SUMMARY
Lantibiotics are ribosomally synthesized and posttranslationally modified antimicrobial peptides. The recently discovered lantibiotic epilancin 15X produced by Staphylococcus epidermidis 15X154 contains an unusual N-terminal lactate group. To understand its biosynthesis, the epilancin 15X biosynthetic gene cluster was identified. The N-terminal lactate is produced by dehydration of a Ser residue in the first position of the core peptide by ElxB, followed by proteolytic removal of the leader peptide by ElxP, and hydrolysis of the resulting new N-terminal dehydroalanine. The pyruvate group thus formed is reduced to lactate by an NADPH dependent oxidoreductase designated ElxO. The enzymatic activity of ElxB, ElxP, and ElxO were investigated in vitro or in vivo and the importance of the N-terminal modification for peptide stability against bacterial aminopeptidases was assessed.
INTRODUCTION
Bacterial resistance to known classes of antibacterial agents has been rising at an alarming rate. Currently, 60% to 70% of all Staphylococcus aureus strains isolated in hospitals are multidrug resistant (Taubes, 2008). In order to prevent potential epidemic outbreaks of infectious diseases, new antibacterial drugs not affected by existing resistance mechanisms are much needed. Lantibiotics are ribosomally synthesized and posttranslationally modified polycyclic peptides that contain thioether cross-links and that demonstrate promising activity against pathogenic bacteria (Willey and van der Donk, 2007). Nisin (Figure 1), the most studied lantibiotic, has been used as a food preservative during the last 40 years in more than 80 countries without the development of stable resistance, possibly as a consequence of its multiple modes of action (Breukink and de Kruijff, 2006). The N-terminal portion of nisin recognizes the membrane-bound cell wall precursor lipid II, inhibiting peptidoglycan biosynthesis (Breukink, et al., 1999; Brötz, et al., 1998). Once the molecule is docked to the membrane, the C-terminal portion generates stable pores that result in membrane damage and depolarization (Wiedemann, et al., 2001).
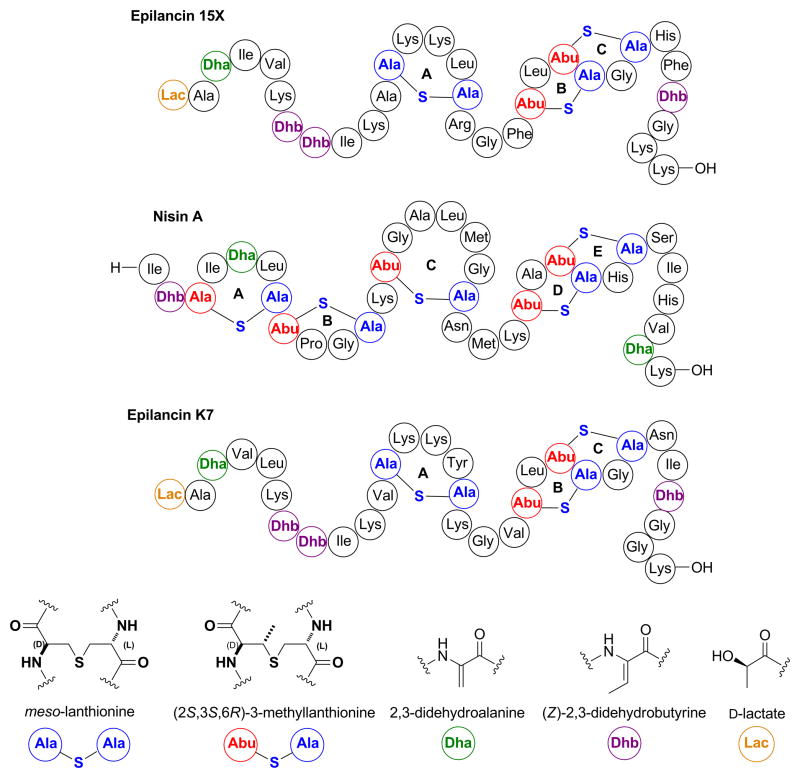
Figure 1.
Structures of the lantibiotics epilancin 15X, nisin A, and epilancin K7 using a previously reported shorthand notation (Chatterjee, et al., 2005). The stereochemical configuration of the N-terminal lactate group in epilancin 15X was determined in this study.
The lantibiotic epilancin 15X (Figure 1) was isolated from Staphylococcus epidermidis 15X154 and is active against several pathogenic bacteria, including methicillin-resistant S. aureus (MRSA) and vancomycin-resistant Enterococci (VRE), with MIC values among the lowest reported for lantibiotics (Ekkelenkamp, et al., 2005; Verhoef, et al., 2005). Epilancin 15X contains one lanthionine (Lan) and two 3-methyllanthionine (MeLan) bridges, one 2,3-dehydroalanine (Dha), three (Z)-2,3-dehydrobutyrine (Dhb) residues, and an unusual N-terminal 2-hydroxypropionyl group (lactate, Lac). The C-terminal B and C rings of epilancin 15X are structurally similar to the D and E rings of nisin A (Figure 1) that are believed to be involved in pore formation (Wiedemann, et al., 2001). However, formation of pores would not explain the low MIC values of epilancin 15X. In the case of nisin, the potent antibacterial activity is a consequence of docking onto lipid II by the A and B rings (Hsu, et al., 2004), which greatly enhances the pore forming ability (Breukink, et al. 1999). However, epilancin 15X does not contain the A and B-rings found in nisin, and lipid II does not appear to be a target of epilancin K7 (Figure 1), a structurally closely related analog of epilancin 15X (van de Kamp, et al., 1995; van de Kamp, et al., 1995). Thus, the N-terminal portion of epilancin 15X and the unusual lactate group may be involved in a currently unknown alternative mechanism. Indeed, additional posttranslational modifications beyond the Lan or MeLan rings are often important for biological activity of lantibiotics. In the case of cinnamycin, a β-hydroxylated aspartate residue and a lysinoalanine ring are important for recognition of its target (Hosoda, et al., 1996). In another example, the lantibiotic microbisporicin, which has a very similar ring topology as epidermin, is two orders of magnitude more potent than the latter compound against several strains of S. aureus, presumably because of hydroxylated proline and chlorinated tryptophan residues that are absent in epidermin (Castiglione, et al., 2008). Thus, the N-terminal lactate group might also be important for the antimicrobial activity of epilancin 15X.
An N-terminal lactate is also present in epicidin 280 and epilancin K7 (van de Kamp, et al., 1995), but its biosynthetic origin has not been determined. In the case of epicidin 280, a putative oxidoreductase EciO may be involved, but this hypothesis has not been confirmed experimentally (Heidrich, et al., 1998). Herein, we report the gene cluster involved in the biosynthesis of epilancin 15X. Additionally, we demonstrate that ElxO is an NADP(H)-dependent alcohol dehydrogenase that catalyzes the conversion of an N-terminal pyruvate to lactate and that the lactate group plays a protective role against proteolytic degradation. We also report the in vitro reconstitution of the enzymatic activity of ElxP, the first such example for a member of the serine-type lantibiotic proteases.
