Abstract
Retinal degeneration slow (RDS) is a photoreceptor specific tetraspanin membrane protein. It is expressed in the rim region of rod outer segment (OS) discs and cone OS lamellae. Mutations in RDS cause both rod and cone-dominant retinal degenerations. We have recently shown that RDS functions differently in rods vs. cones, and have used the cone-dominant nrl−/− and rod-dominant wild-type (WT) murine retinas to study these differences and help understand the mechanism of rod and cone OS biogenesis. We hypothesize that the differential role of RDS in rods vs. cones is in part related to differences in RDS binding partners. RDS has been shown to bind to the GARP portion of the β subunit of the rod-cyclic nucleotide gated (CNG) channel. This interaction has been hypothesized to play a role in anchoring the disc rim to the rod plasma membrane. In this study we show that RDS does not interact with the cone CNG. Given that cone lamellae are not entirely encased in plasma membrane and therefore may have different anchoring requirements compared with rods, this observation may help explain some of the differential behavior of RDS in rods vs. cones.
Over 70 different disease causing mutations in the photoreceptor specific protein retinal degeneration slow (RDS) have been identified. Usually, mutations in photoreceptor-specific genes cause predictable disease phenotypes, i.e. rod specific genes associate with forms of rod dominant diseases (such as retinitis pigmentosa, RP) while mutations in cone specific genes tend to cause cone-dominant diseases (such as macular degeneration, MD). In contrast, different mutations in RDS have been linked to both rod-dominant and cone-dominant disease. The spectrum of RDS-associated retinal disease phenotypes ranges from traditional RP to MD to butterfly macular dystrophy to cone-rod degeneration depending on the mutation (http://www.retina-international.com/sci-news/rdsmut.htm).
The naturally occurring retinal degeneration slow (rds−/−) mutant mouse has proved an excellent model for studying the phenotypic divergence in RDS-associated retinal degenerations. These mice do not form OSs and exhibit slow, progressive death of photoreceptors coupled with panretinal degeneration (Chaitin et al. 1988; Sanyal et al. 1980; Sanyal and Zeilmaker 1984). In the rds+/− mouse, OSs form but do not assemble into properly organized, stacked discs (Hawkins et al. 1985; Sanyal et al. 1986). This haploinsufficiency phenotype is characterized by an early onset slow rod degeneration followed by late onset slow cone degeneration (Cheng et al. 1997). The deformed discs are capable of subnormal levels of phototransduction, but the lack of a proper OS causes degeneration throughout the retina.
Unfortunately, the wild-type (WT) mouse retina is 95–97% rods rendering it difficult to study the mechanisms underlying cone degeneration associated with mutations in RDS. Even in cone only models such as the early ages of the rhodopsin knockout mouse (rho−/−), the small number of cones makes analysis very difficult. The recent development of a cone-dominant mouse model containing a null mutation in the Neural Retinal Leucine Zipper (NRL) transcription factor gene (Daniele et al. 2005; Mears et al. 2001; Nikonov et al. 2005) has provided an elegant way to overcome the rod-bias problem. When retinal progenitor cells fail to receive the NRL regulatory cue, they divert from a developmental fate to rods and form blue-responsive cones instead (Farjo et al. 1997; Farjo et al. 1993; Mitton et al. 2000; Rehemtulla et al. 1996; Swaroop et al. 1992). The nrl−/− mouse is an ideal model for studying MD since it makes possible the examination of cones in the absence of rods. We have recently taken advantage of the nrl−/− mouse to study the behavior of RDS in cones. Unlike the WT rods, cones of the nrl−/− retina lacking RDS (nrl−/−/rds−/−) retain OS structures and the capacity for photopic visual function. The cone OSs of the nrl−/−/rds−/− are severely malformed and have no lamellae, but continue to express OS proteins such as S-opsin. This situation is in marked contrast to the WT and underscores the different role of RDS in rods and cones.
