecular diagnostic of steroid 21-hydroxylase deficiency. J Mol Diagnostic. 2001;3:49–54. doi: 10.1016/S1525-1578(10)60651-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll MC, Campbell RD, Bentley DR, Porter RR. A molecular map of the human major histocompatibility complex class III region linking complement genes C4, C2 and factor B. Nature. 1984;307:237–241. doi: 10.1038/307237a0. [DOI] [PubMed] [Google Scholar]
- White PC, Grossberger D, Onufer BJ, Chaplin DD, New MI, Dupont B, Strominger JL. Two genes encoding steroid 21-hydroxylase are located near the genes encoding the fourth complement in man. Proc Natl Acad Sci USA. 1985;82:1089–1093. doi: 10.1073/pnas.82.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi Y, Yoshioka H, Yamane M, Gotoh O, Fujii-Kuriyama Y. Complete nucleotide sequence of two steroid 21-hydroxylase genes tandemly arranged in human chromosome: a pseudogene and a genuine gene. Proc Natl Acad Sci USA. 1986;83:2841–2845. doi: 10.1073/pnas.83.9.2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PC, New MI, Dupont B. Structure of human steroid 21-hydroxylase genes. Proc Natl Acad Sci USA. 1986;83:5111–5115. doi: 10.1073/pnas.83.14.5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Mendoza AR, Welch TR, Zipf WB, Yu CY. Modular variations of the human major histocompatibility complex class III genes for serine/threonine kinase RP, complement component C4, steroid 21-hydroxylase CYP21, and tenascin TNX (the RCCX module). A mechanism for gene deletions and disease associations. J Biol Chem. 1999;274:12147–12156. doi: 10.1074/jbc.274.17.12147. [DOI] [PubMed] [Google Scholar]
- Dangel AW, Mendoza AR, Baker BJ, Daniel CM, Carroll MC, Wu LC, Yu CY. The dichotomous size variation of human complement C4 genes is mediated by a novel family of endogenous retroviruses, which also establishes species-specific genomic patterns among Old World primates. Immunogenetics. 1994;40:425–436. doi: 10.1007/BF00177825. [DOI] [PubMed] [Google Scholar]
- Tassabehji M, Strachan T, Anderson M, Campbell RD, Collier S, Lako M. Identification of a novel family of human endogenous retroviruses and characterization of one family member, HERV-K(C4), located in the complement C4 gene cluster. Nucleic Acids Res. 1994;22:5211–5217. doi: 10.1093/nar/22.24.5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu X, Rittner C, Schneider PM. Length polymorphism of the human complement component C4 gene is due to an ancient retroviral integration. Exp Clin Immunogenet. 1995;12:74–81. [PubMed] [Google Scholar]
- Blanchong CA, Zhou B, Rupert KL, Chung EK, Jones KN, Sotos JF, Zipf WB, Rennebohm RM, Yu Yung C. Deficiencies of human complement component C4A and C4B and heterozygosity in length variants of RP-C4-CYP21-TNX (RCCX) modules in caucasians. The load of RCCX genetic diversity on major histocompatibility complex-associated disease. J Exp Med. 2000;191:2183–2196. doi: 10.1084/jem.191.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider PM, Carroll MC, Alper CA, Rittner C, Whitehead AS, Yunis EJ, Colten HR. Polymorphism of the human complement C4 and steroid 21-hydroxylase genes. Restriction fragment length polymorphisms revealing structural deletions, homoduplications, and size variants. J Clin Invest. 1986;78:650–657. doi: 10.1172/JCI112623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppens PF, Hoogenboezem T, Degenhart HJ. Duplication of the CYP21A2 gene complicates mutation analysis of steroid 21-hydroxylase deficiency: characteristics of three unusual haplotypes. Hum Genet. 2002;111:405–10. doi: 10.1007/s00439-002-0810-7. [DOI] [PubMed] [Google Scholar]