RESULTS
Cloning and sequencing of the epilancin 15X biosynthetic gene cluster
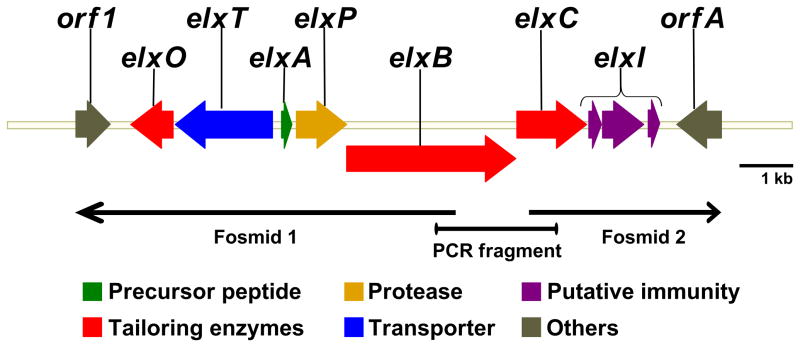
To identify and sequence the epilancin 15X biosynthetic gene cluster, a fosmid library of S. epidermidis 15X154 genomic DNA was constructed in Escherichia coli. The fosmid library was screened by PCR using degenerate primers to amplify a fragment of elxA (the gene encoding the precursor peptide) and elxC (the gene encoding a lanthionine cyclase). The primers were designed based on the amino acid sequence of epilancin 15X and conserved amino acid sequences in the cyclase enzymes. Two positive clones containing non-overlapping DNA fragments were isolated and the fosmids were sequenced using transposon insertions. Specific primers annealing with regions of S. epidermidis 15X154 genomic DNA were then used to amplify by PCR a bridging 1.2 kb DNA fragment that was sequenced to obtain the biosynthetic gene cluster (Figure 2). The open reading frames (ORFs) were analyzed with the Basic Local Alignment Search Tool (BLAST) (Altschul, et al., 1990) and six ORFs encoding putative proteins with high sequence identity to enzymes involved in the production and transport of lantibiotics were identified: elxA, elxB, elxC, elxP, elxT, and elxO (Table 1, Figure 2, Figure S1). Additionally, three genes with no homology to characterized lantibiotic genes and presumably involved in immunity were identified and designated as elxI1, elxI2, and elxI3. With the exception of elxO, the role of the other genes in epilancin 15X biosynthesis can be predicted based on homology to previously characterized lantibiotic genes. The gene encoding the precursor peptide, elxA, encodes a serine residue at the first position of the core peptide (the region of the precursor peptide that is modified to the mature lantibiotic (Oman and van der Donk, 2010)), which corresponds to lactate in epilancin 15X. Thus, Ser1 must be posttranslationally modified by an undetermined enzyme.
Figure 2.
Epilancin 15X gene cluster. For amino acid sequences, see Figure S1.
Table 1.
Open reading frames analysis of the epilancin 15X gene cluster using BLASTp at the NCBI website. See also Figure S5.
| Predicted ORF | Number of AA1 | Protein homology2 | Identity3 | Expect4 |
|---|---|---|---|---|
| elxA | 55 | ElkA, epilancin K7 precursor peptide, S. epidermidis K17, (AAA79236) ( 55 aa) | 38/55 (69%) | 7e-15 |
| elxB | 986 | PepB, Pep5 dehydratase, S. epidermidis 5, (CAA90025) (967 aa) | 327/994 (32%) | 7e-104 |
| elxC | 402 | PepC, Pep5 cyclase, S. epidermidis 5, (CAA90026) (398 aa) | 142/379 (37%) | 8e-56 |
| elxO | 248 | EciO, oxidoreductase, S. epidermidis BN280, (CAA74346) (247 aa) | (126/248) 50% | 8e-64 |
| elxP | 297 | EciP, epicidin 280 protease, S. epidermidis 5, (CAA74349) (300 aa) | 123/286 (43%) | 3e-48 |
| elxT | 573 | PepT, Pep5 ABC transporter, S. epidermidis 5, (CAA90021) (571 aa) | 354/571 (61%) | 0.0 |
| elxI1 | 72 | Hypothetical protein SE2390, S. epidermidis ATCC 12228, (NP_765945) (76 aa) | 43/65 (66%) | 7e-18 |
| elxI2 | 241 | CAAX amino protease, S. epidermidis M23864:W1, (ZP_04817536) (248 aa) | 70/178 (39%) | 8e-24 |
| elxI3 | 71 | Hypothetical protein, S. aureus subsp. aureus USA300_TCH959, (ZP_04865952) (75 aa) | 43/71 (60%) | 6e-17 |
| orf1 | 177 | Recombinase, S. aureus subsp. aureus TCH70, (ACZ58811) (182 aa) | 154/176 (87%) | 4e-85 |
| orfA | 261 | Membrane spanning protein, S. hominis SK119, (ZP_04060547) (257 aa) | 249/257 (96%) | 1e-110 |
AA: amino acid.
Results are from a BLASTp search of the GenBank protein database on January 2010.
Identities in the aligned region.
Expectation value.
Cloning and overexpression of elxO
To study the role of ElxO, the corresponding gene was cloned into a pET- 28b vector to generate pHis6-ElxO that encodes an N-terminal hexahistidine fusion protein (His6-ElxO). His6-ElxO was heterologously produced in E. coli Rosetta 2 cells and the enzyme was purified by immobilized metal ion affinity chromatography (IMAC) with Ni2+, resulting in 60 mg of purified protein per liter of cell culture. The enzyme migrated as a protein of approximately 30 kDa by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis, close to the predicted monomeric molecular weight of His6-ElxO (29.7 kDa). Native molecular weight analysis using gel filtration chromatography showed that His6-ElxO exists as a dimer (59 kDa, observed).
In vitro reconstitution of the enzymatic activity of His6-ElxO and determination of the stereochemistry of the reaction
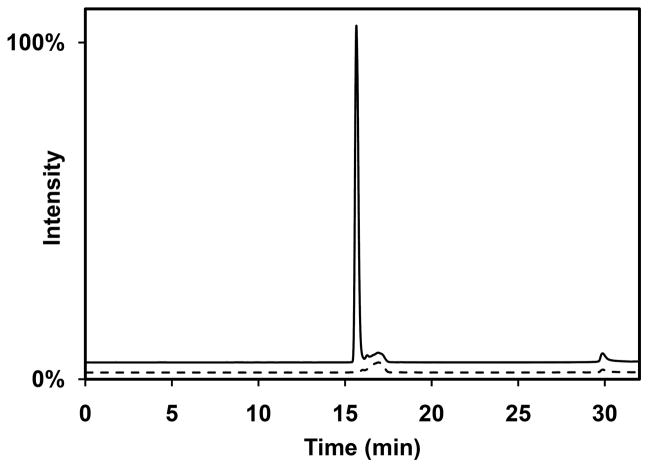
On the basis of its amino acid sequence (Figure S1D), ElxO is a member of the short-chain dehydrogenase/reductase (SDR) superfamily that catalyzes the interconversion of alcohols to aldehydes or ketones using NAD(P)(H) as a cofactor. Dehydro-epilancin 15X, containing an N-terminal pyruvate group, could therefore be the substrate for ElxO. Since dehydro-epilancin 15X was not available, the small peptide AAIVK was synthesized by Fmoc-based solid phase peptide synthesis (SPPS) followed by coupling of pyruvic acid to produce the ketone containing substrate Pyr-AAIVK. This peptide resembles the N-terminal portion of dehydro-epilancin 15X, with Dha at position 3 replaced with Ala for simplicity. Incubation of Pyr-AAIVK with ElxO resulted in a decrease in absorbance at 340 nm over time with NADPH but not NADH. The reaction sample was also analyzed by liquid chromatography mass spectrometry (LC-MS) and a new peak with slightly shorter retention time and with m/z 573.4, corresponding to Lac-AAIVK, was observed (Figure 3).
Figure 3.
Enzymatic assay of His6-ElxO with Pyr-AAIVK. Shown are single ion chromatograms at m/z = 573.4, corresponding to the expected [M+H]+ ion for Lac-AAIVK, for the reaction mixture (solid line) and a control sample (dashed line). The peak at 16 min observed in the reaction sample but not in the control confirms the enzymatic reduction of the peptide. The peak at 17 min in the control sample is derived from higher molecular weight isotopologues of the substrate (monoisotopic m/z = 571.4). See also Figure S2.
To determine the stereochemical configuration of the N-terminal Lac, the two possible reaction products (D-Lac-AAIVK and L-Lac-AAIVK) were synthesized by Fmoc-based SPPS using D- or L-lactic acid during the last coupling step. The enzymatic product of Pyr-AAIVK, after incubation with His6-ElxO and NADPH, was combined with D-Lac-AAIVK or L-Lac-AAIVK and analyzed by high performance liquid chromatography (HPLC) (Figure S2A-D). The enzymatic product of His6-ElxO co-eluted with D-Lac-AAIVK, but not with L-Lac-AAIVK, demonstrating that ElxO catalyzes the formation of an N-terminal D-lactate ((R)-2-hydroxypropionate).
Production of epilancin 15X and stereochemical characterization of the N-terminal Lac group
The production of lanthionine-containing polypeptide antibiotics by staphylococci is highly dependent on the composition of the media (Horner, et al., 1990; Horner, et al., 1989). To identify optimal conditions for the production of epilancin 15X, a set of cultures were grown in which the concentrations of meat extract, NaCl, and NH4Cl were systematically varied. Analysis of culture supernatants for bioactivity using an agar diffusion assay with Staphylococcus carnosus TM300 as indicator strain demonstrated that a medium containing 10% Lab-Lemco meat extract, 2% NaCl, 20 mM NH4Cl, 3% malt extract, and 0.4% Ca(OH)2 produced the highest concentration of epilancin 15X. Purification yielded about 3.0 mg of bacteriocin per liter of culture, compared with a yield of 0.5 mg per liter reported previously (Ekkelenkamp, et al., 2005).
