Abstract
Ideally shotgun proteomics would facilitate the identification of an entire proteome with 100% protein sequence coverage. In reality, the large dynamic range and complexity of cellular proteomes results in oversampling of abundant proteins, while peptides from low abundance proteins are undersampled or remain undetected. We tested the proteome equalization technology, ProteoMiner, in conjunction with Multidimensional Protein Identification Technology (MudPIT) to determine how the equalization of protein dynamic range could improve shotgun proteomics methods for the analysis of cellular proteomes. Our results suggest low abundance protein identifications were improved by two mechanisms: (1) depletion of high abundance proteins freed ion trap sampling space usually occupied by high abundance peptides and (2) enrichment of low abundance proteins increased the probability of sampling their corresponding more abundant peptides. Both mechanisms also contributed to dramatic increases in the quantity of peptides identified and the quality of MS/MS spectra acquired due to increases in precursor intensity of peptides from low abundance proteins. From our large data set of identified proteins, we categorized the dominant physicochemical factors which facilitate proteome equalization with a hexapeptide library. These results illustrate that equalization of the dynamic range of the cellular proteome is a promising methodology to improve low abundance protein identification confidence, reproducibility, and sequence coverage in shotgun proteomics experiments, opening a new avenue of research for improving proteome coverage.
Keywords: shotgun proteomics, peptide identification, proteome coverage, protein abundance dynamic range
Many diseases are caused by the dysfunction of low abundance proteins and post-translational modifications.1–3 In order to speed the discovery of low abundance proteins involved in disease, a high-sensitivity, comprehensive proteomics method is essential. The state-of-the-art analysis method which fulfills these requirements is mass spectrometry-based shotgun proteomics. Thousands of proteins can be identified and quantified within a single analysis, leading to many relevant biological discoveries.4,5 However, the reproducible, simultaneous, and comprehensive analysis of all proteins within the cell using mass spectrometry is currently limited by the large concentration dynamic range of the cellular proteome (> 109).
Shotgun proteomics combines technologies to identify peptides produced by proteolytic digestion of proteins.6,7 After the introduction of SEQUEST, an automated search algorithm for tandem mass spectra of peptides,8 a dramatic leap in sensitivity and comprehensiveness was first introduced through automated, online fractionation of peptides using strong cation exchange and reverse-phase liquid chromatography called MudPIT.9,10 Further variations to methodologies of separations at the peptide level have produced complementary peptide identifications, yet only modest improvements in protein identifications.11–17 Advances in the sensitivity, speed, and mass accuracy of ion trap mass spectrometers consistently yields improvements in the number of protein identifications per time.18–25 These trends appear to indicate that current proteomics technology is not separation-limited at the peptide level, but instead mass spectrometer-limited. Fundamentally, the finite ion capacity of ion trap mass spectrometers used in many shotgun proteomics experiments limit the isolation, fragmentation, and detection of low abundance peptide ions, and thus the identification of low abundance proteins. We expect continued improvements in mass spectrometers to achieve greater proteomic coverage of the substantial dynamic range of the proteome. Until that time is reached, another complementary alternative is adjusting the concentration dynamic range of the proteome to the detectable dynamic range of mass spectrometers.
A robust method has emerged for minimizing the protein concentration dynamic range of bodily fluid samples through the use of a combinatorial hexapeptide-bead library, marketed as ProteoMiner by Bio-Rad.26 The random generation of hexapeptides creates a substantial ligand library (64 million) for which proteins can selectively bind.27,28 A complex protein sample is incubated in excess of the protein mass binding capacity of the bead-conjugated hexapeptide ligands and unbound proteins are washed away. The protein to ligand excess creates conditions where proteins saturate the ligands. Thus the final concentration of the proteins in the sample are defined by the amount of the ligand present. In the case of high abundance proteins this is expected to result in their depletion, while for low-abundance proteins, enrichment.
Plasma is one of the most difficult samples to analyze with shotgun proteomics. Not only are proteins from every cell type potentially present, but the protein concentration dynamic range is 12 orders of magnitude.29 The ProteoMiner technology has likely worked particularly well for depletion of high abundance proteins in plasma since only three proteins (hemoglobin, albumin, and immunoglobin G) make up the top 103 of the protein concentration dynamic range. Recently, the use of ProteoMiner in conjunction with glycopeptide and phosphopeptide enrichment aided in the analysis of post-translational modifications in saliva using LC-MS/MS.30,31 Herein we examine the use of a hexapeptide library to equalize the protein concentration dynamic range of a human cell lysate to improve several shotgun proteomic metrics and identify binding trends to the hexapeptide library to better understand the mechanisms of enrichment and depletion. For such, we tested the ProteoMiner technology on HeLa cell lysates and analyzed the samples using MudPIT.
METHODS AND MATERIALS
Reagents and Chemicals
Unless otherwise noted all chemicals were purchased from Thermo Fisher Scientific (Waltham, MA). Deionized water (18.2 MΩ, Barnstead, Dubuque, IA) was used for all preparations.
HeLa cell growth and lysis
Cells were cultured in 10 cm plates and DMEM with 10% FBS. Harvesting was performed at ~80% confluence, each plate contained about 3–3.5 million cells. Cells were scraped into ice cold PBS and frozen in dry ice immediately after a short spin and supernatant aspiration. Cells were lysed to 25 mg/mL in 50 mM Tris(hydroxyethylamine) pH 7.8, 150 mM NaCl, 1 mM EDTA, 2 mM PMSF, 1% SDS, and 1X protease inhibitor (Roche).
ProteoMiner Enrichment
Instructions from the ProteoMiner Small-Capacity Kit (Bio-Rad, Hercules, CA, cat #163–3006) were followed. Briefly, ProteoMiner columns containing 20 μL beads were washed thrice with 1X PBS with 5 min incubations and 1,000 × g spins. Lysates were diluted to 0.1% SDS using 1X PBS. ProteoMiner beads were transferred to a higher volume centrifuge tube by repetitively creating bead-sample slurries until all the beads were transferred. The final bead-sample slurry was incubated for two hours at room temperature with rotation. The unbound proteins were separated from the ProteoMiner beads by repetitively transferring smaller volumes of the bead-sample slurry back to the ProteoMiner columns and briefly spinning at 1,000 × g. Additionally, the remaining ProteoMiner beads with bound proteins were again washed thrice with 1X PBS.
2-D gel analysis
Proteins were eluted off of the ProteoMiner beads thrice by incubating with 100 μl of elution buffer (9 M urea, 2% CHAPS, 100 mM acetic acid) and spun at 1,000 × g for 30 sec. The fractions were pooled and analyzed by protein assay. Proteins eluted from ProteoMiner beads were precipitated with the 2-D Cleanup Kit (Bio-Rad) to remove any ionic contaminants and solubilized in 2-D rehydration buffer (7 M Urea, 2 M thiourea, 2% CHAPS, 50 mM dithiothreitol, 2 mM tributylphosphine, 0.2% Bio-Lyte ampholyte pH 5–8 and bromophenol blue to desired color). For the 1st dimension of 2-D electrophoresis, 100 μg of protein was loaded onto an 11 cm ReadyStrip IPG strip, pH 5–8 (Bio-Rad). Isoelectric focusing was performed using a PROTEAN IEF Cell (Bio-Rad) at 8000 volts for 35,000 volt-hours. For the 2nd dimension, the IPG strip was transferred onto a Criterion Tris-HCl 8–16% gradient gel (Bio-Rad) and run for 1 hour at 200 volts. Gels were stained with Flamingo fluorescent gel stain (Bio-Rad) and imaged on the Molecular Imager PharosFX system (Bio-Rad). The resultant gel images were analyzed with PDQuest 2-D Analysis Software (Bio-Rad).
