Abstract
A search for genes potentially regulated by STAT5 identified leukemia inhibitory factor (LIF) as a good candidate. Using various experimental approaches, we have validated LIF as a direct transcriptional target of STAT5 in myeloid cell lines: STAT5 binds to LIF promoter, and LIF expression is increased after activation of the JAK2/STAT5 pathway. We also found that LIF expression is significantly increased in patients with chronic myeloproliferative neoplasms with and without activating mutations of the pathway, indicating that LIF might play an important role in STAT5-mediated oncogenesis.
Keywords: STAT5, LIF, myeloproliferative neoplasms
Introduction
The Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling pathway plays a crucial role in myeloid differentiation. Binding of cytokines and growth factors to membrane receptors activates a JAK nonreceptor tyrosine kinase that phosphorylates a specific STAT transcription factor. Once phosphorylated, STATs migrate to the nucleus and bind to specific consensus sequences in the promoter of target genes involved in various cellular processes such as proliferation and apoptosis. This pathway is frequently altered in BCR/ABL1–negative chronic myeloproliferative neoplasms (CMPNs) by activating mutations in JAK2.1 However, constitutive activation of the pathway by the same mutation (V617F) leads to the development of MPNs with distinct phenotypic features.2 Although some variables (such as V617F allele burden, other mutations in JAK2, or mutations in other genes such as TET2) have been proposed to explain this fact, none of them fully accounts for the observed phenotypic differences in this group of diseases. This suggests that downstream effectors of JAK-STAT signaling could play a significant role in STAT5-mediated oncogenesis and might account for this unexplained phenotypic variability.
In order to identify such downstream effectors, we performed a genome-wide bioinformatic search for the presence of STAT5-binding sites in human promoters. This identified leukemia inhibitory factor (LIF) as a potential candidate gene to be transcriptionally regulated by phosphorylated STAT5. Because LIF codes for a multifunctional cytokine that plays an important role in hematopoietic differentiation3 and in the maintenance of the pluripotent state of stem cells,4 expression changes of this gene might be important in the pathogenesis of CMPNs. Thus, we set out to confirm the status of LIF as a direct transcriptional target of STAT5.
Results and Discussion
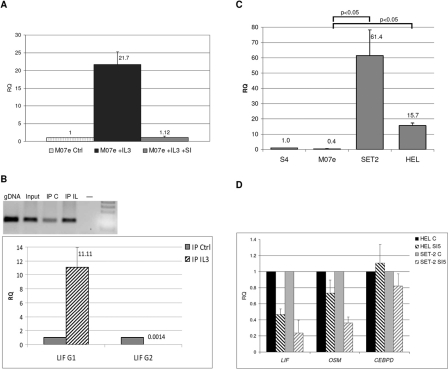
In order to validate the status of LIF as a bona fide STAT5-regulated gene, we used qRT-PCR to measure LIF expression changes in myeloid cell lines after activation of the JAK2/STAT5 pathway with interleukin-3 (IL-3). M07e cells were starved of growth factors for 16 hours and treated either with IL-3 alone or with a combination of IL-3 and a specific STAT5 inhibitor (cat. no. 573108, Calbiochem, San Diego, CA). As shown in Figure 1A, LIF expression was upregulated by IL-3 more than 20-fold, and this effect was reversed by treatment with STAT5 inhibitor. Similar changes were observed for STAT5 phosphorylation and for expression of other STAT5-regulated genes (Suppl. Results).
Figure 1.