To determine the stereochemical configuration of the N-terminal Lac in epilancin 15X, a sample of purified lantibiotic was treated with trypsin generating Lac-ADhaIVK, among other peptide fragments. The resulting peptide mixture and synthetic samples of D-Lac-ADhaIVK and L-Lac-ADhaIVK, produced by solid phase peptide synthesis, were analyzed by LC-MS (Figure S2E-H). The N-terminal proteolytic fragment of epilancin 15X co-eluted with D-Lac-ADhaIVK, confirming the stereochemical configuration of the N-terminal Lac in epilancin 15X.
Cloning and overexpression of elxP
The gene elxP was cloned initially into a pET28b vector to generate an N-terminal hexahistidine fusion of ElxP (His6-ElxP). However, attempts to overexpress the protein in E. coli were unsuccessful, since the rate of growth of the host was greatly reduced after induction with IPTG, suggesting that ElxP is toxic to the heterologous host. To overcome toxicity problems and improve solubility, elxP was cloned into pHis6-MPB-ElxP that encodes for a fusion protein containing an N-terminal pelB signal, followed by a hexahistidine tag for purification, and a maltose binding protein (MBP) tag for solubility, separated from ElxP by a Tobacco Etch Virus (TEV) protease cleavage site (Figure S3A). Heterologous expression trials in E. coli Rosetta 2 by using the plating method (Suter-Crazzolara and Unsicker, 1995) afforded successful production of His6-MBP-ElxP. The enzyme was purified by IMAC with Ni2+, resulting in about 9 mg of purified protein per liter of cell culture. After treatment of His6-MBP-ElxP with TEV protease and SDS-PAGE analysis, protein bands at about 34 kDa and 45 kDa, corresponding to ElxP (predicted mass 34.3 kDa) and His6-MBP (predicted mass of 45.5 kDa), respectively, were observed.
In vitro reconstitution of ElxP
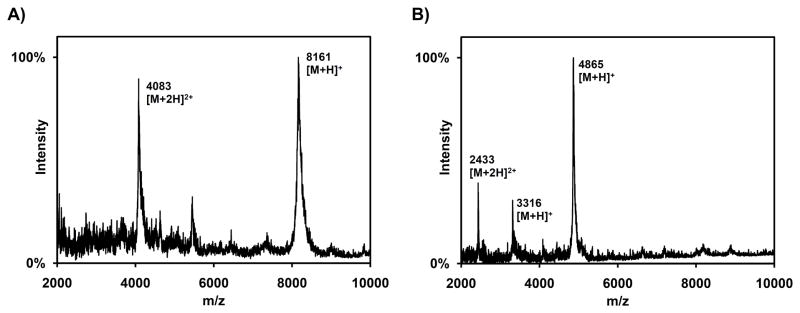
Based on its amino acid sequence (Figure S1C), ElxP is a serine protease that may cleave the leader peptide from the fully cyclized precursor peptide. Since modified ElxA was not available, linear His6-ElxA was tested as substrate, obtained by expression from a pET-28b vector in E. coli Rosetta 2 cells and purification by IMAC with Ni2+ and subsequent HPLC. Linear His6-ElxA was incubated with ElxP or His6-MBP-ElxP at pH 7.5. The reaction products were analyzed by MALDI-TOF MS, confirming the formation of two proteolytic products in both cases (Figure 4). The peak at m/z = 4865 corresponds to the N-terminal hexahistidine tagged leader peptide (residues −1 to −43) resulting from amide bond hydrolysis at the predicted Gln( −1) Ser(1) cleavage site, whereas the peak at m/z = 3316 corresponds to the C-terminal unmodified core peptide.
Figure 4.
MALDI-TOF MS analysis of proteolytic digestion of His6-ElxA by His6-MBP-ElxP. Similar results were obtained when ElxP was used. A) Analysis of His6-ElxA (calculated mass = 8162.3 Da) after incubation without enzyme in reaction buffer. B) Analysis of peptide mixture after incubation with enzyme under the same conditions. Peaks corresponding to the leader peptide (calculated mass = 4864.3 Da) and to the core peptide (calculated mass = 3316.0 Da) are observed. See also Figure S3.
Cloning and coexpression of elxA and elxB
Based on bioinformatic analysis (Table 1), ElxB catalyzes the dehydration of the precursor peptide ElxA. To confirm the role of ElxB in epilancin 15X biosynthesis, the genes elxA and a synthetic codon optimized version of elxB were cloned into a pRSFDuet-1 vector, as previously described for nisin (Shi, et al., 2011), to generate pHis6-ElxA.ElxB that encodes for an N-terminal hexahistidine fusion of ElxA (His6-ElxA) and for untagged ElxB. Upon coexpression in E. coli BL21(DE3), followed by peptide purification and cleavage of the leader region with His6-MBP-ElxP, a mixture of partially dehydrated peptides was observed by MALDI-TOF MS (Figure S3B). Attempts to obtain fully dehydrated peptide or to reconstitute the enzymatic activity of ElxB in vitro were not successful.
Incubation of peptides with an aminopeptidase
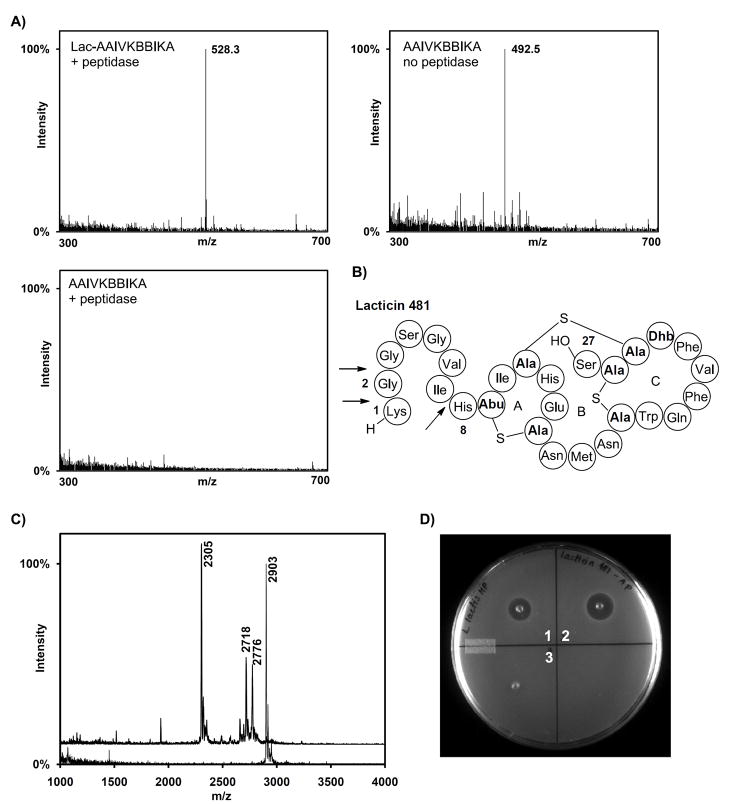
A sample of epilancin 15X was incubated with a commercially available aminopeptidase from Aeromonas proteolytica. Interestingly, no proteolysis products were observed by MALDI-TOF MS, suggesting that the lactate group protects the lantibiotic against degradation (Figure S4). To further evaluate the possibility of the lactate group conferring stability against aminopeptidases, the peptide Lac-AAIVKBBIKA (where B stands for L-2-aminobutyric acid) mimicking the N-terminal portion of dehydro-epilancin 15X and the corresponding peptide without the Lac group were generated. The two peptides were incubated with the aminopeptidase from A. proteolytica, followed by analysis by electrospray ionization mass spectrometry (ESI-MS) (Figure 5A). Whereas the peptide lacking the lactate group was completely degraded, the peptide containing the N-terminal lactate was detected intact.
Figure 5.
