Abstract
Cellular signaling pathways exhibit complex response profiles with features such as thresholds and steep activation (i.e. ultrasensitivity). In a reconstituted mitotic spindle orientation pathway, activation of Drosophila Pins (LGN in mammals) by Gαi is ultrasensitive (apparent Hill coefficient of 3.1), such that Pins recruitment of the microtubule-binding protein Mud (NuMA) occurs over a very narrow G αi concentration range. Ultrasensitivity is required for Pins function in neuroblasts as a non-ultrasensitive Pins mutant fails to robustly couple spindle position to cell polarity. Pins contains three Gαi binding GoLoco domains (GLs); Gαi binding to GL3 activates Pins whereas GLs 1 and 2 shape the response profile. Although cooperative binding is one mechanism for generating ultrasensitivity, we find GLs 1 and 2 act as “decoys” that compete against activation at GL3. Many signaling proteins contain multiple protein interaction domains and the decoy mechanism may be a common method for generating ultrasensitivity in regulatory pathways.
Introduction
Cellular inputs are coupled to specific physiological outputs by networks of dynamically interacting signaling proteins (Kholodenko, 2006). These proteins are often highly modular, composed of multiple protein -protein interaction or catalytic domains in the same polypeptide (Pawson and Nash, 2003). Regulatory pathways composed of signaling proteins underlie many of the complex decision -making behaviors implemented by cells. Two properties that are commonly found in such pathways are thresholding and ultrasensitivity (Tyson et al., 2003). Thresholding limits output activity until a specific input level is reached, a property which is likely useful for preventing spurious activity in the presence of biological noise (Ferrell, 1999). Ultrasensitivity, in which small variation in input levels leads to a large change in output, can convert graded inputs into more switch-like outputs and be used to generate more complex behaviors such as bistability and hysteresis, the basis of all or none decisions and cellular memory (Burrill and Silver; Goldbeter and Koshland, 1981; Tyson et al., 2003). Oxygen binding to Hemoglobin is a classic example of ultrasensitivity (Koshland et al., 1966). Although thresholding and ultrasensitivity are fundamental features of cellular signaling, binary protein interactions typically exhibit a hyperbolic response profile requiring large changes in input levels for maximal output (Figure 1A). A fundamental question in cellular signaling is how complex input-output relationships are built from individual protein -protein interactions. In particular, are alternative mechanisms besides cooperativity used to generate ultrasensitivity in protein interaction based regulation?
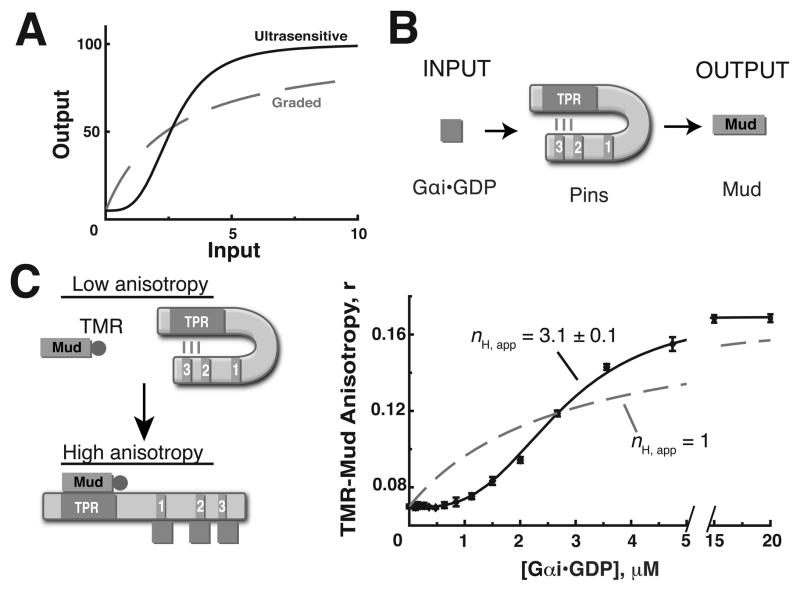
Figure 1. Gαi activation of Pins is ultrasensitive.