- Lee HH. Chimeric CYP21P/CYP21 and TNXA/TNXB genes in the RCCX module. Mol Gen Met. 2005;84:4–8. doi: 10.1016/j.ymgme.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Carroll MC, Palsdottir A, Belt KT, Porter RR. Deletion of complement C4 and steroid 21-hydroxylase genes in the HLA class III region. EMBO J. 1985;4:2547–2552. doi: 10.1002/j.1460-2075.1985.tb03969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werkmeister JW, New MI, Dupont B, White PC. Frequent deletion and duplication of the steroid 21-hydroxylase genes. Am J Hum Genet. 1986;39:461–469. [PMC free article] [PubMed] [Google Scholar]
- White PC, Vilek A, Dupont B, New MI. Characterization of frequent deletions causing steroid 21-hydroxylase deficiency. Proc Natl Acad Sci USA. 1988;85:4436–4440. doi: 10.1073/pnas.85.12.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel Y, David M, Forest MG, Betuel H, Hauptman G, Andre J, Bertrand J, Miller WL. Gene conversions and rearrangements cause discordance between inheritance of forms of 21-hydroxylase deficiency and HLA types. J Clin Endocrinol Metab. 1989;68:592–599. doi: 10.1210/jcem-68-3-592. [DOI] [PubMed] [Google Scholar]
- Donohoue PA, Jospe N, Migeon CJ, Van Dop C. Two distinct areas of unequal crossingover within the steroid 21-hydroxylase genes produce absence of CYP21B. Genomics. 1989;5:397–406. doi: 10.1016/0888-7543(89)90002-5. [DOI] [PubMed] [Google Scholar]
- Lee HH. Diversity of the CYP21P-like gene in CYP21 deficiency. DNA Cell Biol. 2005;24:1–9. doi: 10.1089/dna.2005.24.1. [DOI] [PubMed] [Google Scholar]
- Paldosttir A, Fossdal R, Arnason A, Edwards JH, Jennson O. Heterogeneity of human C4 gene size, a large intron (6.6 kb) is present in all C4A genes and some C4B genes. Immunogenetics. 1987;25:299–304. doi: 10.1007/BF00404422. [DOI] [PubMed] [Google Scholar]
- Haglund-Stengler B, Martin Ritzén E, Gustafsson J, Luthman H. Haplotypes of the steroid 21-hydroxylase gene region encoding mild steroid 21-hydroxylase deficiency. Proc Natl Acad Sci USA. 1991;88:8352–8356. doi: 10.1073/pnas.88.19.8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torresani T, Biason-Lauber A. Congenital adrenal hyperplasia: diagnostic advances. J Inherit Metab Dis. 2007;30:563–575. doi: 10.1007/s10545-007-0696-6. [DOI] [PubMed] [Google Scholar]
- Lee HH, Chang JG, Tsai CH, Tsai FJ, Chao HT, Chung B. Analysis of the chimeric CYP21P/CYP21 gene in steroid 21-hydroxylase deficiency. Clin Chem. 2000;46:606–611. [PubMed] [Google Scholar]
- Lee HH, Niu DM, Lin RW, Chan P, Lin CY. Structural analysis of the chimeric CYP21P/CYP21 gene in steroid 21-hydroxylase deficiency. J Hum Genet. 2002;47:517–522. doi: 10.1007/s100380200077. [DOI] [PubMed] [Google Scholar]
- Friães A, Rêgo AT, Aragüés JM, Moura LF, Mirante A, Mascarenhas MR, Kay TT, Lopes LA, Rodrigues JC, Guerra S, Dias T, Teles AG, Gonçalves J. CYP21A2 mutations in Portuguese patients with congenital adrenal hyperplasia: identification of two novel mutations and characterization of four different partial gene conversions. Mol Genet Metab. 2006;88:58–65. doi: 10.1016/j.ymgme.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Concolino P, Mello E, Minucci A, Giardina E, Zuppi C, Toscano V, Capoluongo E. A new CYP21A1P/CYP21A2 chimeric gene identified in an Italian woman suffering from classical congenital adrenal hyperplasia form. BMC Med Genet. 2009;22:10–72. doi: 10.1186/1471-2350-10-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Araujo M, Sanches MR, Suzuki LA, Guerra G Jr, Farah SB, De Mello MP. Molecular Analysis of CYP21 and C4 Genes in Brazilian Families the Classical Congenital Adrenal Hyperplasia. Brazilian J Med Biol Res. 1996;29:1–13. [PubMed] [Google Scholar]