The question remains though, what causes RDS to behave differently in rods vs. cones? Expression of RDS is limited to the OS rim region of both rod and cone photoreceptor disc rims/lamellae (Arikawa et al. 1992; Moritz et al. 2002). RDS forms tetramers in the photoreceptor inner segment which then traffic to the OS and further assemble via the second intracellular (D2) loop disulfide bonds into octamers and other higher order complexes. RDS is known to form non-covalent heterotetramers with its homologue, a protein called rod outer segment membrane protein 1 (ROM-1) in addition to homotetramers (Chakraborty et al. 2008; Goldberg and Molday 2000; Goldberg et al. 1995). However, differential interactions with ROM-1 are unlikely to be responsible for rod-cone differences in RDS behavior. Both rods and cones express ROM-1 and our biochemical studies on ROM-1 in the nrl−/− retina show that RDS/ROM-1 are biochemically similar in cones and rods (Chakraborty et al. 2008).
Little is known though about other potential RDS interacting partners in rods and cones. Tetraspanin proteins typically assemble with themselves and other proteins into a large functional protein complex known as the tetraspanin web which is similar to but distinct from lipid rafts (Hemler 2003; Levy and Shoham 2005; Stipp et al. 2003). This web consists of proteins that are covalently and non-covalently bound (similar to RDS and ROM-1) and of proteins that are more loosely associated. The function of tetraspanins is often dictated or modulated by the composition of this tetraspanin web, so it makes sense that differential behavior of RDS in rods vs. cones might result from different interacting partners in the two cell types.
In addition to ROM-1, RDS in the rod dominant retina is known to interact with at least two other proteins. The first protein, melanoregulin, is a protein associated with membrane fusion. Kathy Boesze Battaglia’s group has shown that in concert with melanoregulin, RDS helps mediate membrane fusion and thus possibly OS disc sealing at the base of the OS (Boesze-Battaglia 2000; Boesze-Battaglia et al. 2007). The expression of melanoregulin and its potential interactions with RDS in cones have not been studied, but represent a particularly interesting area for future experimentation.
The second known RDS interacting protein is the β subunit of the rod cyclic nucleotide-gated (CNG) channel. The CNG channel functions as a heterotetramer comprised of α and β subunits (Kaupp et al. 1989). It is responsible for the resting dark current in rod photoreceptors and closes upon hydrolysis of cGMP by phosphodiesterase after light induced G-protein coupled receptor/second messenger signaling (Kaupp et al. 1989; Pugh 2000). The N-terminus of the β subunit has a large glutamic acid/proline rich domain called GARP (Colville and Molday 1996). There are also two free protein forms of this GARP domain, GARP-1 and the slightly longer GARP-2 (Colville and Molday 1996; Korschen et al. 1999).
These proteins are intrinsically disordered and have been known to act as scaffolding type proteins in other tissues (Batra-Safferling et al. 2006). Molday’s group has shown that both free GARP and the GARP domain of the CNG channel β subunit can interact with RDS (Poetsch et al. 2001) in bovine rod OSs. They hypothesize that CNG-GARP-RDS interactions might be responsible for the anchoring filaments that have been observed between the disc rim and plasma membrane of rods (Poetsch et al. 2001; Roof and Heuser 1982). They further hypothesize that interactions between free GARP and RDS might serve to connect adjacent discs. Although there is no direct proof supporting this hypothesis, advanced structural analysis of GARP has shown that its size and shape are consistent with this role (Batra-Safferling et al. 2006).
Cones also have a cGMP gated channel responsible for maintenance of the dark, “off” current, but it is not the same channel as that expressed in rods. It is thought that differences in the regulation/kinetics of the cone vs. rod CNG channels help explain the differences in phototransduction kinetics between the two cell types. Cone channels have a ten-fold lower ligand sensitivity than rod channels and are between 30 and 100 times less sensitive to light (Picones and Korenbrot 1995; Pugh 2000). It has recently been shown that the cone channel also functions as a heterotetramers composed of α and β subunits (Matveev et al. 2008). The β subunit of the cone channel does not have a GARP domain, so we hypothesized that RDS would not interact with the cone channel thus partially explaining the differential role of RDS in rods vs. cones.