(A) Examples of ultrasensitive (black, solid) and graded (gray, dashed ) pathway response profiles.
(B) The Gαi-Pins-Mud spindle orientation pathway. Pins is activated by the upstream signal Gαi• GDP (input) and subsequently binds the microtubule associated protein Mud (output).
(C) Left: Schematic diagram of in vitro Pins activation reconstitution. Initially, the system is in a low anisotropy state because Pins is repressed and unable to interact with tetramethylrhodamine (TMR) labeled Mud peptide. Pins is activated upon Gαi binding to the GoLoco domains, leading to increased anisotropy as Pins binds TMR-Mud through the TPRs. Right: Quantification of pathway response shows activation of Pins by Gαi is ultrasensitive. 1 μM WT Pins was incubated with 0.5 μM TMR-Mud in the presence of increasing concentrations of Gαi. The data was fit using the Hill equation as described in the methods. The activation profile is well fit with an apparent Hill coefficient nH,app = 3.1 ± 0.1, but poorly fit assuming a hyperbolic curve with nH, app = 1. Error bars represent one standard deviation from three independent experiments. See also Figure S1.
We have investigated ultrasensitivity and thresholding in the regulatory pathway that controls mitotic spindle orientation. This fundamental cellular process is important for development and adult physiology because the site of cleavage furrowing, and subsequently the position of the two daughter cells, are determined by mitotic spindle orientation (Doe, 2008). For example, epithelial cells divide in a planar fashion with their spindle aligned along the sheet plane such that the two daughter cells remain in the plane (Morrison and Kimble, 2006). During epidermal stratification, cells in the basement layer switch between proliferative and differentiating divisions by either dividing with their spind le parallel or orthogonal with the plane of the epithelium (Lechler and Fuchs, 2005). Such asymmetric divisions are one mechanism used to generate cellular diversity (Gönczy, 2008). Drosophila neuroblasts (NBs) divide asymmetrically to generate a self-renewed NB and a ganglion mother cell that divides once more to generate two neurons (Yu et al., 2006). This process requires polarization of cortical factors that specify the two cell fates and rapid alignment of the spindle with the polarity axis such that the cleavage plane precisely bisects the determinants into the two daughter cells (Atwood and Prehoda, 2009; Siller and Doe, 2009). Understanding spindle orientation regulation has implications for cancer biology as failure to align the spindle in NBs can increase the stem cell pool (Cabernard and Doe, 2009).
In metazoans the spindle is positioned by conserved, cortically localized factors that anchor astral microtubules (Siller and Doe, 2009). These factors include the heterotrimeric G -protein α subunit Gαi, Partner of Inscuteable (Pins; GPR-1/2 in C. elegans, LGN in mammals), and Mushroom body defect (Mud; Lin5 in C. elegans, NuMA in mammals) (Bowman et al., 2006; Izumi et al., 2006; Siller et al., 2006; Srinivasan et al., 2003; Yu et al., 2000; Yu et al., 2003). Gαi is an upstream component that localizes to the apical NB cortex where it recruits Pins. Pins is an adapter protein that links signals from GDP-loaded Gαi (hereafter “Gαi”) to mitotic spindle orientation through Mud (Srinivasan et al., 2003; Du and Macara, 2004; Nipper et al., 2007; Figure 1B). The ability of Pins to recruit Mud is regulated by an evolutionarily conserved autoinhibitory intramolecular interaction between the Pins N -terminal tetratricopeptide repeats (TPRs) and C-terminal GoLoco domains (GLs) that inhibits Mud -Pins binding (Du and Macara, 2004, Nipper et al., 2007). Gαi weakens the Pins intramolecular interaction, thereby increasing the affinity of Pins for Mud (Nipper et al., 2007, Figure S1A).
In NBs, spindle orientation is remarkably dynamic, as Gαi, Pins, and Mud are transiently polarized at the cortex during mitosis (Izumi et al., 2006; Siller et al., 2006). Although the overall structure of the regulatory pathway is fairly well understood, the quantitative aspects that inevitably support the dynamic spindle orienting behavior in systems such as NBs have not been investigated. To address this gap in our understanding, we reconstituted the G αi-Pins-Mud regulatory pathway in vitro and, as described below, found that it is highly ultrasensitive. This allowed us to investigate the molecular origins of ultrasensitivity and furthermore, by systematically altering Pins to make it non -ultrasensitive, to examine the role of this property in vivo.