MudPIT analysis
Proteins (~200 μg) were digested off of the ProteoMiner beads by first denaturing and reducing in 120 μL 8 M urea, 100 mM Tris(hydroxyethylamine) pH 8.5, and 5 mM tris(2-carboxyethyl)phosphine for 30 min. Cysteine residues were acetylated with 10 mM iodoacetamide for 15 min in the dark. The sample was diluted to 2 M urea with 100 mM Tris(hydroxyethylamine) pH 8.5. Trypsin (4μg as 0.5 μg/μL) was added at a 1:50 protease:protein ratio (~200 μg ProteoMiner bead protein capacity) along with CaCl2 to 1 mM for an overnight digestion at 37°C. Peptides were spun off of the ProteoMiner column at 1,000 × g, split into two aliquots, and stored at −80°C until the day of analysis. On the day of analysis peptide samples were acidified to 5% formic acid and spun at 18,000 × g. The same digestion procedure was carried out for digestion of 100 μg HeLa protein lysate Control samples.
Capillary columns were prepared in-house for LC-MS/MS analysis from particle slurries in methanol. An analytical RPLC column was generated by pulling a 100 μm ID/360 μm OD capillary (Polymicro Technologies, Inc, Phoenix, AZ) to 5 μm ID tip. Reverse phase particles (Jupiter C18, 4 μm dia., 90 Å pores, Phenomenex, Torrance, CA) were packed directly into the pulled column at 800 psi until 15 cm long. The column was further packed, washed, and equilibrated at 100 bar with buffer B followed by buffer A. A MudPIT trapping column was prepared by creating a Kasil frit at one end of an undeactivated 250 μm ID/360 μm OD capillary (Agilent Technologies, Inc., Santa Clara, CA), then successively packed with 2.5 cm strong cation exchange particles (Luna SCX, 5 μm dia., 100 Å pores, Phenomenex) and 2.5 cm reverse phase particles (Aqua C18, 5 μm dia., 125 Å pores, Phenomenex). The Kasil frit was prepared by briefly dipping a 20 cm capillary in well-mixed 300 μL Kasil 1624 (PQ Corporation, Malvern, PA) and 100 μL formamide, curing at 100°C for 4 hrs, and cutting the frit to ~2 mm in length. The MudPIT trapping column was equilibrated using buffer A for 15 min at 400 bar. Peptide samples (~100 μg) were loaded onto columns at 400 bar. MudPIT and analytical columns were assembled using a zero-dead volume union (Upchurch Scientific, Oak Harbor, WA).
LC-MS/MS analysis was performed using an Agilent 1100 HPLC pump and Thermo LTQ XL using an in-house built electrospray stage. Electrospray was performed directly from the analytical column by applying the ESI voltage at a tee (150 μm ID, Upchurch Scientific) directly downstream of a 1:1000 split flow used to reduce the flow rate to 250 nL/min through the columns. 12-step MudPIT experiments were performed where each step corresponds to 0, 10, 15, 20, 25, 30, 40, 50, 60, 70, 85, and 100% buffer C being run for 5 min at the beginning of a 2 hr gradient. The repetitive 2 hr gradients were from 100 % buffer A to 60% buffer B over 70 min, up to 100% B over 20 min, held at 100% B for 10 min, then back to 100% A for a 10 min column re-equilibration. Buffer A was 5% acetonitrile 0.1% formic acid, B was 80% acetonitrile 0.1% formic acid, and C was 500 mM ammonium acetate. Data-dependent acquisition of MS/MS spectra on the LTQ were performed with the following settings: inlet capillary temperature − 200 °C, full scan automatic gain control target − 20K ions, MSn scan automatic gain control target − 10K ions, ESI voltage − 2.5 kV, maximum injection time − 100 ms, dynamic exclusion time − 60 sec, and 5 MS/MS per MS on the most intense precursor ions.
Tandem mass spectra were extracted from raw files using RawExtract 1.9.932 and were searched against a combined UniProt Swiss-Prot and VarSplic database with reversed sequences using ProLuCID.33 The search space included all fully-, semi-, and non-tryptic peptide candidates. Carbamidomethylation of cysteine was considered as a static modification. The validity of peptide spectrum matches were assessed computationally as follows using an in-house software termed Search Engine Processor (SE Pro). Identifications were grouped by charge state (+1, +2, and ≥ +3) and then by tryptic status (fully tryptic, half-tryptic, or non-tryptic), resulting in nine distinct subgroups. For each result, ProLuCID XCorr, DeltaCN and ZScore values were used to generate a Bayesian discriminator. Outlier points in the two distributions having a Mahalanobis distance greater than 4 were discarded. The identifications were sorted in a non-decreasing order according to the discriminator score. A cut-off score was established to accept a false discovery rate of 1% based on the number of reverse and scrambled decoy proteins and similarly 0.1% based on decoy peptides. This procedure was independently performed on each data subset, resulting in a false-positive rate that was independent of tryptic status or charge state. Additionally, a minimum sequence length of six amino acid residues was required.
Data analysis
Identification comparisons of proteins and peptides between Control and ProteoMiner runs were performed using PatternLab for Proteomics.34 Spectral counts of proteins and peptides between Control and ProteoMiner runs were extracted using Census.35 Peptide precursor intensities and XCorr values were extracted and integrated from MS spectra and filtered SE Pro results using an in-house written script. Calculations and comparisons of peptide precursor intensities and XCorr values were performed using Microsoft Excel. Unique peptide sequences and protein isoforms were identified and compared using a modified sparse matrix generator from PatternLab that only considers unique peptides. Isoelectric point, molecular weight, and Kyte-Doolittle score of proteins were calculated in parallel using an in-house written script. Gene ontology and protein class comparisons were made using PANTHER.36 Recurrent protein domains were found by searching proteins with Pfam37 enriched at least two-fold by spectral counts from the use of ProteoMiner. In silico digestions of the UniProt Swiss-Prot database were performed using an in-house program called Axe.
RESULTS AND DISCUSSION
ProteoMiner-2D Gel Results
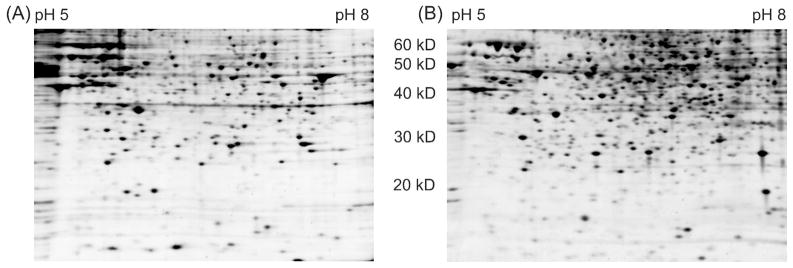
ProteoMiner relies on the competitive binding of proteins from a lysate to at least one of the 64 million randomly generated hexapeptide ligands synthesized on beads. Thus, protein quantity, concentration, and dynamic range are all critical parameters that define the number of proteins that can be pulled out of a protein sample based on their affinity to the hexapeptide library under competitive binding conditions. All three of these parameters are expected to be significantly different between a clinical sample (i.e. plasma or saliva) and a biological sample such as a cell lysate. Thus, we first used 2-D gel electrophoresis with silver staining to evaluate whether the protein-hexapeptide incubation conditions were adequate for a cell lysate by estimating the improvement in detectable proteins after using ProteoMiner (Figure 1). From simply counting protein spots on the fluorescently stained gels, a ~40% increase in proteins were detected, increasing from 315 to 435 spots. The increase in detectable spots was attributed to enrichment of lower abundance proteins. Similarly, the largest and darkest protein spots in the Control were smaller and lighter in the ProteoMiner gel due to depletion of the most abundant proteins. Given the increased functional complexity of the contents of a cell lysate compared to a clinical sample, such as plasma or urine, we rationalized that there are many more protein-protein interactions which could interfere with protein binding to hexapeptide ligands. We found that a low concentration of detergent (0.1% SDS) was required to see an increase in protein spots. The detergent likely serves multiple functions: (1) disrupts stable protein interactions of protein complexes, (2) disrupts non-specific protein interactions of high abundance proteins to others, (3) denatures proteins and further exposes high affinity epitopes, and (4) minimizes non-specific hydrophobic protein-hexapeptide interactions.