LIF is a direct transcriptional target of STAT5. (A) LIF expression is increased in M07e cells after activation of the JAK2-STAT5 pathway with IL-3. This was prevented by the addition of a specific STAT5 inhibitor (SI). Untreated control cells (RQ = 1) were used to normalize expression values. (B) Chromatin immunoprecipitation shows increased binding of STAT5 to a specific DNA motif in LIF promoter in IL-3–treated M07e cells (IP IL-3) compared to nonstimulated cells (IP Ctrl). Quantitative qPCR assays (bottom) confirm increased binding to a motif (G1) located 740 bp upstream of the transcription start site (TSS) but not to another putative binding motif (G2) located 154 bp upstream of the TSS. Input DNA (RQ = 1) was used to normalize values. An agarose gel of the PCR products (only for motif G1) is shown at the top. (C) LIF expression is increased in cell lines carrying the V617F activating mutation of JAK2 (HEL and SET-2) compared to cell line M07 and to peripheral blood from a healthy control (P < 0.05). (D) Treatment of JAK2-mutated cell lines HEL and SET-2 with a specific STAT5 inhibitor reduces the basal expression levels of LIF and OSM without affecting the expression of the STAT3-regulated gene CEBPD. For each gene, results show fold inhibition compared to untreated cells (RQ = 1).
To confirm whether STAT5 binds to the promoter of LIF, we compared the amount of DNA-bound STAT5 before and after IL-3–mediated induction of the JAK2/STAT5 pathway in M07e cells. To this end, we performed chromatin immunoprecipitation (ChIP) with a specific STAT5 antibody and measured the amount of STAT5-bound DNA by quantitative PCR. We obtained an 11-fold increase in STAT5 binding to one site in the promoter of LIF (a TTCCCAGAA motif located 740 nucleotides upstream from the transcription start site) in IL-3–treated cells compared to untreated cells (Fig. 1B). Taken together, these results indicate that LIF behaves as a direct transcriptional target of STAT5 in this experimental model.
We next wanted to confirm whether LIF is regulated by activation of the JAK2/STAT5 pathway in other cell line models. To this end, we measured LIF expression in cell lines harboring the activating V617F mutation in JAK2. Compared to M07e, which does not carry this mutation, LIF expression was increased 16-fold in HEL cells and 61-fold in SET-2 cells, as shown in Figure 1C. Furthermore, we studied the effect of suppressing the activity of the pathway in these 2 cell lines using the specific inhibitor. Incubation of HEL and SET-2 cells with STAT5 inhibitor decreased LIF mRNA levels (2.1-fold decrease in HEL and 4.2-fold decrease in SET-2), as shown in Figure 1D. We also measured expression of CEBPD, a gene known to be regulated by STAT3 that lacks STAT5-binding sites in its promoter. As a further confirmation of the specificity of this inhibitor for STAT5, CEBPD expression was not affected by treatment of HEL and SET-2 cells (Fig. 1D). These data confirm that LIF expression is directly regulated by the induction of JAK2/STAT5 signaling in myeloid cell lines.
Our results are in agreement with several datasets available from the literature, in which gene expression has been monitored during myeloid differentiation. In one study,5 cord blood CD34+ hematopoietic progenitor cells were differentiated through erythroid, megakaryocytic, granulocytic, and monocytic pathways at different time points. Their data support the notion that LIF is upregulated in erythroid, megakaryocytic, and granulocytic lineages, with particularly marked expression changes in the latter. Another recent study6 used an estrogen receptor–STAT5 fusion to induce STAT5 and monitor gene expression in human hematopoietic stem cells and in myeloid progenitor cells. In this dataset, LIF expression was significantly upregulated in GMP and HSC after STAT5 induction.
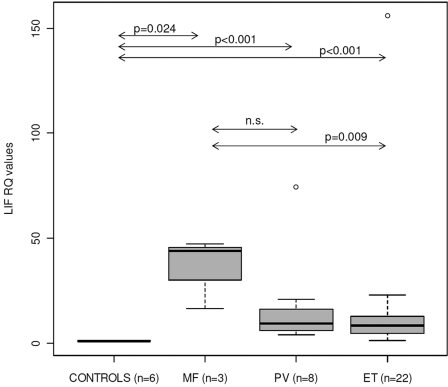
In order to explore the potential clinical impact of our findings, we measured LIF expression levels by qRT-PCR in peripheral blood samples from 33 patients with CMPNs and 6 healthy controls. In agreement with our previous findings, LIF expression levels were significantly increased in CMPN samples (Fig. 2). Furthermore, significant differences were found between idiopathic myelofibrosis (MF) and essential thrombocytemia (ET) samples. Because all patient samples had been analyzed for mutations in JAK2 (V617F and exon 12 mutations) and in MPL (exon 10 mutations), we could also compare LIF expression levels between mutated and nonmutated samples. However, all polycytemia vera (PV) samples were mutated, and we only had 3 MF samples. Therefore, we could only compare mutated versus nonmutated ET samples, finding significantly higher LIF expression levels in ET mutated samples (P = 0.048, Mann-Whitney test). Interestingly, expression levels of OSM, a canonical STAT5 target gene, were also significantly increased in patient samples but did not discriminate between MF and ET patients or between mutated and nonmutated ET samples (not shown).