Results
Pins activation by Gαi is ultrasensitive
In order to quantify the relationship of Pins output (Mud binding) to Pins input (Gαi concentration), we reconstituted the Gαi-Pins-Mud pathway in vitro from purified components. A tetramethylrhodamine labeled Mud peptide (TMR-Mud residues 1936–1951, isoform B) containing the region that binds the Pins TPRs allows detection of Pins-Mud association via the fluorescence anisotropy of the conjugated TMR (Figure 1C). Because Pins is autoinhibited, its intrinsic affinity for Mud is low such that very little complex forms in the absence of G αi. Consistent with this, we observed little change in anisotropy upon addition of Pins to TMR-Mud (see activation at 0 μM Gαi in Figure 2B). When Gαi is titrated into the system, Pins becomes activated and binds Mud, resulting in a large increase in TMR-Mud anisotropy (Figure 1C). Activation is highly ultrasensitive as the entire transition occurs between 1 and 5 μM Gαi (Figure 1C, S1). We analyzed the Pins activation profile using the Hill equation as the Hill coefficient (nH) is commonly used to measure ultrasensitivity (Ferrell, 1996). We denote the resulting Hill coefficient as “apparent” (nH,app ) because the Hill equation is a model for cooperative systems (cooperativity can lead to ultrasensitivity, but ultrasensitivity is a general term used to describe sigmoidal responses; Ferrell, 1999). The nH, app for this binding isotherm is 3.1 ± 0.1, which would be very large for a cooperative system (i.e. perfectly cooperative) considering that Pins contains three Gαi binding sites (Figure 1C).
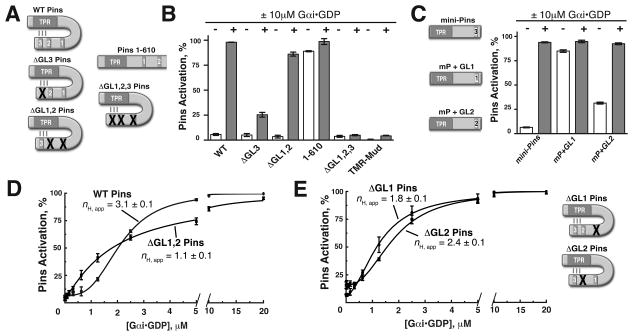
Figure 2. Pins GoLoco 3 is coupled to output while GoLocos 1 and 2 shape the pathway response curve.
(A) Schematic of the Pins mutants used in this study. Inactivating point mutations to various GL domains (R to F of conserved E/DQR triad) are represented by a red “X” over the mutated domain.
(B) GL3 is the key regulatory element for pathway activation and repression. Bar graphs represent Pins activation (%) in the absence (−, white) or presence (+, gray) of 10μM Gαi for Pins constructs shown in (A). ΔGL3 (R631F) is only weakly activated, while the ΔGL1,2 (R486F and R570F, respectively) is nearly fully activated. Deletion of the GL3 domain (Pins amino acids 1–610) breaks autoinhibition, while the ΔGL1,2,3 Pins is unable to be activated by Gαi.
(C) Activation of minimally repressed Pins (“mini-Pins”) consisting of a fusion of the GL3 region to the Pins TPRs (aa 42–396:590–639). Mini-Pins is able to reconstitute autoinhibition of the TPRs and can be activated by Gαi. Substitution of GL1 for GL3 in this context (mP + GL1) is unable to restore autoinhibition, while GL2 (m P +GL2) has an intermediate effect.
(D) GLs 1 and 2 are required for ultrasensitivity. Inactivating mutations to GLs 1 and 2 in ΔGL1,2 Pins (R486F and R570F respectively) abolishes ultrasensitivity in the system as the profile is graded nH,app = 1 (red curve).