Figure 1.
2D gels of (A) Control and (B) ProteoMiner treated HeLa cell lysates.
ProteoMiner-MudPIT Results
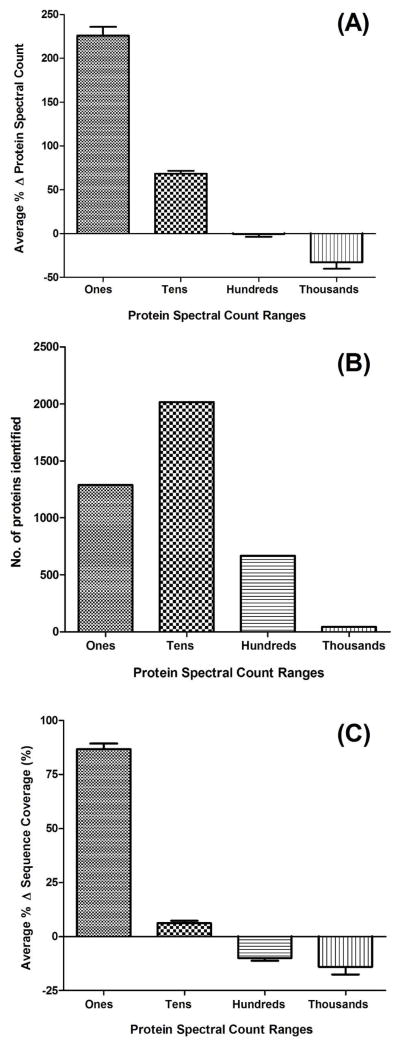
ProteoMiner used in combination with fluorescently stained 2D gels yielded a promising increase in protein identifications, so we performed the same experiment with MudPIT as a more comprehensive protein identification readout. Through simultaneous depletion of high abundance proteins and enrichment of low abundance proteins, ProteoMiner treatment of a cell lysate was expected to equalize protein abundance. In order to test this without a visualization method such as a 2D gel, we used the spectral counts of proteins as a measure of their relative abundances.38 The comparison of protein abundance change between Control and ProteoMiner runs using their percent spectral count changes is shown in Figure 2A. When we grouped proteins by their relative abundance (protein spectral count order of magnitude) in Control runs, an obvious equalization trend was observed. Proteins identified by 1000 or more spectral counts (“Thousands”) in Control runs, on average were identified with 32% fewer peptide spectra in ProteoMiner runs. Conversely, proteins identified by less than 10 spectral counts in Control runs (“Ones”) were on average identified by 226% more spectra in ProteoMiner runs. The results for proteins identified by 10 to 99 spectral counts (“Tens”) had a similar trend with a 68% increase in spectral counts from using ProteoMiner. Proteins identified by “Hundreds” of spectral counts (100–999) showed no significant change, indicating that ProteoMiner acts to equalize the protein dynamic range to an abundance for which MudPIT can identify proteins by greater than 100 spectral counts. Specific examples of the top ten most depleted and enriched proteins are listed in Tables 1 and 2, respectively. In Figure 2B the relative importance of protein concentration equalization is illustrated based on initial protein abundances. That is, the proteins that have the largest gains in spectral counts (“Ones” and “Tens”) are also the more frequently identified. Thus, a relatively small decrease (32%) in spectral counts of 46 high abundance proteins dramatically increased the spectral counts of 1288 (“Ones”) and 2016 (“Tens”) proteins by, on average, 226 and 68%, respectively. Although protein identification probability is not routinely quantified based on spectral counts,39 these increases in protein spectral and sequence count both improve protein identification probability. Additionally, the spectral count changes directly translated to changes in protein sequence coverage, illustrated in Figure 2C. Similar to the trend from spectral count changes, small decreases in a small number of high abundance proteins’ sequence coverage significantly increased the relative sequence coverage of lower abundance proteins. Comparable observations have been found with immunoaffinity depletion of serum using antibody columns to remove the most abundant proteins.40
Figure 2.
(A) Protein abundance equalization visualization. Proteins were grouped based on spectral counts in Control runs as an approximation of abundance. The average percent change in protein spectral counts are plotted with standard error means for each spectral count order of magnitude. (B) Protein spectral count frequency visualization. Proteins were again grouped by spectral count abundance and counted. (C) Protein sequence coverage visualization. The average percent change in sequence coverage is plotted with standard error means for each spectral count order of magnitude.
Table 1.
The 10 most enriched low abundance proteins based on percent change in spectral counts between Control and ProteoMiner runs.
| Low Abundance Protein | Control | ProteoMiner | % Change |
|---|---|---|---|
| Origin recognition complex subunit 4 (ORC4L) | 2 | 64 | 3100 |
| Transcription elongation factor B polypeptide 3 (TCEB3) | 4 | 125 | 3025 |
| Isoform 1 of AT-rich interactive domain-containing protein 1B (ARID1B) | 1 | 31 | 3000 |
| Pentatricopeptide repeat-containing protein 1 (PTCD1) | 1 | 27 | 2600 |
| Isoform Long of Plakophilin-4 (PKP4) | 1 | 26 | 2500 |
| Isoform 1 of Serine/threonine-protein kinase PAK 4 (PAK-4) | 3 | 67 | 2133 |
| DNA polymerase subunit gamma-1 (POLG) | 2 | 43 | 2050 |
| Nucleolar protein 12 (NOL12) | 1 | 21 | 2000 |
| Isoform 1 of UPF0461 protein C5orf24 (C5orf24) | 1 | 21 | 2000 |
| Isoform 1 of Serine/threonine-protein kinase TAO1 (TAOK1) | 1 | 21 | 2000 |
Table 2.
The 10 most depleted high abundance proteins based on decrease in spectral counts between Control and ProteoMiner runs.
| Highest Abundance Protein | Control | ProteoMiner | ΔSpec Count |
|---|---|---|---|
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | 4290 | 127 | −4163 |
| Actin, cytoplasmic 1 (ACTB) | 6908 | 3169 | −3739 |
| Isoform alpha-enolase of Alpha-enolase (ENO1) | 3617 | 381 | −3236 |
| Elongation factor 1-alpha 1 (EEF1A1) | 4519 | 2224 | −3236 |
| 60 kDa heat shock protein, mitochondrial (HSPD1) | 2164 | 252 | −1912 |
| Peptidyl-prolyl cis-trans isomerase A (PPIA) | 1907 | 309 | −1598 |
| Tubulin alpha-4A chain (TUBA4A) | 2841 | 1268 | −1573 |
| Fructose-bisphosphate aldolase A (ALDOA) | 1723 | 194 | −1529 |
| Isoform 1 of Heat shock cognate 71 kDa protein (HSPA8) | 2696 | 1498 | −1198 |
| Isoform 1 of Carbamoyl-phosphate synthase, mitochondrial (CPS1) | 3212 | 2094 | −1118 |
These trends are further illustrated, independently of protein abundance, by distribution plots of sequence count and sequence coverage in Supplemental Figure 1A and 1B, respectively. Sequence count is indicative of the number of peptides that contribute to identifying a protein sequence. Although the maximum sequence count of 10 remained constant between Control and ProteoMiner runs, shown in Supplemental Figure 1A, the sequence count shifted to higher values from the use of ProteoMiner. Again, the sequence count results directly correlated to sequence coverage. Thus, a similar trend is seen in the sequence coverage plot shown in Supplemental Figure 1B, with an overall shift to higher sequence coverages from the use of ProteoMiner.