Degradation of peptides by A. proteolytica aminopeptidase. A) ESI-MS analysis of the peptides Lac-AAIVKBBIKA (top left), and AAIVKBBIKA (bottom) after incubation with A. proteolytica aminopeptidase. Only the peptide lacking the Lac group was completely degraded by the aminopeptidase (bottom right) compared with a control sample lacking enzyme (top right). The observed m/z peaks correspond to the [M+2H]2+ ions. B) Structure of the lantibiotic lacticin 481, using the shorthand notation described in Figure 1. C) MALDI-TOF MS analysis of lacticin 481 (calculated m/z = 2902) after incubation with A. proteolytica aminopeptidase (top spectrum) and a control sample lacking peptidase (bottom spectrum). Peaks corresponding to lacticin(8–27) (calculated m/z = 2304), lacticin(2–27) (calculated m/z = 2774) and lacticin(3–27) (calculated m/z = 2717) were observed. D) Agar diffusion bioactivity assay of lacticin 481 after treatment with the peptidase (spot 1) and control sample without peptidase (spot 2) using L. lactis subsp. cremoris HP as indicator strain. A control sample containing the peptidase but no lantibiotic was also tested (spot 3). See also Figure S4.
In contrast, when lacticin 481 (Figure 5B), a lantibiotic lacking N-terminal modifications, was incubated with the aminopeptidase under similar conditions, a peptide missing the first seven N-terminal residues was obtained as the main product (Figure 5C). Thus, the aminopeptidase was able to remove N-terminal unmodified amino acids with the exception of the His preceding the first MeLan ring that protects the lantibiotic from additional proteolysis. Lacticin 481(2–27) and lacticin 481(3–27) were also observed, suggesting that the GG motif at position 2 is processed at a lower rate and may partially protect the bacteriocin against the action of aminopeptidases. The proteolyzed lacticin 481 and control samples were tested by agar diffusion assays using the indicator strain Lactococcus lactis subsp. cremoris HP (Figure 5D), demonstrating a decrease in antibacterial activity of the proteolyzed peptides. These results are consistent with previous reports that have shown that the linear N-terminal portion of lacticin 481 is important for bioactivity (Levengood, et al., 2009; Uguen, et al., 2005).
DISCUSSION
Lantibiotics are produced from ribosomally synthesized linear precursor peptides that consist of an N-terminal leader region and a C-terminal core peptide. The mature lantibiotic is generated from the core peptide after several posttranslational modifications. The reactions involved in the formation of the characteristic Lan and MeLan rings formation have been investigated (Goto, et al., 2010; Li, et al., 2006; Xie, et al., 2004), but only a few enzymes introducing other posttranslational modifications have been studied (Kupke, et al., 1994; Majer, et al., 2002). This investigation focused on the mechanism of formation of N-terminal lactate groups and on the enzymatic cleavage of the leader peptide.
The epilancin 15X gene cluster contains five genes involved in biosynthesis (elxABCOP), one gene involved in the export of the mature peptide (elxT), and three genes potentially involved in immunity (elxI1, elxI2, and elxI3) (Table 1 and Figure 2). The cluster organization resembles that of the lantibiotics Pep5 (Meyer, et al., 1995) and epicidin 280 (Heidrich, et al., 1998) produced by different strains of S. epidermidis, suggesting that these clusters have evolved from a common ancestor. The predicted peptide ElxA has high amino acid sequence similarity to the epilancin K7 precursor peptide ElkA (van de Kamp, et al., 1995) (Figure S1A). ElxA contains an N-terminal leader sequence of 24 amino acids and a C-terminal core peptide region comprised of 31 amino acids, as predicted from the chemical structure of epilancin 15X. The leader region also contains the conserved motif F-(N/D)-L-(N/D/E) and a Pro at position −2 that are characteristic of class I lantibiotics (Figure S1A) (van der Meer, et al., 1994).
Downstream of elxA, an ORF designated as elxP was identified. The encoded protein ElxP possesses high amino acid sequence similarity to EciP, the protease involved in the biosynthesis of epicidin 280 (Heidrich, et al., 1998), and to other subtilisin-like serine proteases. Importantly, the residues of the predicted catalytic triad and oxyanion hole (Asp27, His62, Ser240, and Asn154) are conserved in ElxP (Figure S1C). The lack of an N-terminal sec-signal sequence and a C-terminal cell wall anchor sequence (LPXTG) suggests that ElxP is localized inside the cytoplasm, possibly as part of a membrane-bound biosynthetic complex. Thus, ElxP likely removes the leader peptide before the mature peptide is transported outside the cell, in contrast to other class I lantibiotic proteases, such as NisP or EpiP, that are located extracellularly and remove the leader region once the peptide has been secreted (Figure S1C).
In a previous attempt to study LanP proteases, an E. coli host carrying a plasmid encoding for NisP was able to express the protease at low concentrations based on SDS-PAGE analysis using 35S-Met (van der Meer, et al., 1993). Although NisP was not purified, the E. coli cell extracts cleaved the nisin A cyclized precursor peptide, producing a biologically active compound (van der Meer, et al., 1993). Additional in vivo studies indicated that NisP is able to cleave the leader region only from fully processed precursor peptide, but not from uncyclized dehydrated or unmodified NisA (Kuipers, et al., 2004). Similar results were obtained from in vivo studies of the lantibiotic Pep5 (Meyer, et al., 1995). In contrast, culture supernatants of S. carnosus TM300 expressing EpiP processed unmodified EpiA to the expected proteolytic products (Geissler, et al., 1996). In the present study, ElxP was successfully expressed in E. coli and its enzymatic activity was reconstituted in vitro. The protease was able to process unmodified His6-ElxA, indicating that neither the Lan/MeLan ring nor the dehydrated residues, including Dha at position 1, are strictly required for enzyme recognition and proteolytic processing.
Downstream of elxP, an ORF designated as elxB encodes a protein with homology to PepB, the enzyme that catalyzes the dehydration of Ser or Thr residues in the Pep5 precursor peptide (Meyer, et al., 1995). Analysis of ElxB with SignalP 3.0 (Emanuelsson, et al., 2007) suggests that this protein may contain an N-terminal cell membrane anchor signal. The activity of ElxB homologs of class I lantibiotics has never been reconstituted in vitro (Xie, et al., 2002) and the cofactors or metals involved in catalysis are currently unknown. In this work, we were also unable to reconstitute ElxB activity in vitro. However, coexpression of His6-ElxA and ElxB in E. coli produced a partially dehydrated peptide confirming the role of the protein in the biosynthesis of epilancin 15X. Closer inspection of the ElxB sequence indicates that it contains an almost conserved Walker A motif (GXXXXGKT/S: GLLENWKT) and a conserved Walker B motif (hhhhD: IIFPD, where h stands for hydrophobic residue) (Figure S1B). In addition, the three potential binding sites characteristic of GTP binding proteins are also present (DXXG: DFLG, NKXG: NTID/NDID/NLND/NRND, SAX: SAT) (Kjeldgaard, et al., 1996) suggesting that GTP or another nucleotide may be required for dehydration by LanB proteins, similar to class II–IV lanthionine synthetases (Chatterjee, et al., 2005; Goto, et al., 2010; Müller, et al., 2010; Xie, et al., 2004). The ORF elxC encodes a protein with high amino acid sequence similarity to PepC, the cyclase responsible for Lan and MeLan ring formation in Pep5 (Meyer, et al., 1995). Additionally, ElxC contains the conserved residues comprising a zinc ion binding site (Cys269, Cys318, and His319) and the residues involved in acid-base catalysis (His205 and Asp142) that are characteristic of this family of proteins (Li and van der Donk, 2007; Li, et al., 2006).