(E) Contributions of GL1 and 2 to ultrasensitivty. Single mutation of either GL1 or GL2 has differing effects on ultrasensitivity. ΔGL1 (R486F, nH,app = 1.8 ± 0.1) decreases threshold and steepness while ΔGL2 (R570F, nH,app = 2.4 ± 0.1) partially decreases thresholding and steepness. Error bars represent one standard deviation from three independent experiments. See also Figure S2.
Ultrasensitivity is a common property of signaling pathways, yet its importance for biological function and its molecular origins are not fully understood. In the following sections we use the reconstituted in vitro system to identify components required for ultrasensitivity, the molecular mechanisms by which ultrasensitivity is achieved, and the function of ultrasensitivity in NB spindle orientation.
Pins GoLoco 3 is linked to activation, GoLocos 1 and 2 shape the activation profile
What are the elements of Pins required for ultrasensitivity? We first examined which of the three Gαi-binding GoLoco motifs (GLs) are required for Pins activation by G αi using a series of inactivating point mutations (Figure 2A). Although we previously used gel filtration to examine Gαi binding (Nipper et al., 2007), here we used the fluorescence anisotropy assay described above, which allowed us to more precisely quantify the role of each GL. Of the three GLs, we found that only inactivation of GL3 (by a single point mutation to a conserved arginine residue, R631F; Adhikari and Sprang, 2003; Kimple et al., 2002; henceforth ΔGL3) caused a dramatic reduction in the amount of Pins activation (Figure 2B). This suggests that G αi binding at GL3 is required for Pins activation. Consistent with this, Pins with inactivating mutations to GLs 1 and 2 (R486F and R570F for ΔGL1, 2), such that Gαi can only bind to GL3, is nearly fully activated at the same Gαi concentration (86%, Figure 2B). Furthermore, deletion of GL3 (Pins amino acids 1–610) caused Pins to bind Mud in the absence of Gαi (Figure 2B), suggesting that GL3 is structurally coupled to the TPRs and required for Pins autoinhibition. The triple Pins mutant with no functional GL domains shows very little activation (5%, Figure 2B), similar to TMR-Mud control. We conclude Gαi binding to GL3 activates Pins for subsequent Mud binding.
The sequences of the three Pins GLs are highly similar (Figure S2A), leading us to examine why GL3 is unique in its ability to couple Gαi input with Mud binding output. A fusion of the Pins TPRs to the GL3 domain (including a 20 residue linker outside of the GL proper), which we term “mini-Pins” (amino acids 42–396 followed by 590–639) recapitulates autoinhibition and activation (Figure 2C). An analogous version of LGN, the mammalian Pins homologue, also recapitulates autoinhibition (Du and Macara, 2004) and can be activated by Gαi to bind the Mud homologue, NuMA (Figure S2B) suggesting conservation of this regulatory module. To determine whether positional information or small sequence differences between each of the GLs are important for coupling to the TPRs, we examined whether each could substitute for GL3 in the context of the mini-Pins. While GL1 is able to bind Gαi (Figure S2C), it is unable to replace GL3. However, GL2 is nearly able to functionally replace GL3 in this context (Figure 2C). This result suggests that both sequences within and adjacent to the GL domain contribute to the specificity of GL3 interactions with the TPRs. Consistent with this finding, deletion of the linker sequence N -terminal to GL3 in full-length Pins disrupts autoinhibition (Figure S2D).
What is the function of GLs 1 and 2 if not to couple Gαi binding to Pins activation? While GLs 1 or 2 are not required for Pins activation, we hypothesized that they may be important for shaping the response profile. To test this possibility, we measured activation of ΔGL1,2 Pins and found that, although this protein can be nearly fully activated, the response profile has lost all of its sigmoidal character (nH,app = 1.1 ± 0.1; Figure 2D). Individual loss of GL 1 or 2 activity leads to intermediate effects: ΔGL1 reduces nH,app and thresholding whereas ΔGL2 has only minor effects on nH,app and thresholding (Figure 2E), suggesting a complex interplay between the two binding sites (see below ). We conclude that GLs 1 and 2 are not coupled to Pins activation directly, but are required for ultrasensitivity.