MudPIT can routinely detect thousands of proteins in a single 24 hr analysis, approximately an order of magnitude higher than a stained 2D gel. Thus, we were somewhat skeptical that we could achieve the same 40% percent increase in protein identifications using ProteoMiner in combination with MudPIT. However, even with a modest 10% increase in new protein identifications from the use of ProteoMiner with MudPIT relative to Control runs, we still increased new protein identifications by a factor of three over the 2D gel results (316 versus 120 new proteins). A summary of the results from the MudPIT experiments are listed in table 3. The most dramatic improvement we observed from the ProteoMiner-MudPIT combination was a ~30% increase in both the average number of peptides and unique peptides identified among triplicate Control and ProteoMiner runs. Notably, the increased number of peptides identified was achieved from the same number of acquired MS/MS spectra and peptide spectral matches (see Table 3). These results begin to indicate that through the equalization of protein abundance from ProteoMiner the MS/MS sampling space is more efficiently utilized.
Table 3.
Comparison of proteomic metrics between Control and ProteoMiner MudPIT runs.
| Control | ProteoMiner | |||||||
|---|---|---|---|---|---|---|---|---|
| Total | Average | SDa | Total | Average | SDa | Change | P Valueb | |
| Protein IDs | 4,254 | 3,618 | 106 | 4,570 | 3,965 | 178 | 9.6% | 0.01 |
| Peptide IDs | 38,747 | 29,238 | 546 | 50,097 | 37,705 | 1,696 | 29.0% | 0.01 |
| Unique Peptides | 16,690 | 11,346 | 321 | 21,396 | 14,983 | 276 | 32.1% | 6.3E–6 |
| Spectral IDs | 255,899 | 85,300 | 18,026 | 251,916 | 83,972 | 4,713 | − 1.6% | 0.98 |
| MS/MS Spectra | 706,198 | 235,399 | 17,003 | 714,820 | 238,273 | 11,320 | 1.2% | 0.76 |
| Precursor Intensity | 5.74E5 | 7.10E5 | 23.7% | 0.07 | ||||
| Sequence Count | 8.2 | 9.9 | 20.7% | |||||
| Sequence Coverage | 18.3% | 21.5% | 62.1% | |||||
SD is standard deviation.
Calculated using a two-sample, two-tail t-test assuming unequal variances.
Protein and Peptide Replicate Reproducibility Improvements
Due to undersampling of peptides selected for MS/MS in data-dependent acquisition, reproducible identification of low abundance proteins between replicate analyses is often a challenge.38 Even as mass spectrometer sensitivity and dynamic range are improved, this trend remains constant for the lowest abundance proteins detected within a shotgun proteomics experiment. The equalization of protein dynamic range through the use of ProteoMiner prior to MudPIT begins to actively address this problem. These results are illustrated in Table 4. Comparison of the percent increase in proteins identified in duplicate and triplicate analyses with and without ProteoMiner, reveals that ProteoMiner significantly increases the number of proteins that can be reproducibly identified. A similar comparison at the peptide level, also in Table 4, shows a more dramatic trend. That is, more reproducible identification of peptides leads to more reproducible identification of proteins.
Table 4.
Reproducibility of peptide and protein identifications
| Replicate Criterion | Peptide | Protein | ||||
|---|---|---|---|---|---|---|
| Control | ProteoMiner | Increase | Control | ProteoMiner | Increase | |
| 1 experiment | 27,888 | 37,051 | 32.9% | 8,169 | 8,787 | 7.6% |
| 2 experiments | 17,225 | 25,899 | 50.4% | 5,645 | 6,204 | 9.9% |
| 3 experiments | 10,716 | 17,552 | 63.8% | 4,199 | 4,628 | 9.3% |
ProteoMiner-MudPIT Peptide Identification Improvements
Ultimately, the aforementioned increases in protein spectral and sequence counts, sequence coverage, and identification reproducibility are due to identification of ~30% more peptides using ProteoMiner. This increase in peptide identifications was facilitated by equalization of the protein dynamic range. However, since we are directly identifying peptides to infer protein presence, we wondered what aspects of peptide identification improvements facilitated this enhancement in protein detection. Theoretically, the reduction of higher abundance proteins, and thus their corresponding peptides, significantly opens the liquid chromatography separation and ion trap sampling spaces. The opening of separation and sampling space reduces ion suppression during the electrospray and precursor ion selection processes, respectively, making sampling of low abundance peptides more probable and thus more reproducible between experiments. The most indicative attribute that could solidify this concept is increases in peptide precursor ion intensity. If fewer high abundance peptides are present to suppress low abundance peptide signals, then an overall increase in peptide precursor ion intensity should be observed. Indeed this was the case as illustrated in Supplemental Figure 1C. The majority of the difference in the two distributions can be attributed to a larger number of peptide identifications using ProteoMiner since their maxima are nearly the same (see Table 3). However, the ProteoMiner peptide precursor ion intensity distribution is slightly shifted to higher values, partially disguised by the logarithmic x-axis.
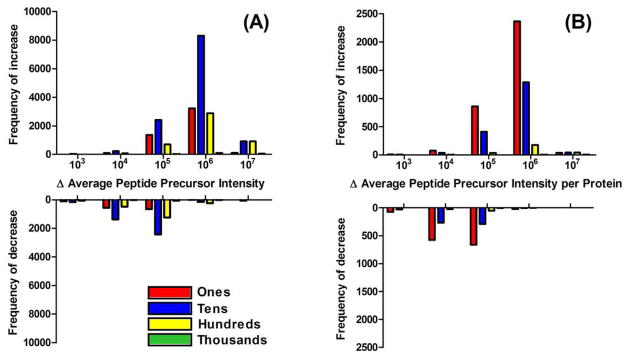
In order to better illustrate the overall increase in precursor ion intensity from using ProteoMiner we plotted the frequency of change in the average peptide precursor intensity for a peptide (Figure 3A) and the peptides identifying a protein (Figure 3B). As can be seen in the figures, the magnitudes and frequencies of increases in the average peptide precursor intensity at the peptide and protein level both outweigh the magnitudes and frequencies of decreases in peptide precursor intensity. Additionally, we plotted these results as a function of peptide and protein abundance (spectral counts), indicating that peptides identified by 10–99 spectral counts most frequently had their precursor intensities increase by 106, followed in frequency by peptides identified by 1–9 spectral counts, then 100–999 spectral counts. Similarly, in Figure 3B, the most frequent average protein precursor intensity increase was 106 for proteins identified by 1–9 spectral counts, followed by 10–99 spectral counts and a 105 improvement of proteins also identified by 1–9 spectral counts. Both of the peptide precursor intensity increases averaged at the peptide and protein levels are astonishing. Ultimately, the most important result in a shotgun proteomics experiment focused on identification and expression profiling correspond to the improvements at the protein level. Along these lines, higher intensity precursor ion intensities of peptides for low abundance proteins, identified in the Control by 1–9 spectral counts, are at the core for the overall improvements at the protein level. Considering a recent publication confirmed our hypothesis that peptide precursor intensity is the limiting factor in most shotgun proteomics experiments,41 we believe the increases in peptide precursor intensities we observed with ProteoMiner begin to address a significant problem in shotgun proteomics and provides a basis for future improvements.