Figure 2.
LIF expression is significantly increased in MPN patients. Relative quantification by real-time RT-PCR in peripheral blood samples from 6 healthy controls and 33 MPN patients (3 myelofibrosis, 8 polycytemia vera, and 22 essential thrombocytemia). Boxplots show the median (horizontal line), interquartile range (box), and range (whiskers). All results were normalized to the average of the 6 control samples. Significance of the Mann-Whitney test is shown for several comparisons.
In conclusion, we show that LIF is directly regulated by STAT5 and that it is overexpressed in myeloid cell lines with increased activity of the JAK2/STAT5 signaling pathway. Furthermore, LIF expression is significantly increased in CMPN patients, particularly MF cases and samples with activating mutations in JAK2 or in MPL. It is interesting that LIF codes for a cytokine that binds the heterodimeric gp130/LIFR receptor and phosphorylates STAT3. In this respect, LIF behaves like OSM, a STAT5-regulated gene coding for a cytokine whose receptor signals via STAT3. This, in murine leukemia cells, leads to increased expression of the receptor for IL-3,7 which signals via JAK2/STAT5. Thus, LIF might represent a crucial link in the coordination of STAT3 and STAT5 signaling. Similarly, it could have a role in determining the balance between STAT5 and STAT1 activation, which has recently been proposed to be one of the major determinants of clinical phenotype in MPNs with mutations in JAK2.8 The fact that LIF and OSM expression are increased in MPN samples negative for mutations in JAK2 and MPL lends further support to the existence of additional genetic or epigenetic events that contribute to activate the JAK2/STAT5 pathway. Furthermore, LIF (but not OSM) seems to discriminate between MF and PV or ET patients, suggesting that effectors of JAK2/STAT5 signaling (such as STAT5-regulated genes) might explain the phenotypic variability observed in patients with constitutive activation of this signaling pathway. The finding that a multifunctional cytokine involved in the maintenance of pluripotency is a transcriptional target of STAT5 and is upregulated in MPNs might improve our understanding of the mechanisms governing STAT5-mediated oncogenesis.
Materials and Methods
Cell lines and patient samples
Binding of STAT5 and activation/inhibition studies were performed in cell lines where JAK2/STAT5 activity can be induced with IL-3 or in cell lines with constitutively active signaling that can be suppressed by specific inhibitors. We used M07e cells, a myeloid cell line strictly dependent on IL-3, the HEL cell line (human erythroleukemia cells that harbor the V617F activating mutation in JAK2 gene), and SET-2 cells (a cell line heterozygous for the V617F mutation in JAK2, originally derived from a patient with ET at megakaryoblastic transformation). For stimulation studies, M07e cells were starved of growth factors for 16 hours and then treated with 30 ng/mL IL-3 for 45 minutes. For inhibition experiments, cells were treated with 100 uM STAT5 inhibitor (cat. no. 573108, Calbiochem) for 1 hour.
LIF expression was also assayed in peripheral blood samples obtained (with informed consent) from 33 patients with CMPNs, all of whom were positive for the V617F mutation in JAK2 (12 males and 21 females). Eight patients had the diagnosis of PV, 3 had MF, and 22 had ET.