Pins ultrasensitivity is required for robust alignment of the mitotic spindle in Drosophila neuroblasts
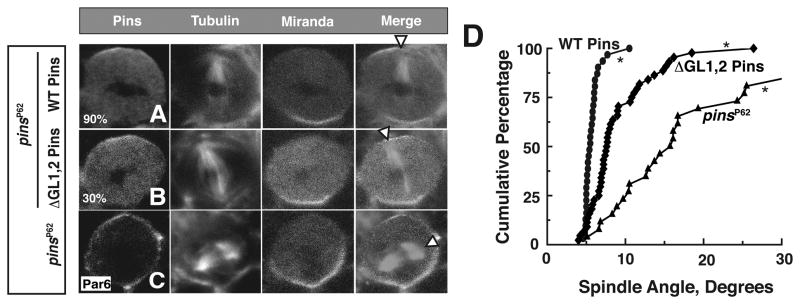
Ultrasensitivity is a common feature of cell signaling pathways, yet its importance in signal transduction has rarely been examined. The ΔGL1,2 Pins mutant is activated by Gαi but without ultrasensitivity (Figure 2D). We tested if ultrasensitivity is important for Pins function by attempting to rescue the spindle orientation defects of the pinsp 62 null allele (Yu et al., 2000) with ΔGL1,2 Pins. We stained larval brain NBs for Pins, tubulin and Miranda (a basal polarity marker) and scored the spindle angle relative to the Pins apical crescent (Figure 3A –C). The mitotic spindle is closely aligned with the apical-basal polarity axis in wild -type NBs (Yu et al., 2000), but pinsp 62 cells have misaligned spindles (Figure 3C, quantified in D; avg. angle = 22.2±19.9, Siegrist and Doe, 2005). Expression of wild -type (WT) Pins via the NB-specific worniu-Gal4 driver (Albertson and Doe, 2003) rescues the mitotic spindle orientation defect of pinsp 62 (Figure 3A, D avg. angle = 5.7±1.1; Nipper et al., 2007). On average, neuroblasts expressing ΔGL1,2 Pins had more cytoplasmic Pins than those expressing WT Pins (Figure S3). This qualitative difference could be due in part to a reported role for Gαi in cortical Pins recruitment (Yu et al., 2003). To ensure that a localization defect did not influence our analysis of spindle orientation, we only analyzed neuroblasts with cortical ΔGL1,2 Pins levels comparable to WT Pins. We ensured that this level was sufficient for function by staining for the apical marker Inscuteable whose localization requires cortical Pins activity (Yu et al., 2000; Figure S3). Addtionally, the presence of cytoplasmic Pins had no effect on spindle orientation phenotype (Figure 3B,D and Figure S3C). In neuroblasts expressing ΔGL1,2 Pins at levels com parable to WT and sufficient to localize Inscuteable, we observed that spindle alignment was not as robust as WT (Figure 3B,D; mean angle = 9.4 ± 4.4), and not as severe as the pins null. We conclude ultrasensitivity in the Gαi-Pins-Mud pathway is required for robust spindle orientation.
Figure 3. Pins ultrasensitivity is required for robust spindle alignment in vivo.
(A–C) Metaphase larval brain neuroblasts (NBs) were fixed and stained for Pins, tubulin and Miranda. White arrow heads denote spindle vector. (A,B) NBs expressing WT Pins or ΔGL1,2 Pins in the genetic background of the pinsP62 null allele. (C) pinsP62 NB negative control, Par6 was used to mark the apical cortex.
(D) Cumulative percentage of spindle angle measurements for each experimental condition relative to the center of the apical Pins crescent. pinsP62 NB spindle angles were determined relative to the apical Par6 signal. Average spindle angles: WT Pins (n = 31) 5 ± 1, ΔGL1,2 Pins (n = 48) 9 ± 4, pinsP62 (n = 26) 22 ± 20. Spindle angle was measured using ImageJ software. Asterisks denote differences are statistically significant by 1-way ANOVA. See also Figure S3.