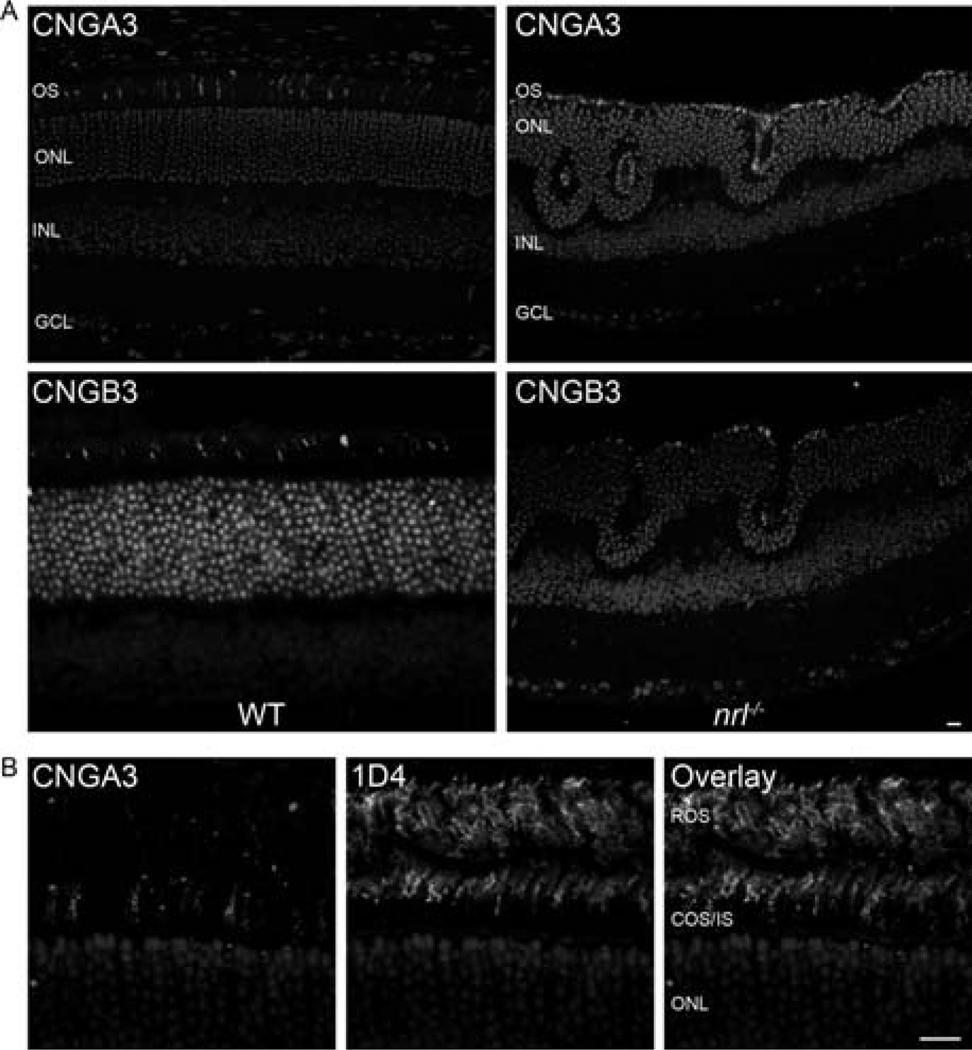
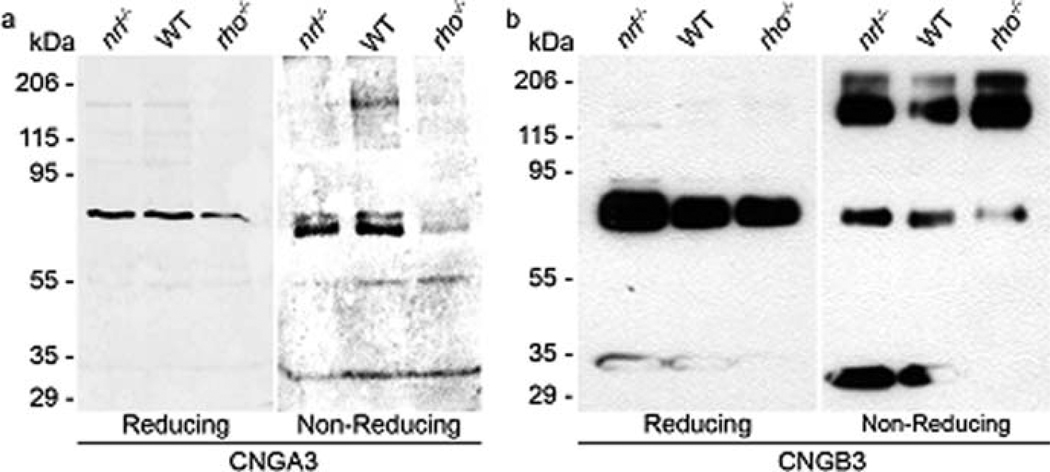
Our first step was to determine expression of the cone CNG channel in WT and nrl−/− retina using polyclonal antibodies against the cone CNG channel subunit CNGA3 and CNGB3 (Matveev et al. 2008). Immunohistochemical analysis of retinal sections taken from 1-month-old animals labeled with the CNGA3 and CNGB3 antibodies demonstrates that both subunits of the cone CNG channel are expressed in the cone OSs of WT and nrl−/− retinas (Fig. 8.1a).We do not detect any protein expression in other retinal cell types, or in other portions of the photoreceptor. The labeling pattern in the WT retina is consistent with cone-only expression, but to confirm this, co-labeling studies were undertaken with rod opsin (mAB 1D4, generously shared by Dr. Robert Molday) and CNGA3. The left panel of Fig. 8.1b shows CNGA3 labeling while the middle panel is rhodopsin (1D4) labeling. The labeling patterns are quite distinct and the two proteins do not co-localize (overlay, right) indicating that cone CNG is not expressed in rods. To confirm the size of the cone CNG subunits, whole retinal extracts from P30 retinas underwent reducing and non-reducing SDS-PAGE/Western blotting. Rod-dominant WT extracts were used as controls while nrl−/− was used as a cone-dominant model. Historically, the only available cone-dominant model has been the young rhodopsin knockout (rho−/−) retina, so those retinas were