Chromatin immunoprecipitation
Control cells (untreated) and cells in which JAK2/STAT5 signaling was induced with IL-3 were incubated with 1.5% formaldehyde (Fluka, St. Louis, MO), cell pellets were washed, and nuclear extracts were sonicated using a Bioruptor apparatus (Diagenode, Liege, Belgium) for 13 minutes (30-second pulses) on ice. Supernatants were incubated overnight with a polyclonal STAT5-specific antibody (C-17, Santa Cruz Biotechnology Inc., Santa Cruz, CA) under rocking conditions at 4°C, while an aliquot was set aside before inmunoprecipitation (“input” DNA sample). After incubation for 3 hours with protein A sepharose (GE Healthcare, Waukesha, WI), specific complexes were washed and recovered from beads with an extraction buffer. Finally cross-links were reversed, and both immunoprecipitated and input DNA samples were recovered by phenol:chloroform extraction, ethanol precipitation, and redissolved in nuclease-free water. DNA recovered from the procedure was amplified with GenomePlex WGA and purified with the GenElute PCR Clean-Up Kit (both from Sigma-Aldrich, St. Louis, MO). In order to quantify the enrichment of specific regions in IL-3–treated cells, relative to noninduced cells, immunoprecipitated and input DNA were used as templates in qPCR reactions using specific primers flanking putative STAT5-binding motifs in the promoter of LIF, with SYBR GreenER qPCR SuperMix Universal (Invitrogen, Paisley, UK) in an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA). The amount of PCR product in immunoprecipitated samples was normalized to input DNA in both experimental conditions (untreated or IL-3–treated M07e cells).
Quantification of gene expression
For LIF qRT-PCR assays, total RNA was isolated with TRIzol reagent (Invitrogen) and quantitated using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE). One microgram of total RNA was converted to cDNA using SuperScript III First-Strand Synthesis SuperMix (Invitrogen), and 25 ng of cDNA was amplified using specific primers (LIF_F: 5′-CAACTGGCACAGCTCAATGG-3′ and LIF_R: 5′-CTCCCCCTGGGCTGTGTA-3′) with SYBR GreenER qPCR SuperMix Universal (Invitrogen) in an ABI 7500 Real-Time PCR System (Applied Biosystems). As endogenous control, we used TBP, a housekeeping gene that showed consistent expression levels across different experimental conditions.
Supplementary Material
Footnotes
Supplementary material for this article is available on the Genes & Cancer website at http://ganc.sagepub.com/supplemental.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
This work was supported by the Spanish Ministry of Science and Innovation [grant number SAF 2007-62473]; the PIUNA Program of the University of Navarra; the Mutua Madrileña Foundation; and the Caja Navarra Foundation through the program “You Choose, You Decide” [project 10.830].
References
- 1. Kota J, Caceres N, Constantinescu SN. Aberrant signal transduction pathways in myeloproliferative neoplasms. Leukemia. 2008;22:1828-40 [DOI] [PubMed] [Google Scholar]
- 2. Goldman JM, Green AR, Holyoake T, et al. Chronic myeloproliferative diseases with and without the Ph chromosome: some unresolved issues. Leukemia. 2009;23:1708-15 [DOI] [PubMed] [Google Scholar]
- 3. Trouillas M, Saucourt C, Guillotin B, et al. The LIF cytokine: towards adulthood. Eur Cytokine Netw. 2009;20:51-62 [DOI] [PubMed] [Google Scholar]
- 4. Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118-22 [DOI] [PubMed] [Google Scholar]
- 5. Felli N, Cianetti L, Pelosi E, et al. Hematopoietic differentiation: a coordinated dynamical process towards attractor stable states. BMC Syst Biol. 2010;4:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fatrai S, Wierenga AT, Daenen SM, Vellenga E, Schuringa JJ. Identification of HIF2{alpha} as an important STAT5 target gene in human hematopoietic stem cells. Blood. 2011;117:3320-30 [DOI] [PubMed] [Google Scholar]
- 7. Iwamoto T, Senga T, Adachi K, Hamaguchi M. Stat3-dependent induction of interleukin-3 receptor expression in leukemia inhibitory factor-stimulated M1 mouse leukemia cells. Cytokine. 2004;25:136-9 [DOI] [PubMed] [Google Scholar]
- 8. Chen E, Beer PA, Godfrey AL, et al. Distinct clinical phenotypes associated with JAK2V617F reflect differential STAT1 signaling. Cancer Cell. 2010;18:524-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.