Ultrasensitivity is required to generate maximal Gαi-Pins-Mud pathway output
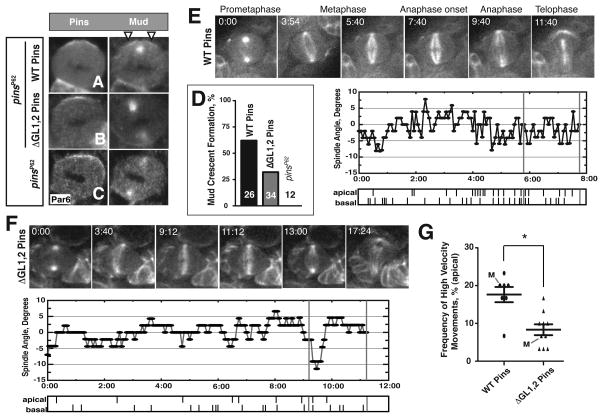
Why might ultrasensitivity be required for spindle alignment? Ultrasensitivity allows for activation over a small input range once a threshold is reached (Figure 1A; Goldbeter and Koshland, 1981). Thus the reduction in spindle orienting activity of ΔGL1,2 Pins could arise from reduced Pins output or ectopic output from loss of thresholding. To distinguish between these possibilities we assayed apical Mud recruitment, which requires activated Pins (Izumi et al., 2006; Siller et al., 2006; Bowman et al., 2006; Nipper et al., 2007). In ΔGL1,2 Pins NBs with apical Pins enrichment, Mud crescents were observed significantly less often than in NBs expressing WT Pins (32% vs. 62%, Figure 4D), but more often than in pinsp62 null cells (0%). These cells also had an identical spindle phenotype as determined by Mud -stained centrosome position relative to the apical Pins crescent (Figure S4A). We conclude ΔGL1,2 Pins NBs have decreased spindle orienting activity from a reduced ability of Pins to recruit Mud to the cortex.
Figure 4. ΔGL1,2 Pins neuroblasts have reduced Gαi-Pins-Mud pathway output.
(A–C) Metaphase larval brain neuroblasts (NBs) were fixed and stained for Pins and Mud. (A,B) NBs expressing WT Pins or ΔGL1,2 Pins in the genetic background of the pinsP62 null allele. (C) pinsP62 NB negative control, Par6 was used to mark the apical cortex. White arrow heads mark the apical Mud crescent.
(D) Quantification of percent of metaphase NBs imaged that showed a detectable Mud crescent. Mud crescents were scored as a ratio of Mud intensity at the apical cortex greater than or equal to two fold that of the cytoplasm (see supplemental methods). Pixel intensities were measured in ImageJ.
(E,F) Top: Image time course from representative movies capturing dividing NBs expressing WT or ΔGL1,2 Pins in pinsP62 background. GFP-Jupiter marks the mitotic spindle. Time is given in minutes relative to prometaphase, the moment when the spindle poles nucleated microtubules that penetrated the cell center. Bottom: Spindle angle relative to position at anaphase onset starting from prometaphase is plotted for each representative movie. Tick marks denote a rapid spindle movement for the apical or basal spindle pole respectively. The two-minute period prior to anaphase onset analyzed in each movie is marked by horizontal grey lines.
(G) NBs expressing ΔGL1,2 Pins have reduced apical spindle pole dynamics. High velocity spindle movements were scored during a two-minute period prior to anaphase onset (see methods). Each point on plot represents an independent measurement. The data taken from the movies shown in “E” and “F” and in the supplemental data are marked with an “M”. Error bars represent one standard deviation. Asterisks mark statistically significant differences. See also Figure S4.
In NBs and mammalian cells, the apical Gαi-Pins-Mud complex is required for spindle rocking immediately prior to anaphase by coupling cortical polarity to the Lis 1-Dynein complex (Siller and Doe, 2008). Because we observed reduced Mud recruitment in NBs expressing the grad ed ΔGL1,2 Pins mutant, we hypothesized these cells would have decreased spindle dynamics relative to NBs expressing WT Pins. We observed spindle movements using live cell imaging in mitotic NBs expressing a GFP fusion to the spindle associated protein Jupiter (Siller et al., 2005). WT and ΔGL1,2 Pins larval brain NBs were imaged from late prophase through telophase (Supplemental Movies 1 and 2, respectively). High velocity spindle movements were scored in the two-minute period prior to anaphase onset (Figures 4E–G, see methods). While dynamics at the basal pole of each cell were indistinguishable during this period (Figure S4B), NBs expressing WT Pins had a greater frequency of high velocity spindle movements at the apical pole than ΔGL1,2 Pins cells (Figure 4G). We conclude the observed spindle orientation defect of non-ultrasensitive ΔGL1,2 Pins mutant cells is due to decreased Mud activity at the apical cortex from reduced Pins output.