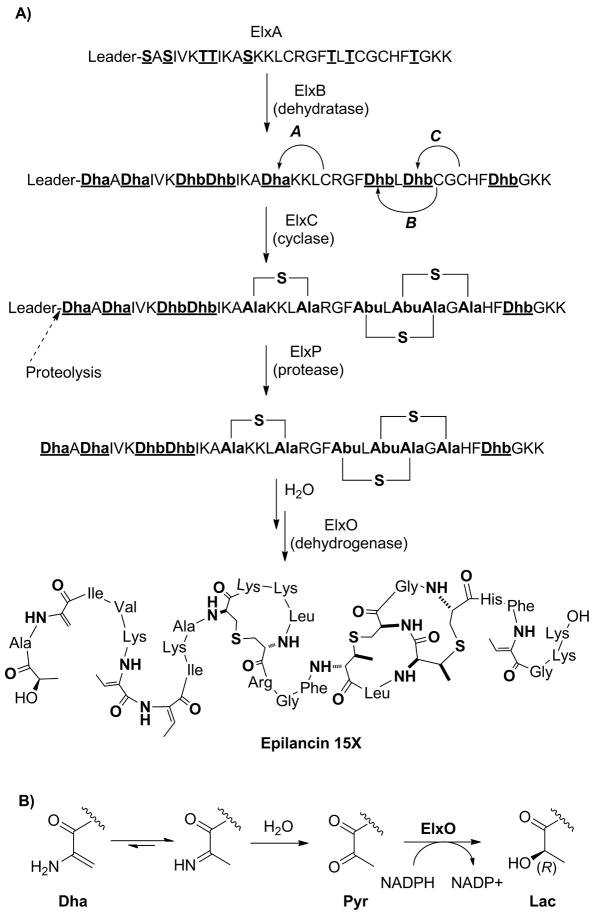
Based on the experimental results and on bioinformatic analysis, the first steps of epilancin 15X biosynthesis can be postulated (Figure 6). The precursor peptide ElxA is modified by the dehydratase ElxB and the cyclase ElxC to produce the crosslinked peptide. Then, the leader peptide is removed by the protease ElxP producing an N-terminal enamine (dehydroalanine, Dha) present in equilibrium with the corresponding imine that can be hydrolyzed to produce dehydro-epilancin 15X. Although enamine hydrolysis is fast (Sollenberger and Martin, 1970), enzymatic assistance (e.g. by ElxP) cannot be ruled out at present. The reduction of the N-terminal ketone to the respective alcohol will complete the synthesis of mature epilancin 15X.
Figure 6.
Proposed biosynthetic steps involved in the production of epilancin 15X and its N-terminal D-lactate group. Note that it is not known if dehydration is completed before cyclization commences.
The ORF elxO encodes a protein with homology to EciO, an oxidoreductase hypothesized to be involved in the reduction of pyruvate to lactate in the biosynthesis of epicidin 280 (Heidrich, et al., 1998). ElxO contains a predicted N-terminal NAD(P)(H) binding site (GXXGXG: GGFKGIGK) and the catalytic triad residues (Ser139, Tyr152, and Lys156) of the short-chain dehydrogenase/reductase (SDR) protein superfamily (Figure S1D) (Jörnvall, et al., 1995; Tanaka, et al., 2001). The absence of the ‘proximal Asp residue’ (Ser33 in ElxO) and the presence of the ‘proximal basic residues’ (Lys12 and Arg34), responsible for cofactor specificity, correctly predicted that ElxO is an NADPH-dependent enzyme and that it belongs to the cP3 subfamily (Kallberg, et al., 2002; Tanaka, et al., 2001). The stereochemical course of the reaction established here allows assignment of the configuration of the N-terminal lactate group of epilancin 15X as (R). Because His6-ElxO was able to reduce a hexamer peptide, the thioether rings or other structural motifs in dehydro-epilancin 15X are not required for enzyme recognition.
The ORF elxT encodes a putative protein with homology to PepT (Meyer, et al., 1995). The C-terminal domain of ElxT contains an ATP-binding site characterized by the conserved Walker A motif (GXXXXGKT/S: GPSGAGKT) or P-loop, the Walker B motif (hhhhD: ILLLD), and the C motif or “signature” motif (LSGGQ) specific to ABC transporters (Figure S1E). Additionally, analysis of ElxT with TMHMM 2.0 (Emanuelsson, et al., 2007) indicates that its N-terminal portion contains six transmembrane helices. Thus, ElxT is likely involved in lantibiotic secretion.
Epilancin 15X has potent activity against staphylococci, including strains of S. epidermidis (Verhoef, et al., 2005), suggesting that the producer strain must have an effective self-resistance mechanism. Such immunity is particularly important for epilancin 15X because it is activated by leader peptide removal within the cytoplasm unlike most lantibiotics for which leader peptide cleavage occurs after or concomitant with secretion. Downstream of elxC, three ORFs elxI1, elxI2, and elxI3 were identified. The genes elxI1 and elxI3 potentially encode for 72 aa and 71 aa paralog peptides (47% identity) with no sequence homology to previously characterized proteins. Analysis by TMHMM 2.0 (Emanuelsson, et al., 2007) indicates that ElxI1 and ElxI3 contain two highly hydrophobic α-helical domains followed by strongly hydrophilic C-terminal segments (Figure S5). Thus, although ElxI1 and ElxI3 contain no signal peptides, these small proteins may be localized at the cytoplasmic membrane. A similar pattern of helical domains followed by a hydrophilic region is found in small proteins encoded by the gene clusters of the closely related lantibiotics Pep5 (Hoffmann, et al., 2004) and epicidin 280 (Heidrich, et al., 1998), the structurally unrelated lactosin S (Skaugen, et al., 1997), the nonlantibiotic bacteriocin divergicin A (Worobo, et al., 1995), and the circular bacteriocins AS-48, acidocin B, butyrivibriocin AR10, and circularin A (Maqueda, et al., 2008). In the case of Pep5, the protein was designated PepI and was shown to be a determinant for self-immunity of the producer strain (Reis, et al., 1994). Interestingly, in all of the lantibiotics mentioned above, intracellular peptidases remove the leader peptide and the mature bacteriocin is produced inside the cell. PepI has been suggested to bind to a (currently unknown) target molecule, avoiding docking of the lantibiotic Pep5 onto the target (Hoffmann, et al., 2004). Despite the absence of significant sequence homology between PepI and ElxI1/3, the topological similarity suggests that these peptides may protect the host organism in a similar fashion (Figure S5).
Finally, the putative immunity protein ElxI2 has high sequence similarity to Abi proteins, membrane-bound metalloproteases that are involved in self-immunity to plantaricin EF and JK, sakacin 23K, or streptolysin S (Datta, et al., 2005; Kjos, et al., 2010). ElxI2 is predicted to contain seven transmembrane domains, including the final four α-helices that form the Abi domain (Figure S1F). Three highly conserved motifs (EEXXXR: EEILYR, FXXXH: FSLIH, and His226) likely constitute the active site of the protease (Pei and Grishin, 2001). Thus, ElxI2 may protect the host against the bacteriocin by direct degradation of the peptide.
Whether elxI1, elxI2, and elxI3 are part of the cluster and are involved in the immunity mechanism is at present not certain. Closer analysis of the non-coding sequences upstream of elxC, elxI1, and elxI2 suggests the presence of only one relevant inverted repeat between elxC and elxI1, partially overlapping elxC, and with a calculated free energy of −10.2 kcal/mol. This repeat may work as a weak rho-independent transcriptional terminator that allows partial read-through, indicating that elxC and elxI1–3 may be part of a single operon and the same gene cluster. Downstream of elxI3, a non-coding region of 329 bp is followed by the ORF orfA in a different operon that encodes a putative ABC transporter with no sequence homology to any known lantibiotic proteins, but with significant homology to transporters from staphylococci. Thus, OrfA is not likely to be related to epilancin 15X biosynthesis. Flanking the putative epilancin 15X gene cluster on the other side, an ORF designated orf1 was identified. Orf1 is a putative recombinase, not likely to be involved in epilancin 15X biosynthesis, transport, or immunity. Thus, the epilancin 15X cluster likely spans a 9.2 kb region in the S. epidermidis 15X154 genome and includes the genes from elxO to elxI3 (Figure 2).
The role of the N-terminal Lac in epilancin 15X is currently unknown. However, N-terminal modifications are common in lantibiotics and include (methyl)lanthionines, disulfides, pyruvate and lactate groups, 2-oxobutyrate groups, and acylations. The N-terminal disulfide in Halα (one of the two peptides in haloduracin) was shown not to be important for antimicrobial activity, but to protect the peptide from exoproteases (Cooper, et al., 2008; McClerren, et al., 2006). Similarly, the N-terminal lanthionine in lacticin 3147 A1 is not required for antimicrobial activity (Cotter, et al., 2006), but may protect lacticin 3147 A1 from proteolysis. A similar role for the N-terminal lactate group in epilancin 15X is supported in this study.