Figure 3.
Histograms of the changes in average peptide precursor intensities at the (A) peptide and (B) protein level.
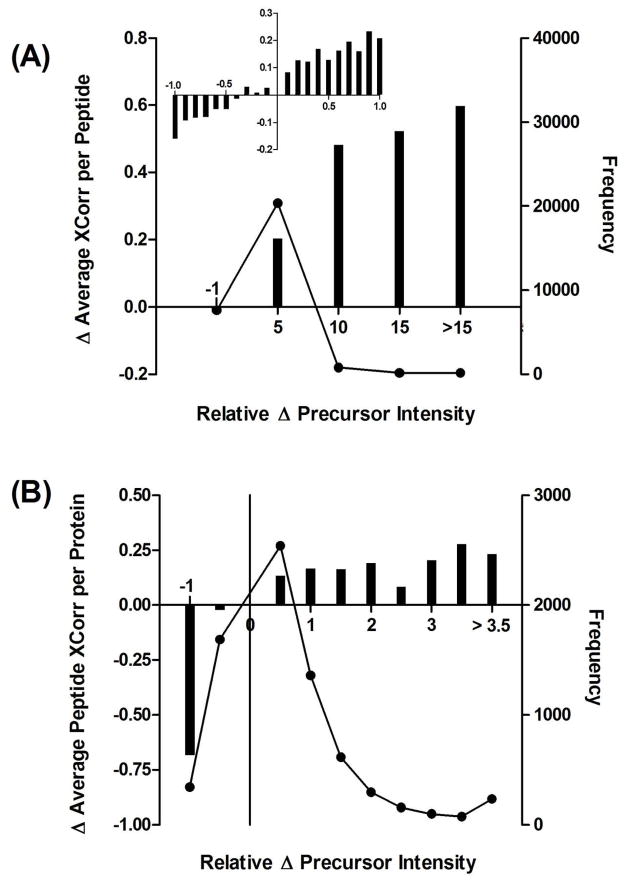
It is logical that higher precursor ion intensities must be beneficial to identification of more peptides, but we interrogated the data further to find a correlation that would directly indicate why more peptides were identified using ProteoMiner. Supplemental Figure 1D shows that there was also a systematic increase in the XCorr of peptides identified using ProteoMiner, so we wondered if the two improvements were related. What we found was a direct correlation between the relative peptide precursor intensity increase and the absolute increase in XCorr values. These results are plotted in Figures 4A and 4B. We compared the changes in the average peptide XCorr at the peptide and protein level in relation to their frequency. At the peptide level, the most frequent improvement was a 5-fold increase in average precursor intensity that correlated to a 0.2 increase in XCorr. Similarly, at the protein level the most frequent increase in the average peptide XCorr was 0.1 from a 50% increase in average precursor intensity. Thus, equalization of protein dynamic range using ProteoMiner increases peptide precursor intensities, leading to more confident identification of peptides and thus proteins.
Figure 4.
Correlation of the relative precursor intensity increases from ProteoMiner equalization to increases in (A) average XCorr per peptide and (B) average peptide XCorr per protein. The change in peptide XCorr are plotted as bars against the left y-axis and the frequency of peptide identifications with the associated precursor intensity change as connected dots against the right y-axis.
Increased sequence coverage yields improved differentiation of isoforms and redundant proteins
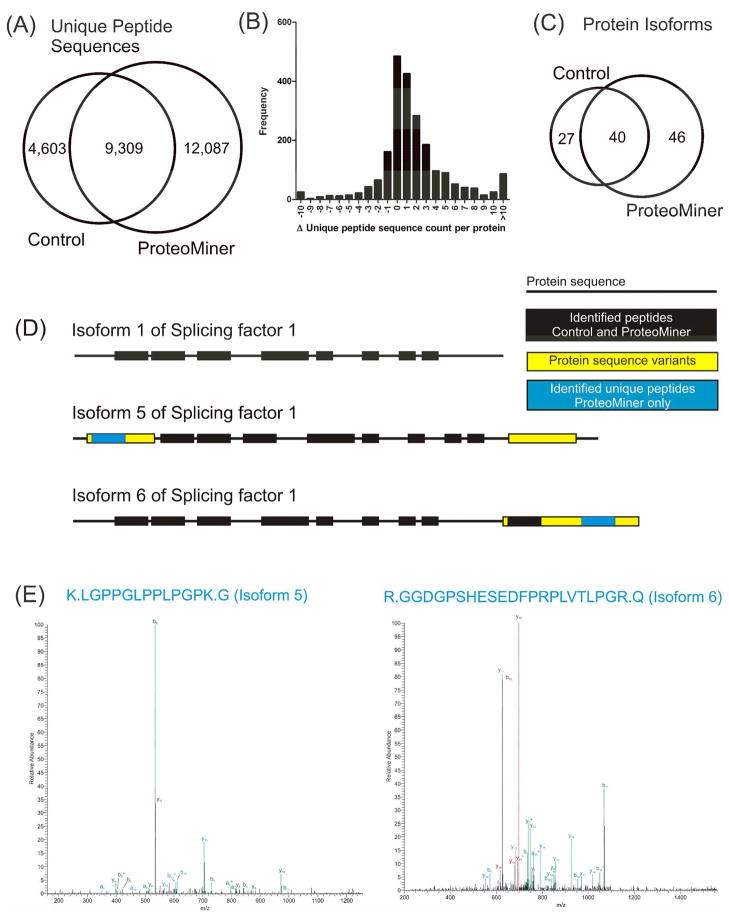
Due to alternative splicing and genetic duplication, many proteins have largely related sequences, yet different functions. Differentiation of these protein isoforms and their roles in biology and disease creates a formidable challenge in any proteomic analysis. In the context of shotgun proteomics, at least one peptide with a unique sequence from a particular protein isoform must be identified to have any confidence that it is present within the sample.42 However, common peptide sequences among redundant proteins are more easily identified since they are innately more abundant. Identification of the less abundant unique peptides is similar to identification of low abundance proteins, meaning their identification is governed by the inherent undersampling of data-dependent acquisition in shotgun proteomics.38 Thus, methods which increase the probability of sampling these unique peptides would be highly beneficial to proteomic analysis. We found that protein abundance equalization using ProteoMiner is a promising option for differentiating protein isoforms. Figure 5A shows that we identified ca. 7500 more unique peptide sequences using ProteoMiner. The distribution of the changes in unique peptide sequences at the protein level is illustrated in Figure 5B. The majority of unique sequences per protein increased by one to three sequence counts, but many also increased by more than 10 counts. The increases in unique sequence counts from the use of ProteoMiner led to identification of ~20 more protein isoforms than Control as shown in Figure 5C. Of the 46 protein isoforms identified only in ProteoMiner we selected an example to illustrate this concept. Figure 5D represents the isoform sequences of Splicing factor 1, the peptides identified from Control and ProteoMiner MudPIT runs, and the identified peptides unique to isoform 5 and 6. As can be seen, two peptides were identified only in ProteoMiner runs that directly led to differentiation of isoforms 5 and 6 from each other and isoform 1. The annotated MS/MS spectra for the peptides that differentiate the isoforms are shown in Figure 5E.