Pins ultrasensitivity originates from “decoy” binding at GLs 1 and 2
What is the molecular mechanism by which Pins is activated in an ultrasensitive manner? Cooperativity can be a source of ultrasensitivity (Ferrell, 1996). In this case, Gαi binding between the Pins GL domains would be thermodynamically coupled to yield a sigmoidal activation profile. In fact, the Pins activation profile strongly resembles the behavior of cooperative systems such as hemoglobin, the classic example of cooperativity (nH = 2.8; Koshland et al., 1966). In the case of Pins, a cooperative mechanism predicts GL3 exists in a low affinity (T) state but Gαi binding to GLs 1 and/or 2 causes a conversion to a high affinity (R) state.
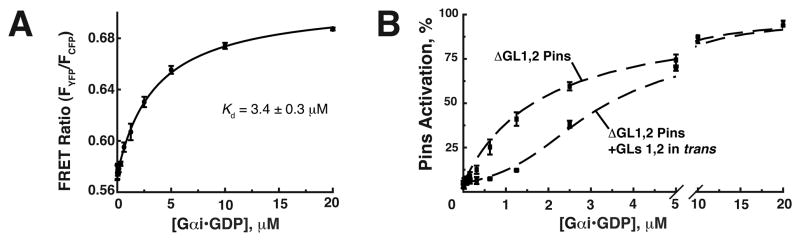
We tested if cooperativity is responsible for Pins ultrasensitivity in two ways. First, we measured the affinity of Gαi for GL3 when GL1 and 2 are not bound to Gαi (i.e. ΔGL1,2 Pins), which should prevent transition into the high affinity state. If cooperativity is responsible for ultrasensitive Pins activation, the GL3 affinity should be significantly lower than the midpoint of the activation threshold observed for the wild type protein (otherwise activation would occur at lower Gαi concentration). By analogy, initial binding events in hemoglobin are of lower affinity than the observed dissociation constant (Kd) as the system transitions into a higher affinity state. Previously, our lab used a Pins FRET sensor to observe Gαi binding to Pins (Nipper et al., 2007). We engineered a YFP-ΔGL1,2 Pins-CFP fusion protein to monitor the conformational changes of Pins as Gαi binds to GL3. Using this method, we measured the affinity of GL3 for Gαi to be 3.4 ± 0.3 μM (Figure 5A). This affinity is near the activation transition mid point, inconsistent with cooperativity being responsible for Pins ultrasensitivity.
Figure 5. Pins ultrasensitivity is inconsistent with a cooperative mechanism.
(A) Forster resonance energy transfer (FRET) was used to approximate the Kd of GL3 for Gαi. 100nM of the YFP-ΔGL1,2Pins-CFP FRET fusion protein was incubated with increasing amounts of Gαi. The binding curve was fit to a Kd = 3.4 ± 0.3μM.
(B) Gαi does not need to bind Pins in cis to generate ultrasensitivity. Addition of GLs 1 and 2 in trans (Pins R259A, ΔGL3) restores ultrasensitivity to the graded ΔGL1,2 Pins nH,app = 2.3 ± 0.1.
We further tested the cooperative model by determining if GLs 1 and 2 must be in cis with GL3 for ultrasensitivity, a requirement of a cooperative mechanism. We measured ΔGL1, 2 Pins activation in the presence of a ΔGL3 Pins that also contains a mutation in the Pins TPRs rendering it unable to bind Mud (R259A). The ΔGL3 Pins protein can bind Gαi at GLs 1 and 2, but cannot enhance the affinity at the GL3 domain of ΔGL1,2 Pins, and thus not influence activation through cooperativity. As shown in Figure 5B, the WT activation profile is largely recapitulated in this experiment (nH,app = 2.3 ± 0.1), indicating that GLs 1 and 2 do not need to act in cis to generate Pins ultrasensitivity (we believe the small difference between the two results from the minor contribution of GL2 binding to activation, see ΔGL3 Pins activation data in Figure 2B). We conclude Pins ultrasensitivity does not result from cooperative interactions between Gαi binding sites.