SIGNIFICANCE
The recently discovered lantibiotic epilancin 15X produced by S. epidermidis 15X154 has potent antimicrobial activity against drug-resistant strains of S. aureus. Epilancin 15X is structurally simple compared with other lantibiotics and yet is very active. The compound contains an unusual N-terminal D-lactate group that could be essential for biological activity. This study demonstrates that this moiety confers stability against proteolytic degradation by aminopeptidases, a feature that may be applied for the engineering of novel lantibiotics with enhanced antibacterial activities or different spectra of action. Furthermore, the gene cluster for epilancin 15X was determined and the enzymatic activity of the dehydratase, protease, and oxidoreductase involved in the biosynthesis were demonstrated in vitro or in vivo.
EXPERIMENTAL PROCEDURES
Genomic library construction, screening, and DNA sequencing
For all primer sequences and microorganisms, see the Supplemental Information (Table S1 and S2). Genomic DNA was obtained as described in the Supplemental Information and partially digested with Sau3A1 (New England Biolabs) in the presence of RNAse A (Sigma-Aldrich) using serial dilutions of the enzyme. The DNA solutions were analyzed by field inversion gel electrophoresis on a 1% agarose/TBE gel and the fraction containing ~20–60 kb DNA fragments was treated with shrimp alkaline phosphatase (Roche Diagnostics).
Cosmid pJK050 was digested with NheI (New England Biolabs), treated with shrimp alkaline phosphatase (Roche Diagnostics), and purified. The cosmid was further treated with BamHI (Invitrogen). Digested pJK050 and genomic DNA were ligated with T4 DNA ligase (New England Biolabs) and cosmid constructs were packaged into lambda phage with a MaxPlax Lambda Packaging Extract Kit (Epicentre) according to the manufacturer’s protocol, followed by transfection of E. coli WM4489. Library clones were screened by PCR for the elxA gene using forward primer elxA-F1 and reverse primer elxA-R2, or for the elxC gene with forward primer elxC-596F and reverse primer elxC-781R, Taq polymerase (Invitrogen), and 1x PCR premix A (Epicentre).
Plasmid pAE5 was digested with BglII and the 1084 bp fragment to be used as a transposon was gel purified. For each positive fosmid, the fragment and the positive fosmid were mixed and treated with MuA transposase (Finnzymes) and the dialyzed DNA was used to transform E. coli WM4489. Sets of 192 clones were sequenced from the ends of the transposon insert using primers seqaetf and seqaetr at the Keck Center for Comparative and Functional Genomics at the University of Illinois at Urbana-Champaign. Using S. epidermidis 15X154 as template, the specific primers elxBgapF and elxCgapR were used to amplify and sequence a DNA fragment of 1.2 kb, closing the sequence gap between the fosmids.
Construction of plasmids pHis6-ElxO, pHis6-ElxA, and pHis6-MBP-ElxP
The gene elxO was amplified by PCR from S. epidermidis 15X154 genomic DNA using a forward primer containing an NheI restriction site (elxO.NheI.F) and a reverse primer containing a XhoI site (elxO.XhoI.R). The PCR product and the vector pET28b (Novagen) were digested with restriction endonucleases NheI and XhoI (Invitrogen) and ligated using T4 DNA ligase (New England Biolabs) to produce the plasmid pHis6-ElxO.
The gene elxA was amplified by PCR from S. epidermidis 15X154 genomic DNA using a forward primer containing an NdeI restriction site (elxA.NdeI.F) and a reverse primer containing a EcoRI site (elxA.EcoRI.R). The PCR product and the vector pET28b (Novagen) were digested with restriction endonucleases NdeI and EcoRI (Invitrogen) and ligated using T4 DNA ligase (New England Biolabs) to produce the plasmid pHis6-ElxA.
The gene elxP was amplified by PCR from S. epidermidis 15X154 genomic DNA using a forward primer (elxP.28MBP.F) and a reverse primer (elxP.28MBP.R). The PCR product contained annealing regions to a modified pET28b vector (Novagen) that encodes for a fusion pelB, hexahistidine tagged MBP. The purified PCR product was used as a primer to amplify the vector using Phusion® Hot Start DNA polymerase (New England Biolabs), followed by treatment with DpnI (New England Biolabs) before transformation of E. coli DH5α cells to generate the plasmid pHis6-MBP-ElxP that encodes for ElxP fused at its N-terminus to a TEV protease cleavage site, a MBP tag, a hexahistidine tag, and a pelB signal peptide (Figure S3A). The correct sequences of the inserts were confirmed by sequencing at the Keck Center at the University of Illinois at Urbana-Champaign. E. coli BL21 (DE3) Rosetta 2 cells were transformed with pHis6-ElxO, pHis6-ElxA, or pHis6-MBP-ElxP and the proteins were overexpressed and purified by IMAC (Supplemental Information).
His6-ElxO and ElxP activity assays
His6-ElxO (10 μM) and the peptide Pyr-AAIVK (1 mM) generated by Fmoc-based SPPS (Supplemental Information) were incubated with NADPH (1 mM) in assay buffer (100 mM HEPES, pH 7.5) at room temperature for 6 h. Reaction progress was monitored by UV-vis spectrophotometry, measuring the disappearance of the NADPH peak at 340 nm. Formation of reduced peptide was confirmed by LC-MS using an Agilent 1200 instrument equipped with a single quadrupole multimode ESI/APCI ion source mass spectrometry detector (Agilent) and a Synergi Fusion-RP column (4.6 mm i.d. × 150 mm L, Phenomenex). ElxP or His6-MPB-ElxP (5 μM) and purified His6-ElxA (Li, et al., 2009) (50 μM) were incubated in the presence of an assay buffer (50 mM HEPES, 500 mM NaCl, pH 7.5) at room temperature for 2–3 h. A control sample lacking enzyme was incubated under the same conditions. Cleavage of the peptide in the reaction sample was confirmed by MALDI-TOF MS as described above.
Production and purification of epilancin 15X
A medium composed of Lab-Lemco meat extract (10%, Oxoid), malt extract (3%, BD), ammonium chloride (20 mM), Ca(OH)2 (0.4%), and NaCl (2%) was inoculated with an overnight pre-culture of S. epidermidis 15X154 in LB broth (1/100 dilution). Cells were incubated at 37 °C with shaking for 12 h and harvested by centrifugation. The supernatant was filtered through a 0.22 μm pore size filter and heated at 80 °C for 1 h to deactivate proteases. Solid (NH4)2SO4 was added to the culture supernatant to reach 80% saturation at 4 °C and stirred for 4 h followed by centrifugation. The pellet was resuspended in water and loaded onto a Vydac® C4 reverse phase solid phase extraction column (214SPE1000, Discovery Sciences). The column was washed with 25% acetonitrile in ammonium acetate buffer (20 mM, pH 5.0) to remove impurities and the peptide was eluted with 80% methanol in 0.1% trifluoroacetic acid (TFA)/water. The lantibiotic was further purified by HPLC using an Agilent 1200 instrument (Agilent) equipped with a Vydac® 214TP54 C4 reverse phase column (4.6 mm i.d. × 250 mm L, Discovery Sciences). A gradient of 50–60% B (0.1% TFA in methanol) in A (0.1% TFA in water) over 50 min was used. The fractions corresponding to the major peak were collected. Analysis of the final product by ESI MS on a Waters Synapt instrument showed a mass of 3172.6817 Da (calculated monoisotopic mass of epilancin 15X: 3172.6977 Da).
Incubation of peptides with A. proteolytica aminopeptidase
Epilancin 15X was incubated with A. proteolytica aminopeptidase (10 U/mL, Sigma-Aldrich) in assay buffer (40 mM Tris-HCl, pH 8.0) at room temperature for 24 h. Samples containing purified lacticin 481 with or without protease were incubated under the same conditions for 12 h. The reaction products were analyzed by MALDI-TOF or ESI-MS MS and tested by agar diffusion bioactivity assays using M17 agar medium supplemented with 0.5% glucose and L. lactis subsp. cremoris HP as indicator strain.
The peptide Lac-AAIVKBBIKA, generated from Pyr-AAIVKBBIKA enzymatically with His6-ElxO as described above, and the control peptide AAIVKBBIKA were also incubated with the peptidase under similar conditions for 6 h. The peptides were then purified by solid phase extraction using Discovery® DSC-18 columns (1 mL, 50 mg, Supelco) and 0.1% formic acid in water or 0.1% formic acid in 60% acetonitrile as washing and eluting solvents, respectively, and analyzed by ESI-MS using a ZMD quadrupole instrument (Waters) at the Mass Spectrometry Laboratory of the University of Illinois at Urbana-Champaign.