- Bachega TA, Billerbeck AE, Madureira G, Arnhold IJ, Medeiros MA, Marcondes JA, Longui CA, Nicolau W, Bloise W, Mendonca BB. Low frequency of CYP2B deletions in Brazilian patients with congenital adrenal hyperplasia due to 21-hydroxylas deficiency. Hum Hered. 1999;49:9–14. doi: 10.1159/000022833. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis TE. Molecular cloning, a laboratory manual. New York: Cold Spring Harbor; 1989. [Google Scholar]
- Wilson RC, Wei JQ, Cheng KC, Mercado AB, New MI. Rapid deoxyribonucleic acid analysis by allele-specific polymerase chain raction for detection of mutations in the 21-hydroxylase gene. J Clin Endocrinol Metab. 1995;80:1635–1640. doi: 10.1210/jc.80.5.1635. [DOI] [PubMed] [Google Scholar]
- MRC-Holland - SALSA MLPA kit P050 CAH. http://www.mrc-holland.com/WebForms/WebFormProductDetails.aspx?Tag=tz2fAPIAupLo0TRBqtiGHA||&ProductOID=dVNA4DAAaOA|
- Taylor CF, Charlton RS, Burn J, Sheridan E, Taylor GR. Genomic deletions in MSH2 or MLH1 are a frequent cause of hereditary non-polyposis colorectal cancer: identification of novel and recurrent deletions by MLPA. Hum Mutat. 2003;22:428–433. doi: 10.1002/humu.10291. [DOI] [PubMed] [Google Scholar]
- Parajes S, Quinterio C, Domínguez F, Loidi L. A simple and robust quantitative PCR assay to determine CYP21A2 gene dose in the diagnosis of 21-hydroxylase deficiency. Clin Chem. 2007;53:1577–84. doi: 10.1373/clinchem.2007.087361. [DOI] [PubMed] [Google Scholar]
- Szilagyi A, Blasko B, Szilassy D, Fust G, Sasvari-Szekely M, Ronai Z. Real-time PCR quantification of human complement C4A and C4B genes. BMC Genet. 2006;10:1. doi: 10.1186/1471-2156-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinle S, Lang R, Fischer GF, Vierhapper H, Waldhauser F, Födinger M, Baumgartner-Parzer SM. Duplications of the functional CYP21A2 gene are primarily restricted to Q318X alleles: evidence for a founder effect. J Clin Endocrinol Metab. 2009;94:3954–8. doi: 10.1210/jc.2009-0487. [DOI] [PubMed] [Google Scholar]
- Koppens PF, Hoogenboezem T, Degenhart HJ. A rare TaqI polymorphism in a human complement C4 gene is caused by an additional restriction site in the first intron. Immunol Lett. 1992;34:93–97. doi: 10.1016/0165-2478(92)90233-E. [DOI] [PubMed] [Google Scholar]
- Kharrat M, Tardy V, M'Rad R, Maazoul F, Jemaa LB, Refaï M, Morel Y, Chaabouni H. Molecular genetic analysis of Tunisian patients with a classic form of 21-hydroxylase deficiency: identification of four novel mutations and high prevalence of Q318X mutation. J Clin Endocrinol Metab. 2004;89:368–74. doi: 10.1210/jc.2003-031056. [DOI] [PubMed] [Google Scholar]
- Koppens PF, Hoogenboezem T, Degenhart HJ. CYP21 and CYP21P variability in steroid 21-hydroxylase deficiency patients and in the general population in the Netherlands. Eur J Hum Genet. 2000;8:827–836. doi: 10.1038/sj.ejhg.5200543. [DOI] [PubMed] [Google Scholar]
- Sinnott P, Collier S, Costigan C, Dyer PA, Harris R, Strachan T. Genesis by meiotic unequal crossover of a de novo deletion that contributes to steroid 21-hydroxylase deficiency. Proc Natl Acad Sci USA. 1990;87:2107–2111. doi: 10.1073/pnas.87.6.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra-Junior G, Grumach AS, Lemos-Marini SH, Kirschfink M, Condino Neto A, de Araujo M, De Mello MP. Complement 4 phenotypes and genotypes in Brazilian patients with classical 21-hydroxylase deficiency. Clin Exp Immunol. 2009;155:182–188. doi: 10.1111/j.1365-2249.2008.03838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds AW, Ren XD, Willis AC, Law SK. The reaction mechanism of the internal thioester in the human complement component C4. Nature. 1996;379:177–179. doi: 10.1038/379177a0. [DOI] [PubMed] [Google Scholar]