also included as a control, although they express significantly lower quantities of cone proteins than the nrl−/− retina. Under reducing conditions, both the α and β subunits of cone CNG appear as monomers of approximately 75 kDa (Fig. 8.2a, b left) in all samples. This is consistent with the predicted peptide sizes of 72 kDa (A) and 79 kDa (B) and is in contrast to rod CNG channel subunits which have been shown to migrate abnormally on SDS-PAGE (Poetsch et al. 2001). The involvement of disulfide-linkages in multi-subunit channel assembly was examined by non-reducing SDS-PAGE. A significant amount of CNGB3 dimer was detected at ~170 kDa (Fig. 8.2b, right) although little dimerization of CNGA3 was noticed in these retinas (Fig. 8.2a, right).
Fig. 8.1.
CNGA3 and CNGB3 is expressed in cones OSs of WT and nrl−/− Retinas; (a) Paraffin embedded sections from P30 WT or nrl−/− mice were stained with CNGA3 and CNGB3 polyclonal antibodies and visualized using anti-rabbit Cy3 secondary antibody. Sections were counterstained with DAPI to label nuclei. Expression of cone channel subunits is limited to the OS layer in both WT and nrl−/− retina. Scale bar 20 μm. (b) P30 WT paraffin embedded sections were stained with CNGA3 (left) and mAB 1D4 (against rhodopsin-middle). The two proteins do not co-localize (right) indicating that cone CNG is not expressed in rods. Scale bar 10 μm. OS, outer segment; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer
Fig. 8.2.