Supplementary Material
HIGHLIGHTS.
The gene cluster and biosynthetic pathway of epilancin 15X were identified
The alcohol dehydrogenase ElxO catalyzes formation of the N-terminal D-lactate
In vitro ElxP cleaves the leader region from an unmodified precursor peptide
The N-terminal lactate group confers proteolytic stability against aminopeptidases
Acknowledgments
We thank Prof. Bill Metcalf and Mr. Jun Kai Zhang for helpful advice on molecular genetics and for the strains and plasmids used for generating the genomic library. We also thank Ms. Laura Guest at the Core DNA sequencing facility, Ms. Neha Garg for help in the screening of the genomic library, and Ms. Isabel Neacato for help in cloning. This work was supported by a grant from the National Institutes of Health (GM58822 to WAV). JEV was supported by the National Institutes of Health under Ruth L. Kirschstein National Research Service Award T32 GM070421.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Breukink E, de Kruijff B. Lipid II as a target for antibiotics. Nat Rev Drug Discov. 2006;5:321–332. doi: 10.1038/nrd2004. [DOI] [PubMed] [Google Scholar]
- Breukink E, Wiedemann I, van Kraaij C, Kuipers OP, Sahl H, de Kruijff B. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science. 1999;286:2361–2364. doi: 10.1126/science.286.5448.2361. [DOI] [PubMed] [Google Scholar]
- Brötz H, Josten M, Wiedemann I, Schneider U, Götz F, Bierbaum G, Sahl HG. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol Microbiol. 1998;30:317–327. doi: 10.1046/j.1365-2958.1998.01065.x. [DOI] [PubMed] [Google Scholar]
- Castiglione F, Lazzarini A, Carrano L, Corti E, Ciciliato I, Gastaldo L, Candiani P, Losi D, Marinelli F, Selva E, et al. Determining the structure and mode of action of microbisporicin, a potent lantibiotic active against multiresistant pathogens. Chem Biol. 2008;15:22–31. doi: 10.1016/j.chembiol.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Cooper LE, McClerren AL, Chary A, van der Donk WA. Structure-activity relationship studies of the two-component lantibiotic haloduracin. Chem Biol. 2008;15:1035–1045. doi: 10.1016/j.chembiol.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PD, Deegan LH, Lawton EM, Draper LA, O’Connor PM, Hill C, Ross RP. Complete alanine scanning of the two-component lantibiotic lacticin 3147: generating a blueprint for rational drug design. Mol Microbiol. 2006;62:735–747. doi: 10.1111/j.1365-2958.2006.05398.x. [DOI] [PubMed] [Google Scholar]
- Chatterjee C, Miller LM, Leung YL, Xie L, Yi M, Kelleher NL, van der Donk WA. Lacticin 481 synthetase phosphorylates its substrate during lantibiotic production. J Am Chem Soc. 2005;127:15332–15333. doi: 10.1021/ja0543043. [DOI] [PubMed] [Google Scholar]
- Chatterjee C, Paul M, Xie L, van der Donk WA. Biosynthesis and mode of action of lantibiotics. Chem Rev. 2005;105:633–684. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- Datta V, Myskowski SM, Kwinn LA, Chiem DN, Varki N, Kansal RG, Kotb M, Nizet V. Mutational analysis of the group A streptococcal operon encoding streptolysin S and its virulence role in invasive infection. Mol Microbiol. 2005;56:681–695. doi: 10.1111/j.1365-2958.2005.04583.x. [DOI] [PubMed] [Google Scholar]
- Ekkelenkamp MB, Hanssen M, Danny Hsu ST, de Jong A, Milatovic D, Verhoef J, van Nuland NA. Isolation and structural characterization of epilancin 15X, a novel lantibiotic from a clinical strain of Staphylococcus epidermidis. FEBS Lett. 2005;579:1917–1922. doi: 10.1016/j.febslet.2005.01.083. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Geissler S, Gotz F, Kupke T. Serine protease EpiP from Staphylococcus epidermidis catalyzes the processing of the epidermin precursor peptide. J Bacteriol. 1996;178:284–288. doi: 10.1128/jb.178.1.284-288.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Li B, Claesen J, Shi Y, Bibb MJ, van der Donk WA. Discovery of unique lanthionine synthetases reveals new mechanistic and evolutionary insights. PLoS Biol. 2010;8:e1000339. doi: 10.1371/journal.pbio.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich C, Pag U, Josten M, Metzger J, Jack RW, Bierbaum G, Jung G, Sahl HG. Isolation, characterization, and heterologous expression of the novel lantibiotic epicidin 280 and analysis of its biosynthetic gene cluster. Appl Environ Microbiol. 1998;64:3140–3146. doi: 10.1128/aem.64.9.3140-3146.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Schneider T, Pag U, Sahl HG. Localization and functional analysis of PepI, the immunity peptide of Pep5-producing Staphylococcus epidermidis strain 5. Appl Environ Microb. 2004;70:3263–3271. doi: 10.1128/AEM.70.6.3263-3271.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner T, Ungermann V, Zahner H, Fiedler HP, Utz R, Kellner R, Jung G. Comparative studies on the fermentative production of lantibiotics by staphylococci. Appl Microbiol Biotechnol. 1990;32:511–517. doi: 10.1007/BF00173719. [DOI] [PubMed] [Google Scholar]
- Horner T, Zahner H, Kellner R, Jung G. Fermentation and isolation of epidermin, a lanthionine containing polypeptide antibiotic from Staphylococcus epidermidis. Appl Microbiol Biot. 1989;30:219–225. [Google Scholar]
- Hosoda K, Ohya M, Kohno T, Maeda T, Endo S, Wakamatsu K. Structure determination of an immunopotentiator peptide, cinnamycin, complexed with lysophosphatidylethanolamine by 1H-NMR1. J Biochem. 1996;119:226–230. doi: 10.1093/oxfordjournals.jbchem.a021226. [DOI] [PubMed] [Google Scholar]
- Hsu ST, Breukink E, Tischenko E, Lutters MA, de Kruijff B, Kaptein R, Bonvin AM, van Nuland NA. The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat Struct Mol Biol. 2004;11:963–967. doi: 10.1038/nsmb830. [DOI] [PubMed] [Google Scholar]
- Jörnvall H, Persson B, Krook M, Atrian S, Gonzàlez-Duarte R, Jeffery J, Ghosh D. Short-chain dehydrogenases/reductases (SDR) Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- Kallberg Y, Oppermann U, Jornvall H, Persson B. Short-chain dehydrogenases/reductases (SDRs) - Coenzyme-based functional assignments in completed genomes. Eur J Biochem. 2002;269:4409–4417. doi: 10.1046/j.1432-1033.2002.03130.x. [DOI] [PubMed] [Google Scholar]
- Kjeldgaard M, Nyborg J, Clark BFC. Protein motifs. 10 The GTP binding motif: Variations on a theme. Faseb J. 1996;10:1347–1368. [PubMed] [Google Scholar]
- Kjos M, Snipen L, Salehian Z, Nes IF, Diep DB. The Abi proteins and their involvement in bacteriocin self-immunity. J Bacteriol. 2010;192:2068–2076. doi: 10.1128/JB.01553-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers A, De Boef E, Rink R, Fekken S, Kluskens LD, Driessen AJ, Leenhouts K, Kuipers OP, Moll GN. NisT, the transporter of the lantibiotic nisin, can transport fully modified, dehydrated and unmodified prenisin and fusions of the leader peptide with non-lantibiotic peptides. J Biol Chem. 2004;279:22176–22182. doi: 10.1074/jbc.M312789200. [DOI] [PubMed] [Google Scholar]
- Kupke T, Kempter C, Gnau V, Jung G, Götz F. Mass spectroscopic analysis of a novel enzymatic reaction. Oxidative decarboxylation of the lantibiotic precursor peptide EpiA catalyzed by the flavoprotein EpiD. J Biol Chem. 1994;269:5653–5659. [PubMed] [Google Scholar]