Figure 5.
Improvements in protein isoform identification from the use of ProteoMiner. (A) Comparison of unique peptides identified between Control and ProteoMiner runs. (B) Histogram of changes from Control to ProteoMiner in the unique peptide sequence count at the protein level. (C) Comparison of the protein isoforms identified between Control and ProteoMiner. (D) Illustration of improved protein isoform differentiation from the use of ProteoMiner of Splicing factor 1 isoforms 1, 5, and 6 and the unique peptides that facilitated the differentiation. (E) MS/MS spectra of the unique peptides identified only in ProteoMiner runs which differentiate the isoforms. Fragment ions are annotated as +1 (green), +2 (blue), and +3 (red) charge states. Neutral loss of water (°) and ammonia (*) are also annotated.
ProteoMiner-MudPIT Protein Equalization Mechanism
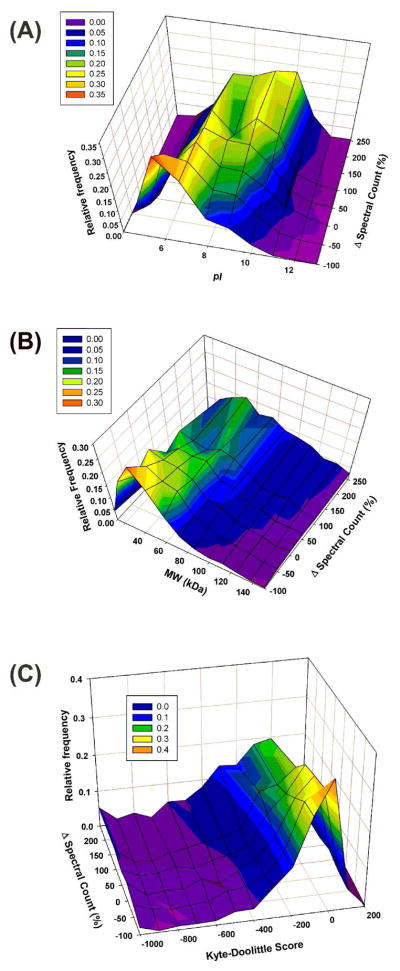
As expected and desired, protein abundance appeared to be the most important factor for protein equalization using ProteoMiner. However, in addition to the differences in the relative concentrations of proteins, their binding constants for specific hexapeptide sequences also determine the extent of equalization of protein abundances. For instance, a lower abundance protein with a stronger binding constant should outcompete a higher abundance protein with a weaker binding constant for that specific ligand. Thus, we wondered what physicochemical properties were most influential in determining the binding constant between proteins and ProteoMiner hexapeptides. These results are illustrated in Figure 6. For all the proteins identified in both Control and ProteoMiner runs, we compared their relative frequencies based on their abundance changes and their theoretical isoelectric points, molecular weights, and hydrophobicities (Kyte-Doolittle score). For all three physicochemical properties, the most highly depleted proteins (ca. −100% Δ spectral count) showed the strongest correlation as red peaks in the surface plots. Although exact values can be assigned to these maxima (MW – 40 kDa, pI – 6, and Kyte-Doolittle Score – 0), we can generalize that small, slightly acidic, amphipathic proteins were preferentially depleted through ProteoMiner equalization. We wondered if these physicochemical properties were only a representation of the abundant protein properties and not of the depletion mechanism. An analysis of the 43 most abundant proteins identified in Control runs (greater than 1000 spectral counts) indicated this was not the case. That is, small, slightly acidic, amphipathic proteins appear to be selectively depleted. The physicochemical properties of all abundant proteins identified in Control runs were, on average, larger in size (MW – 57 kDa), more basic (pI – 7.0), and more hydrophobic (Kyte-Doolittle – −131.8) than the abundant proteins selectively depleted by ProteoMiner treatment. These proteins and their physicochemical properties are listed on the “Abundant Proteins” tab of Supplemental Material 2. Similar concepts are illustrated in the subsequent paragraph which classifies the gene ontology and protein class biases of ProteoMiner. In Figure 6C, the relative maxima remained essentially constant with a Kyte-Doolittle score of zero, indicating that overall protein hydrophobicity may have had little effect on protein enrichment or depletion. We rationalize that the use of 0.1% SDS may have minimized the selectivity of most hydrophobic protein-hexapeptide interactions. However, the use of the Kyte-Doolittle score to calculate protein hydrophobicity instead of identification of hydrophobic protein domains, as commonly used, may not adequately represent this trend. A more informative trend was observed for isoelectric point. As the maxima of the isoelectric point surface plot (Figure 6A) are followed from depleted (−100% Δ spectral count) to enriched (250%) proteins it becomes obvious that protein isoelectric point (i.e. electrostatic or ionic interactions) is the most important physicochemical factor for enrichment of proteins from ProteoMiner equalization. Instead of a single, prominent maxima at a pI of 6 for depleted proteins, a broad range of proteins are uniformly enriched from pI 5 to 11 with a local maximum around a pI of 9. A less prominent trend is observed in the molecular weight plot. The depletion maximum of 40 kDa was shifted slightly to an enrichment maximum between 50–60 kDa dependent on the extent of enrichment. At the biochemical level, these results intuitively make sense if binding cooperativity is considered. Larger proteins with more electrostatic interactions would be expected to bind more tightly to more hexapeptide ligands.
Figure 6.
Visualization and comparison of the effects of theoretical (A) isoelectric point, (B) molecular weight, and (C) hydrophobicity, the physicochemical properties which influence hexapeptide equalization of protein abundance, on the changes in spectral counts from Control to ProteoMiner runs.
Of the top ten most depleted proteins (Table 1), we noticed structural, chaperone, and metabolism-related proteins so we wondered if certain gene ontology classes may be selectively enriched or depleted based on their abundances. These analyses are shown as Supplemental Figures 2–7 and Tables 1–2. Indeed we found the most commonly depleted class of cellular components were structural and abundant, such as actin cytoskeleton, cytoskeleton, and intracellular organelle structures (Supplemental Figure 5). Regarding selective enrichment, proteins categorized as “binding” had the most frequent enrichments (Supplemental Figures 2 and 4) with nucleic acid and RNA binding proteins dominating. These results could be expected since ProteoMiner relies on a binding mechanism for enrichment. This led us to investigate if certain protein domains could be implicated in the ProteoMiner enrichment mechanism. From Supplemental Figure 8, many of the most frequently ProteoMiner-enriched domains were found to contain repeat domains: WD40, Ank, zf-C2H2, HEAT, spectrin, PH, TRP_1, and ARM domains. These domains are composed of pairs of anti-parallel α-helices, β-sheets, or both and are known to be important for facilitating multiple protein-protein interactions and often bind peptide ligands.43 Both of these protein binding characteristics are obviously relevant to the interactions between protein domains and hexapeptides, adding further insight to the specific mechanism of protein enrichment from the use of ProteoMiner. The entire list of enriched protein domains and their frequency are listed in the “Protein Domains” tab in Supplemental Material 2.