CNGB3 is present as both a monomer and dimer, while CNGA3 is present only as a monomer; Retinal extracts from P30 WT (rod-dominant), nrl−/− and rho−/− (cone-dominant) mice underwent reducing (left) and non-reducing (right) SDS-PAGE and Western blotting. (a) CNGA3 is only present as the monomeric form (~75 kDa) while CNGB3 (b) exhibits a substantial quantity of both monomer and dimer
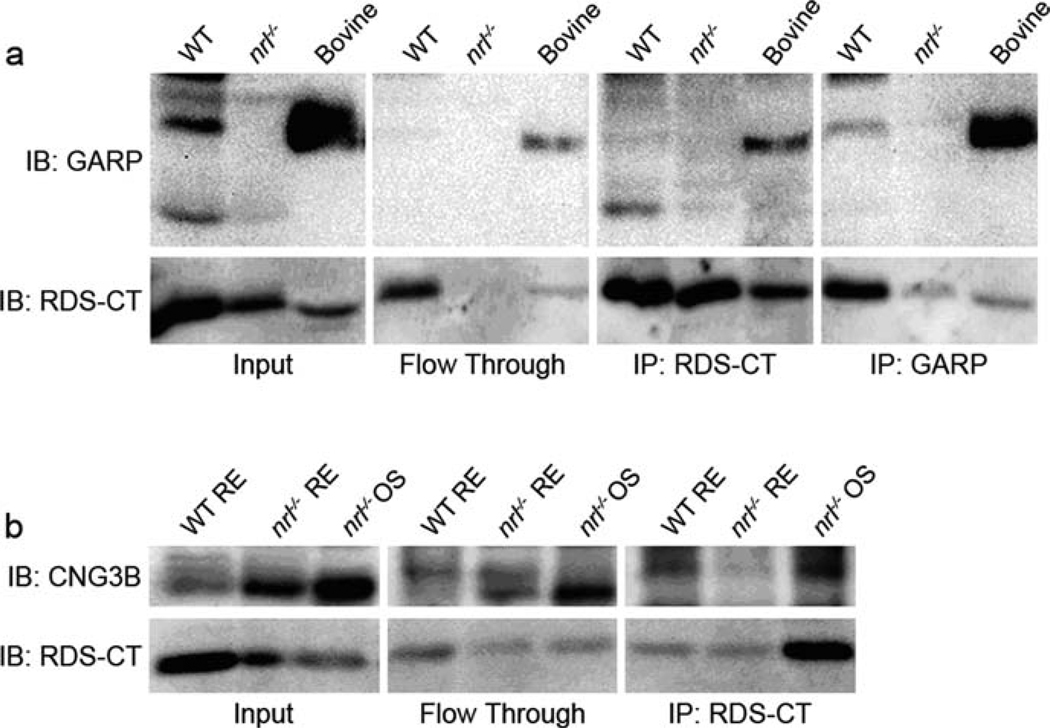
Since previous studies had shown that the bovine rod CNG channel associates with RDS (Poetsch et al. 2001) via the GARP portion of the CNG β-subunit, we wanted to confirm that we could detect this interaction in mice. Retinal extracts from one month old WT mice underwent immunoprecipitation (IP) with either the RDS antibody or the GARP monoclonal antibody 4B1 (generously shared by Dr. Robert Molday, University of British Columbia). Bovine rod OSs were used as a positive control while nrl−/− retinal extracts served as a negative control since they do not express the rod CNG-GARP. As shown in Fig. 8.3a (input), WT retinal extracts express both splice variants of free GARP while the rodless nrl−/− retinas do not. As demonstrated by reciprocal co-IP shown in the left two panels of Fig. 8.3a, in mouse rods, RDS does interact with GARP. To determine whether the cone CNG β-subunit interacts with RDS, similar experiments were undertaken. RDS complexes from WT and nrl−/− retinal extracts or nrl−/− OS preparations were immunoprecipitated using the RDS antibody. OS preparations (Chakraborty et al. 2008) were used as an enriched source of OS proteins such as CNG and RDS. The IP was successful as evidenced by detection of RDS in all three samples (Fig. 8.3b, bottom). However the β subunit of cone CNG was not detected in any of the immunoprecipitants although it was detected in the initial samples (Fig. 8.3b, top left).
Fig. 8.3.
The rod CNG β subunit interacts with RDS while the cone CNG β subunit does not; (a) Retinal extracts from WT and nrl−/− P30 mice and bovine OSs underwent IP using either RDS C-terminal antibody or monoclonal (4B1) GARP antibody. This reciprocal co-IP confirms that rod CNG does interact with RDS. (b) WT and nrl−/− retinal extracts and nrl−/− OSs underwent IP with RDS C-terminal antibody. Although CNGB3 was present in the samples, none was detected in the immunoprecipitants
These results confirm our hypothesis that the cone CNG β-subunit does not interact with RDS, most likely due to the lack of a GARP domain in the cone channel. Furthermore, they clearly identify the first difference in the RDS tetraspanin web in rods vs. cones. It is likely that additional study will identify further RDS interacting partners that are rod or cone specific and will further enhance our understanding of the role of RDS in OS biogenesis.
Acknowledgment
The monoclonal antibodies for this study (4B1 and 1D4) were generously shared with us by Dr. Robert Molday, University of British Columbia. This study was supported by grants from the National Institutes of Health (EY10609 & EY018656), Core Grant for Vision Research EY12190, and the Foundation Fighting Blindness. Dr. Naash is the recipient of a Research to Prevent Blindness James S. Adams Scholar Award.