- Levengood MR, Knerr PJ, Oman TJ, van der Donk WA. In vitro mutasynthesis of lantibiotic analogues containing nonproteinogenic amino acids. J Am Chem Soc. 2009;131:12024–12025. doi: 10.1021/ja903239s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Cooper LE, van der Donk WA. Chapter 21. In vitro studies of lantibiotic biosynthesis. Methods Enzymol. 2009;458:533–558. doi: 10.1016/S0076-6879(09)04821-6. [DOI] [PubMed] [Google Scholar]
- Li B, van der Donk WA. Identification of essential catalytic residues of the cyclase NisC involved in the biosynthesis of nisin. J Biol Chem. 2007;282:21169–21175. doi: 10.1074/jbc.M701802200. [DOI] [PubMed] [Google Scholar]
- Li B, Yu JP, Brunzelle JS, Moll GN, van der Donk WA, Nair SK. Structure and mechanism of the lantibiotic cyclase involved in nisin biosynthesis. Science. 2006;311:1464–1467. doi: 10.1126/science.1121422. [DOI] [PubMed] [Google Scholar]
- Majer F, Schmid DG, Altena K, Bierbaum G, Kupke T. The flavoprotein MrsD catalyzes the oxidative decarboxylation reaction involved in formation of the peptidoglycan biosynthesis inhibitor mersacidin. J Bacteriol. 2002;184:1234–1243. doi: 10.1128/JB.184.5.1234-1243.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maqueda M, Sanchez-Hidalgo M, Fernandez M, Montalban-Lopez M, Valdivia E, Martinez-Bueno M. Genetic features of circular bacteriocins produced by Gram-positive bacteria. FEMS Microbiol Rev. 2008;32:2–22. doi: 10.1111/j.1574-6976.2007.00087.x. [DOI] [PubMed] [Google Scholar]
- McClerren AL, Cooper LE, Quan C, Thomas PM, Kelleher NL, van der Donk WA. Discovery and in vitro biosynthesis of haloduracin, a two-component lantibiotic. Proc Natl Acad Sci U S A. 2006;103:17243–17248. doi: 10.1073/pnas.0606088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C, Bierbaum G, Heidrich C, Reis M, Süling J, Iglesias-Wind MI, Kempter C, Molitor E, Sahl HG. Nucleotide Sequence of the Lantibiotic Pep5 Biosynthetic Gene Cluster and Functional Analysis of PepP and PepC. Eur J Biochem. 1995;232:478–489. doi: 10.1111/j.1432-1033.1995.tb20834.x. [DOI] [PubMed] [Google Scholar]
- Meyer C, Bierbaum G, Heidrich C, Reis M, Suling J, Iglesias-Wind MI, Kempter C, Molitor E, Sahl HG. Nucleotide sequence of the lantibiotic Pep5 biosynthetic gene cluster and functional analysis of PepP and PepC. Evidence for a role of PepC in thioether formation. Eur J Biochem. 1995;232:478–489. doi: 10.1111/j.1432-1033.1995.tb20834.x. [DOI] [PubMed] [Google Scholar]
- Müller WM, Schmiederer T, Ensle P, Süssmuth RD. In vitro biosynthesis of the prepeptide of type-III lantibiotic labyrinthopeptin A2 including formation of a C-C bond as a post-translational modification. Angew Chem Int Ed Engl. 2010;49:2436–2440. doi: 10.1002/anie.200905909. [DOI] [PubMed] [Google Scholar]
- Oman TJ, van der Donk WA. Follow the leader: the use of leader peptides to guide natural product biosynthesis. Nat Chem Biol. 2010;6:9–18. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Grishin NV. Type II CAAX prenyl endopeptidases belong to a novel superfamily of putative membrane-bound metalloproteases. Trends Biochem Sci. 2001;26:275–277. doi: 10.1016/s0968-0004(01)01813-8. [DOI] [PubMed] [Google Scholar]
- Reis M, Eschbach-Bludau M, Iglesias-Wind MI, Kupke T, Sahl HG. Producer immunity towards the lantibiotic Pep5: identification of the immunity gene pepI and localization and functional analysis of its gene product. Appl Environ Microbiol. 1994;60:2876–2883. doi: 10.1128/aem.60.8.2876-2883.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Yang X, Garg N, van der Donk WA. Production of lantipeptides in Escherichia coli. J Am Chem Soc. 2011;133:2338–2341. doi: 10.1021/ja109044r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaugen M, Abildgaard CI, Nes IF. Organization and expression of a gene cluster involved in the biosynthesis of the lantibiotic lactocin S. Mol Gen Genet. 1997;253:674–686. doi: 10.1007/s004380050371. [DOI] [PubMed] [Google Scholar]
- Sollenberger PY, Martin RB. Mechanism of enamine hydrolysis. J Am Chem Soc. 1970;92:4261–4270. [Google Scholar]
- Suter-Crazzolara C, Unsicker K. Improved expression of toxic proteins in E. coli. Biotechniques. 1995;19:202–204. [PubMed] [Google Scholar]
- Tanaka N, Nonaka T, Nakamura KT, Hara A. SDR: Structure, mechanism of action, and substrate recognition. Curr Org Chem. 2001;5:89–111. [Google Scholar]
- Taubes G. The bacteria fight back. Science. 2008;321:356–361. doi: 10.1126/science.321.5887.356. [DOI] [PubMed] [Google Scholar]
- Uguen P, Hindre T, Didelot S, Marty C, Haras D, Le Pennec JP, Vallee-Rehel K, Dufour A. Maturation by LctT is required for biosynthesis of full-length lantibiotic lacticin 481. Appl Environ Microbiol. 2005;71:562–565. doi: 10.1128/AEM.71.1.562-565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Kamp M, Horstink LM, van den Hooven HW, Konings RN, Hilbers CW, Frey A, Sahl HG, Metzger JW, van de Ven FJ. Sequence analysis by NMR spectroscopy of the peptide lantibiotic epilancin K7 from Staphylococcus epidermidis K7. Eur J Biochem. 1995;227:757–771. doi: 10.1111/j.1432-1033.1995.tb20199.x. [DOI] [PubMed] [Google Scholar]
- van de Kamp M, van den Hooven HW, Konings RN, Bierbaum G, Sahl HG, Kuipers OP, Siezen RJ, de Vos WM, Hilbers CW, van de Ven FJ. Elucidation of the primary structure of the lantibiotic epilancin K7 from Staphylococcus epidermidis K7. Cloning and characterisation of the epilancin-K7-encoding gene and NMR analysis of mature epilancin K7. Eur J Biochem. 1995;230:587–600. doi: 10.1111/j.1432-1033.1995.tb20600.x. [DOI] [PubMed] [Google Scholar]
- van der Meer JR, Polman J, Beerthuyzen MM, Siezen RJ, Kuipers OP, de Vos WM. Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J Bacteriol. 1993;175:2578–2588. doi: 10.1128/jb.175.9.2578-2588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer JR, Rollema HS, Siezen RJ, Beerthuyzen MM, Kuipers OP, de Vos WM. Influence of amino acid substitutions in the nisin leader peptide on biosynthesis and secretion of nisin by Lactococcus lactis. J Biol Chem. 1994;269:3555–3562. [PubMed] [Google Scholar]
- Verhoef J, Milatovic D, Ekkelenkamp MB. WO/2005/023852. Antimicrobial compounds. 2005 March 17;
- Wiedemann I, Breukink E, van Kraaij C, Kuipers OP, Bierbaum G, de Kruijff B, Sahl HG. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J Biol Chem. 2001;276:1772–1779. doi: 10.1074/jbc.M006770200. [DOI] [PubMed] [Google Scholar]
- Willey JM, van der Donk WA. Lantibiotics: peptides of diverse structure and function. Annu Rev Microbiol. 2007;61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- Worobo RW, Van Belkum MJ, Sailer M, Roy KL, Vederas JC, Stiles ME. A signal peptide secretion-dependent bacteriocin from Carnobacterium divergens. J Bacteriol. 1995;177:3143–3149. doi: 10.1128/jb.177.11.3143-3149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Chatterjee C, Balsara R, Okeley NM, van der Donk WA. Heterologous expression and purification of SpaB involved in subtilin biosynthesis. Biochem Biophys Res Commun. 2002;295:952–957. doi: 10.1016/s0006-291x(02)00783-0. [DOI] [PubMed] [Google Scholar]
- Xie L, Miller LM, Chatterjee C, Averin O, Kelleher NL, van der Donk WA. Lacticin 481: in vitro reconstitution of lantibiotic synthetase activity. Science. 2004;303:679–681. doi: 10.1126/science.1092600. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.