Previous publications describing the use of ProteoMiner allude to the fact that certain proteins are “lost” from weak binding or lack of complementary to hexapeptide ligands.44,45 These results were obtained through manual identification of proteins using mass spectrometry from 2D gels. We were able to reinvestigate this concept with a different sample type containing many more proteins using a more automated and comprehensive analysis method. A Venn comparison of proteins identified between Control and ProteoMiner runs is shown in Supplemental Figure 9. Among the ~1000 uniquely identified proteins between Control and ProteoMiner runs, we looked for a trend consistent with loss of proteins by characterizing their physicochemical properties and PANTHER protein classes. The results from these analyses are on the “Uniquely Sampled Proteins” tab in Supplemental Material 2. From a comparison of the averages and standard deviations of the physicochemical properties of uniquely sampled proteins between Control and ProteoMiner there doesn’t appear to be a physicochemical property which defines “lost” proteins from the use of ProteoMiner. Similarly, we were unable to find a PANTHER protein class with a strong P-value and high frequency that would indicate loss of a specific protein class by ProteoMiner treatment. However, it should be noted that shotgun proteomics is inherently a protein sampling method.38 This is illustrated for these experiments by the overlap of proteins identified in triplicate Control and ProteoMiner runs shown in Supplemental Figure 10. Thus, a more comprehensive analysis of the proteins identified in Control and not ProteoMiner runs would need to be performed to confidently define critical physicochemical properties or protein classes that identification is prevented by the use of ProteoMiner.
A recent publication reported that hydrophobic interactions of proteins with the ProteoMiner beads were the dominant binding mechanism.46 Additionally, they tested different chromatography reverse phase and ion exchange resins and were able to achieve similar equalization results as with ProteoMiner beads. Their results implied that electrostatic interactions are irrelevant to protein-hexapeptide binding. What the authors failed to recognize is that their methods only facilitated comparison of ~50 abundant proteins, an extremely small portion of the proteome. As a result they were only probing the depletion mechanism of hexapeptide beads and not the enrichment mechanism. Similarly, the comparisons with other resins only probed their depletion capabilities, just half of the mechanism of hexapeptide bead equalization. From the use of MudPIT as a protein readout instead of a 2D gel, we were able to perform a more comprehensive analysis over four orders of magnitude in protein abundance. As a result, the trends in protein physicochemical properties could be better represented and may aid in further improvements to optimization of protein abundance equalization using hexapeptide libraries.
CONCLUSIONS
We demonstrated that an equalization of cellular protein abundance using ProteoMiner increases the quality and quantity of peptide identifications in a shotgun proteomics experiment. Essentially, the large decreases in peptide spectral counts through depletion of a small percentage of high abundance proteins had dramatic effects on the reproducibility and sequence count of low abundance proteins. We were not surprised by this phenomenon since removal of high abundance peptides allows for the ion trap to be filled with more of other lower abundance peptides. The dramatic increases in peptide precursor intensities of all but the highest abundant proteins are a strong indicator of this. Additionally, the largest gains in average precursor intensity were observed for proteins with low abundance (“Tens”, 10–99 spectral counts) in Control runs and not of very low abundance (“Ones”, 1–9 spectral counts). If enrichment of low abundance proteins was the dominant mechanism of improvement from the ProteoMiner-MudPIT combination, the largest gains would have been expected for the very low abundance proteins and not low abundance proteins.
The use of a randomly generated hexapeptide library understandably affords the possibility that a protein may not have a corresponding ligand. By coupling ProteoMiner equalization with the comprehensive nature of MudPIT we found this to be much less of an issue than anticipated. On top of the aforementioned 10% gain in protein identifications, the majority of proteins identified (73%, data not shown) appeared in both experiments, indicating very few proteins were lost through the use of ProteoMiner. We suspect larger protein identification gains were not achieved since we investigated only one protein:bead incubation ratio (100:1). The protein concentration dynamic range for which we enriched using ProteoMiner was well matched to the detection dynamic range of MudPIT. In order to shift this partially redundant overlap of dynamic ranges between ProteoMiner and MudPIT, a further increase in the protein:bead ratio could facilitate protein-hexapeptide interactions and enrichments of even lower abundance proteins that are randomly accessible by current shotgun proteomics techniques. In fact, this type of strategy could be potentially used to “zoom in” on order of magnitude “windows” of the proteome dynamic range.
As mentioned earlier, the ultimate goal of shotgun proteomics would be to identify and sequence the entire cellular proteome. Since even state-of-the-art mass spectrometers used for shotgun proteomics are still primarily undersampling peptides of the lowest abundance proteins we believe that equalization of the proteome dynamic range will be essential to achieve this ultimate goal. We observed the most dramatic sequence coverage improvements for the lowest abundance proteins, indentified by less than 10 spectral counts in Control runs. Until the time when most MS/MS peptide spectra result in the identification of a new protein or an increase in the sequence coverage of a protein, efforts should remain focused on the equalization of protein, and thus peptide, abundance. To typify this challenge, in a 24 hour MudPIT run we identified ~35,000 peptides of which about a third contributed to an increase in sequence coverage of a protein. An in silico digestion of the UniProt Swiss-Prot database yielded 551,753 peptides with no missed trypsin cleavages from 20,248 protein entries and 1,495,634 peptides with one missed cleavage. Both of these peptide counts are sizeable, yet are within current MS/MS sampling rates due to improvements in the sampling speed and sensitivity of current ion trap mass spectrometers.41 However, what cannot be ignored is the fact that a single peptide may be present with 106 copies per cell while another from a very low abundance protein may be present with one copy per cell. Until these peptide abundance differences are addressed, the realization of sequencing the entire proteome may not be achievable.
Supplementary Material
Acknowledgments
The authors would like to acknowledge funding from the National Institute of Health grants R01DK074798 (J.R.Y.), U19AI063603 (J.R.Y.), RFP-NHLBI-HV-10-5 (J.R.Y.), P41RR011823 (J.R.Y.) and the CAPES/Fiocruz 30/2006 grant (P.C.C.). We thank A.J. R. Heck and P.A. Haynes for feedback during their sabbatical visits.
Footnotes
Raw data files can be found at http://fields.scripps.edu/published/ProteoMiner2011/.
Supporting Information Available: This material is available free of charge at http://pubs.acs.org.