References
- Arikawa K, Molday LL, Molday RS, et al. Localization of peripherin/rds in the disk membranes of cone and rod photoreceptors: relationship to disk membrane morphogenesis and retinal degeneration. J Cell Biol. 1992;116:659. doi: 10.1083/jcb.116.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra-Safferling R, Abarca-Heidemann K, Korschen HG, et al. Glutamic acid-rich proteins of rod photoreceptors are natively unfolded. J Biol Chem. 2006;281:1449. doi: 10.1074/jbc.M505012200. [DOI] [PubMed] [Google Scholar]
- Boesze-Battaglia K. Fusion between retinal rod outer segment membranes and model membranes: functional assays and role for peripherin/rds. Methods Enzymol. 2000;316:65. doi: 10.1016/s0076-6879(00)16717-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesze-Battaglia K, Song H, Sokolov M, et al. The tetraspanin protein peripherin-2 forms a complex with melanoregulin, a putative membrane fusion regulator. Biochemistry. 2007;46:1256. doi: 10.1021/bi061466i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaitin MH, Carlsen RB, Samara GJ. Immunogold localization of actin in developing photoreceptor cilia of normal and rds mutant mice. Exp Eye Res. 1988;47:437. doi: 10.1016/0014-4835(88)90054-1. [DOI] [PubMed] [Google Scholar]
- Chakraborty D, Ding XQ, Fliesler SJ, et al. Outer segment oligomerization of rds: evidence from mouse models and subcellular fractionation. Biochemistry. 2008;47:1144. doi: 10.1021/bi701807c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T, Peachey NS, Li S, et al. The effect of peripherin/rds haploinsufficiency on rod and cone photoreceptors. J Neurosci. 1997;17:8118. doi: 10.1523/JNEUROSCI.17-21-08118.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colville CA, Molday RS. Primary structure and expression of the human beta-subunit and related proteins of the rod photoreceptor cGMP-gated channel. J Biol Chem. 1996;271:32968. doi: 10.1074/jbc.271.51.32968. [DOI] [PubMed] [Google Scholar]
- Daniele LL, Lillo C, Lyubarsky AL, et al. Cone-like morphological, molecular, and electrophysiological features of the photoreceptors of the Nrl knockout mouse. Invest Ophthalmol Vis Sci. 2005;46:2156. doi: 10.1167/iovs.04-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farjo Q, Jackson A, Pieke-Dahl S, et al. Human bZIP transcription factor gene NRL: structure, genomic sequence, and fine linkage mapping at 14q11.2 and negative mutation analysis in patients with retinal degeneration. Genomics. 1997;45:395. doi: 10.1006/geno.1997.4964. [DOI] [PubMed] [Google Scholar]
- Farjo Q, Jackson AU, Xu J, et al. Molecular characterization of the murine neural retina leucine zipper gene. Nrl. Genomics. 1993;18:216. doi: 10.1006/geno.1993.1458. [DOI] [PubMed] [Google Scholar]
- Goldberg AF, Molday RS. Expression and characterization of peripherin/rds-rom-1 complexes and mutants implicated in retinal degenerative diseases. Methods Enzymol. 2000;316:671. doi: 10.1016/s0076-6879(00)16756-4. [DOI] [PubMed] [Google Scholar]
- Goldberg AF, Moritz OL, Molday RS. Heterologous expression of photoreceptor peripherin/rds and Rom-1 in COS-1 cells: assembly, interactions, and localization of multisubunit complexes. Biochemistry. 1995;34:14213. doi: 10.1021/bi00043a028. [DOI] [PubMed] [Google Scholar]
- Hawkins RK, Jansen HG, Sanyal S. Development and degeneration of retina in rds mutant mice: photoreceptor abnormalities in the heterozygotes. Exp Eye Res. 1985;41:701. doi: 10.1016/0014-4835(85)90179-4. [DOI] [PubMed] [Google Scholar]
- Hemler ME. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu Rev Cell Dev Biol. 2003;19:397. doi: 10.1146/annurev.cellbio.19.111301.153609. [DOI] [PubMed] [Google Scholar]
- Kaupp UB, Niidome T, Tanabe T, et al. Primary structure and functional expression from complementary DNA of the rod photoreceptor cyclic GMP-gated channel. Nature. 1989;342:762. doi: 10.1038/342762a0. [DOI] [PubMed] [Google Scholar]