- Donohoue PA, Guethlein L, Collins MM, Van Dop C, Migeon CJ, Bias WB, Schmeckpeper BJ. The HLA-A3, Cw6,B47,DR7 extended haplotypes in salt losing 21-hydroxylase deficiency and in the Old Order Amish: identical class I antigens and class II alleles with at least two crossover sites in the class III region. Tissue Antigens. 1995;46:163–172. doi: 10.1111/j.1399-0039.1995.tb03115.x. [DOI] [PubMed] [Google Scholar]
- Chung EK, Yang Y, Rennebohm RM, Lokki ML, Higgins GC, Jones KN, Zhou B, Blanchong CA, Yu CY. Genetic sophistication of human complement components C4A and C4B and RP-C4-CYP21-TNX (RCCX) modules in the major histocompatibility complex. Am J Hum Genet. 2002;71:823–837. doi: 10.1086/342777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu X, Braun-Heimer L, Rittner C, Schneider PM. Identification of the recombination site within the steroid 21-hydroxylase gene (CYP21) of the HLA B47,DR7 haplotype. Exp Clin Immunogenet. 1992;9:80–85. [PubMed] [Google Scholar]
- L'Allemand D, Tardy V, Grüters A, Schnabel D, Krude H, Morel Y. How a patient homozygous for a 30-kb deletion of the C4-CYP 21 genomic region can have a nonclassic form of 21-hydroxylase deficiency. J Clin Endocrinol Metab. 2000;85:4562–4567. doi: 10.1210/jc.85.12.4562. [DOI] [PubMed] [Google Scholar]
- Fasano MB, Winkelstein JA, LaRosa T, Bias WB, McLean RH. A unique recombination event resulting in a C4A*Q0,C4B*Q0 double null haplotype. J Clin Invest. 1992;90:1180–1184. doi: 10.1172/JCI115978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppens P, Smeets H, de Wijs IJ, Degenhart H. Mapping of a de novo unequal crossover causing a deletion of the steroid 21-hydroxylase (CYP21A2) gene and a non-functional hybrid tenascin-X (TNXB) gene. J Med Genet. 2003;40:e53. doi: 10.1136/jmg.40.5.e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soardi FC, Barbaro M, Lau IF, Lemos-Marini SH, Baptista MT, Guerra-Junior G, Wedell A, Lajic S, de Mello MP. Inhibition of CYP21A2 enzyme activity caused by novel missense mutations identified in Brazilian and Scandinavian patients. J Clin Endocrinol Metab. 2008;93:2416–2420. doi: 10.1210/jc.2007-2594. [DOI] [PubMed] [Google Scholar]
- Menassa R, Tardy V, Despert F, Bouvattier-Morel C, Brossier JP, Cartigny M, Morel Y. p.H62L, a rare mutation of the CYP21 gene identified in two forms of 21-hydroxylase deficiency. J Clin Endocrinol Metab. 2008;93:1901–1908. doi: 10.1210/jc.2007-2701. [DOI] [PubMed] [Google Scholar]
- Figueiredo MS, Kerbauy J, Goncalves MS, Arruda VR, Saad ST, Sonati MF, Stoming T, Costa FF. Effect of α-Thalassemia and β-Globin Gene Cluster Haplotypes on the Hematological and Clinical Features of Sickle Anemia in Brazil. Am J Hematol. 1996;53:72–76. doi: 10.1002/(SICI)1096-8652(199610)53:2<72::AID-AJH3>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Killeen AA, Sane KS, Orr HT. Molecular and endocrine characterization of a mutation involving a recombination between the steroid 21-hydroxylase functional gene and pseudogene. J Steroid Biochem Mol Biol. 1991;38:677–686. doi: 10.1016/0960-0760(91)90078-J. [DOI] [PubMed] [Google Scholar]
- Kyllo JH, Collins MM, Donohoue PA. Constitutive human steroid 21-hydroxylase promoter gene and pseudogene activity in steroidogenic and nonsteroidogenic cells with the luciferase gene as a reporter. Endocr Res. 1995;21:777–91. doi: 10.1080/07435809509030491. [DOI] [PubMed] [Google Scholar]
- Sørensen KM, Andersen PS, Larsen LA, Schwartz M, Schouten JP, Nygren AO. Multiplex Ligation-Dependent Probe Amplification Technique for Copy Number Analysis on Small Amounts of DNA Material. Anal Chem. 2008;80:9363–9368. doi: 10.1021/ac801688c. [DOI] [PubMed] [Google Scholar]