References
- 1.Giaccia A, Siim BG, Johnson RS. HIF-1 as a target for drug development. Nat Rev Drug Discov. 2003;2:803–811. doi: 10.1038/nrd1199. [DOI] [PubMed] [Google Scholar]
- 2.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 3.van der Horst A, Burgering BM. Stressing the role of FoxO proteins in lifespan and disease. Nat Rev Mol Cell Biol. 2007;8:440–450. doi: 10.1038/nrm2190. [DOI] [PubMed] [Google Scholar]
- 4.Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol. 2010;11:427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- 5.Cravatt BF, Simon GM, Yates JR., 3rd The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 6.McCormack AL, et al. Direct analysis and identification of proteins in mixtures by LC/MS/MS and database searching at the low-femtomole level. Anal Chem. 1997;69:767–776. doi: 10.1021/ac960799q. [DOI] [PubMed] [Google Scholar]
- 7.McCormack AL, Eng JK, Yates JR., III Peptide sequence analysis on quadrupole mass spectrometers. Methods: A Companion to Methods in Enzymology. 1994;6:274–283. [Google Scholar]
- 8.Eng JK, McCormack AL, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 9.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 10.Wolters DA, Washburn MP, Yates JR., III An automated multidimensional protein identification technology for shotgun proteomics. Analytical Chemistry. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 11.An Y, Cooper JW, Balgley BM, Lee CS. Selective enrichment and ultrasensitive identification of trace peptides in proteome analysis using transient capillary isotachophoresis/zone electrophoresis coupled with nano-ESI-MS. Electrophoresis. 2006;27:3599–3608. doi: 10.1002/elps.200600093. [DOI] [PubMed] [Google Scholar]
- 12.Gilar M, Olivova P, Daly AE, Gebler JC. Two-dimensional separation of peptides using RP-RP-HPLC system with different pH in first and second separation dimensions. J Sep Sci. 2005;28:1694–1703. doi: 10.1002/jssc.200500116. [DOI] [PubMed] [Google Scholar]
- 13.Hubner NC, Ren S, Mann M. Peptide separation with immobilized pI strips is an attractive alternative to in-gel protein digestion for proteome analysis. Proteomics. 2008;8:4862–4872. doi: 10.1002/pmic.200800351. [DOI] [PubMed] [Google Scholar]
- 14.Krijgsveld J, Gauci S, Dormeyer W, Heck AJ. In-gel isoelectric focusing of peptides as a tool for improved protein identification. J Proteome Res. 2006;5:1721–1730. doi: 10.1021/pr0601180. [DOI] [PubMed] [Google Scholar]
- 15.Olsen JV, et al. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol Cell Proteomics. 2009;8:2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen Y, et al. Ultra-high-efficiency strong cation exchange LC/RPLC/MS/MS for high dynamic range characterization of the human plasma proteome. Anal Chem. 2004;76:1134–1144. doi: 10.1021/ac034869m. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, et al. Integrated capillary isoelectric focusing/nano-reversed phase liquid chromatography coupled with ESI-MS for characterization of intact yeast proteins. J Proteome Res. 2005;4:36–42. doi: 10.1021/pr049876l. [DOI] [PubMed] [Google Scholar]
- 18.Blackler AR, Klammer AA, MacCoss MJ, Wu CC. Quantitative comparison of proteomic data quality between a 2D and 3D quadrupole ion trap. Anal Chem. 2006;78:1337–1344. doi: 10.1021/ac051486a. [DOI] [PubMed] [Google Scholar]
- 19.Boersema PJ, Divecha N, Heck AJ, Mohammed S. Evaluation and optimization of ZIC-HILIC-RP as an alternative MudPIT strategy. J Proteome Res. 2007;6:937–946. doi: 10.1021/pr060589m. [DOI] [PubMed] [Google Scholar]
- 20.Chen EI, Hewel J, Felding-Habermann B, Yates JR., 3rd Large scale protein profiling by combination of protein fractionation and multidimensional protein identification technology (MudPIT) Mol Cell Proteomics. 2006;5:53–56. doi: 10.1074/mcp.T500013-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Mayya V, Rezaul K, Cong YS, Han D. Systematic comparison of a two-dimensional ion trap and a three-dimensional ion trap mass spectrometer in proteomics. Mol Cell Proteomics. 2005;4:214–223. doi: 10.1074/mcp.T400015-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saba J, Bonneil E, Pomies C, Eng K, Thibault P. Enhanced sensitivity in proteomics experiments using FAIMS coupled with a hybrid linear ion trap/Orbitrap mass spectrometer. J Proteome Res. 2009;8:3355–3366. doi: 10.1021/pr801106a. [DOI] [PubMed] [Google Scholar]
- 23.Second TP, et al. Dual-pressure linear ion trap mass spectrometer improving the analysis of complex protein mixtures. Anal Chem. 2009;81:7757–7765. doi: 10.1021/ac901278y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yates JR, Cociorva D, Liao L, Zabrouskov V. Performance of a linear ion trap-Orbitrap hybrid for peptide analysis. Anal Chem. 2006;78:493–500. doi: 10.1021/ac0514624. [DOI] [PubMed] [Google Scholar]
- 25.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 26.Pernemalm M, et al. Evaluation of three principally different intact protein prefractionation methods for plasma biomarker discovery. J Proteome Res. 2008;7:2712–2722. doi: 10.1021/pr700821k. [DOI] [PubMed] [Google Scholar]
- 27.Guerrier L, et al. Reducing protein concentration range of biological samples using solid-phase ligand libraries. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;833:33–40. doi: 10.1016/j.jchromb.2005.12.048. [DOI] [PubMed] [Google Scholar]
- 28.Thulasiraman V, et al. Reduction of the concentration difference of proteins in biological liquids using a library of combinatorial ligands. Electrophoresis. 2005;26:3561–3571. doi: 10.1002/elps.200500147. [DOI] [PubMed] [Google Scholar]
- 29.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 30.Bandhakavi S, et al. Hexapeptide Libraries for Enhanced Protein PTM Identification and Relative Abundance Profiling in Whole Human Saliva. J Proteome Res. 2011;10:1052–1061. doi: 10.1021/pr100857t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stone MD, et al. Large-Scale Phosphoproteomics Analysis of Whole Saliva Reveals a Distinct Phosphorylation Pattern. J Proteome Res. 2011 doi: 10.1021/pr1010247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonald WH, et al. MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun Mass Spectrom. 2004;18:2162–2168. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- 33.Xu T, et al. ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Mol Cell Proteomics. 2006;5:S174. [Google Scholar]
- 34.Carvalho PC, Fischer JS, Chen EI, Yates JR, 3rd, Barbosa VC. PatternLab for proteomics: a tool for differential shotgun proteomics. BMC Bioinformatics. 2008;9:316. doi: 10.1186/1471-2105-9-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park SK, Venable JD, Xu T, Yates JR., 3rd A quantitative analysis software tool for mass spectrometry-based proteomics. Nat Methods. 2008;5:319–322. doi: 10.1038/nmeth.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mi H, Thomas P. PANTHER pathway: an ontology-based pathway database coupled with data analysis tools. Methods Mol Biol. 2009;563:123–140. doi: 10.1007/978-1-60761-175-2_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 39.Nesvizhskii AI. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J Proteomics. 2010;73:2092–2123. doi: 10.1016/j.jprot.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tu C, et al. Depletion of abundant plasma proteins and limitations of plasma proteomics. J Proteome Res. 2010;9:4982–4991. doi: 10.1021/pr100646w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michalski A, Cox J, Mann M. More than 100,000 Detectable Peptide Species Elute in Single Shotgun Proteomics Runs but the Majority is Inaccessible to Data-Dependent LC-MS/MS. J Proteome Res. 2011 doi: 10.1021/pr101060v. [DOI] [PubMed] [Google Scholar]
- 42.Fermin D, Basrur V, Yocum AK, Nesvizhskii AI. Abacus: A computational tool for extracting and pre-processing spectral count data for label-free quantitative proteomic analysis. Proteomics. 2011 doi: 10.1002/pmic.201000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andrade MA, Perez-Iratxeta C, Ponting CP. Protein repeats: structures, functions, and evolution. J Struct Biol. 2001;134:117–131. doi: 10.1006/jsbi.2001.4392. [DOI] [PubMed] [Google Scholar]
- 44.Boschetti E, Righetti PG. The art of observing rare protein species in proteomes with peptide ligand libraries. Proteomics. 2009;9:1492–1510. doi: 10.1002/pmic.200800389. [DOI] [PubMed] [Google Scholar]
- 45.Righetti PG, Boschetti E, Lomas L, Citterio A. Protein Equalizer Technology: the quest for a “democratic proteome”. Proteomics. 2006;6:3980–3992. doi: 10.1002/pmic.200500904. [DOI] [PubMed] [Google Scholar]
- 46.Keidel EM, Ribitsch D, Lottspeich F. Equalizer technology--Equal rights for disparate beads. Proteomics. 2010;10:2089–2098. doi: 10.1002/pmic.200900767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.