- Korschen HG, Beyermann M, Muller F, et al. Interaction of glutamic-acid-rich proteins with the cGMP signalling pathway in rod photoreceptors. Nature. 1999;400:761. doi: 10.1038/23468. [DOI] [PubMed] [Google Scholar]
- Levy S, Shoham T. Protein-protein interactions in the tetraspanin web. Physiology (Bethesda) 2005;20:218. doi: 10.1152/physiol.00015.2005. [DOI] [PubMed] [Google Scholar]
- Matveev AV, Quiambao AB, Browning Fitzgerald J, et al. Native cone photoreceptor cyclic nucleotide-gated channel is a heterotetrameric complex comprising both CNGA3 and CNGB3: a study using the cone-dominant retina of Nrl−/− mice. J Neurochem. 2008;106:2042. doi: 10.1111/j.1471-4159.2008.05548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears AJ, Kondo M, Swain PK, et al. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29:447. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- Mitton KP, Swain PK, Chen S, et al. The leucine zipper of NRL interacts with the CRX homeodomain. A possible mechanism of transcriptional synergy in rhodopsin regulation. J Biol Chem. 2000;275:29794. doi: 10.1074/jbc.M003658200. [DOI] [PubMed] [Google Scholar]
- Moritz OL, Peck A, Tam BM. Xenopus laevis red cone opsin and Prph2 promoters allow transgene expression in amphibian cones, or both rods and cones. Gene. 2002;298:173. doi: 10.1016/s0378-1119(02)00923-x. [DOI] [PubMed] [Google Scholar]
- Nikonov SS, Daniele LL, Zhu X, et al. Photoreceptors of Nrl −/− mice coexpress functional S- and M-cone opsins having distinct inactivation mechanisms. J Gen Physiol. 2005;125:287. doi: 10.1085/jgp.200409208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picones A, Korenbrot JI. Spontaneous, ligand-independent activity of the cGMP-gated ion channels in cone photoreceptors of fish. J Physiol. 1995;485 (Pt 3):699. doi: 10.1113/jphysiol.1995.sp020763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poetsch A, Molday LL, Molday RS. The cGMP-gated channel and related glutamic acid-rich proteins interact with peripherin-2 at the rim region of rod photoreceptor disc membranes. J Biol Chem. 2001;276:48009. doi: 10.1074/jbc.M108941200. [DOI] [PubMed] [Google Scholar]
- Pugh EN. Handbook of biological physics. Amsterdam: Elsevier/North-Holland; 2000. [Google Scholar]
- Rehemtulla A, Warwar R, Kumar R, et al. The basic motif-leucine zipper transcription factor Nrl can positively regulate rhodopsin gene expression. Proc Natl Acad Sci USA. 1996;93:191. doi: 10.1073/pnas.93.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof DJ, Heuser JE. Surfaces of rod photoreceptor disk membranes: integral membrane components. J Cell Biol. 1982;95:487. doi: 10.1083/jcb.95.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal S, De Ruiter A, Hawkins RK. Development and degeneration of retina in rds mutant mice: light microscopy. J Comp Neurol. 1980;194:193. doi: 10.1002/cne.901940110. [DOI] [PubMed] [Google Scholar]
- Sanyal S, Dees C, Zeilmaker GH. Development and degeneration of retina in rds mutant mice: observations in chimaeras of heterozygous mutant and normal genotype. J Embryol Exp Morphol. 1986;98:111. [PubMed] [Google Scholar]
- Sanyal S, Zeilmaker GH. Development and degeneration of retina in rds mutant mice: light and electron microscopic observations in experimental chimaeras. Exp Eye Res. 1984;39:231. doi: 10.1016/0014-4835(84)90011-3. [DOI] [PubMed] [Google Scholar]
- Stipp CS, Kolesnikova TV, Hemler ME. Functional domains in tetraspanin proteins. Trends Biochem Sci. 2003;28:106. doi: 10.1016/S0968-0004(02)00014-2. [DOI] [PubMed] [Google Scholar]
- Swaroop A, Xu JZ, Pawar H, et al. A conserved retina-specific gene encodes a basic motif/leucine zipper domain. Proc Natl Acad Sci USA. 1992;89:266. doi: 10.1073/pnas.89.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]