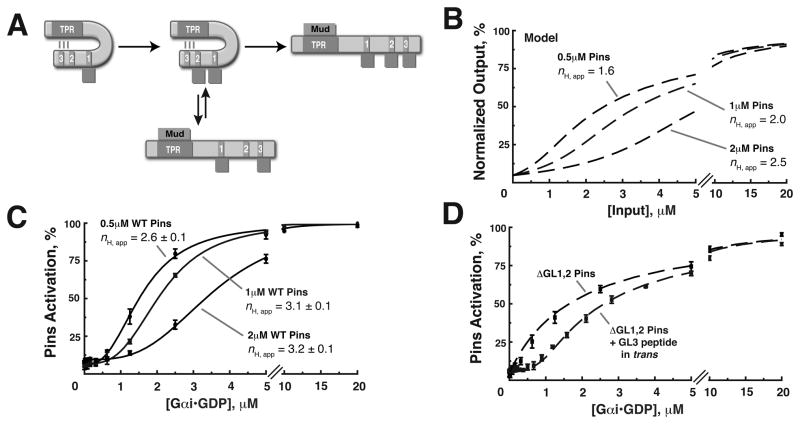
Alternatively ultrasensitivity can result from “decoy” binding. In such a mechanism, decoy sites buffer activator to shape the activation profile. For example, in the MAPK and Wee1 kinase signaling cascades, decoy phosphorylation sites are recognized by the up stream kinase, but are not coupled to a functional output (Ferrell and Machleder, 1998; Kim and Ferrell, 2007). Although such a mechanism has not been reported for protein -protein interaction pathways such as Pins, we tested if decoys might be responsible for Pins ultrasensitivity. In this model, Gαi binding to GLs 1 and 2 would not have any effect beyond competition with GL3. G αi would bind preferentially to GLs 1 and 2, resulting in the observed threshold. Once nearly saturated, Gαi would begin to populate GL3, resulting in rapid activation and the observed transition steepness (Figure 6A).
Figure 6. A “decoy” mechanism generates ultrasensitivity in the Gαi-Pins-Mud spindle orientation pathway.
(A) Schematic diagram of the decoy mechanism: Decoy GLs 1 and 2 generate ultrasensitivity by competing for the Gαi input with the regulatory domain, GL3. At low Gαi inputs, the decoy GLs 1 and 2 are preferentially bound, leading to threshold. At intermediate Gαi concentration, the decoys are nearly saturated, and GL3 begins to be populated. At higher Gαi concentration, the decoys are fully saturated and Gαi binds to GL3, leading to Pins activation and observed steepness.
(B) Predicted effect of varying the concentration of Pins based on the decoy model.
(C) The Pins activation threshold is directly proportional to Pins input concentration. Addition of half (0.5uM) or double (2uM) the amount of Pins leads to a proportional change to the activation threshold, defined as a 5% increase in Pins activation above the initial value and an increase in nH,app.
(D) High affinity decoy domains can lead to thresholding without steepness. Addition of a GL3 peptide (Kd ~ 60nM) in trans to ΔGL1,2 Pins builds a strong threshold to the response and is well approximated by modeling (dashed line). Error bars represent one standard deviation from three independent experiments.
We modeled Gαi activation of Pins using a decoy-based scheme and known parameters of the system (as opposed to fitting the observed activation profile; see Supplemental materials). The model indicates that the concentration of Pins in the system should set the threshold because this variable determines the amount of decoys present (Figure 6B). We observed a dose dependent change in the threshold when the Pins concentration was varied consistent with GLs 1 and 2 acting as decoys (Figure 6C). Decreasing the concentration of Pins by one half or doubling the concentration caused a proportional shift in the threshold (Figure 6C). Additionally, the model predicts that the apparent Hill coefficient will increase as Pins concentration is increased (nH,app = 1.6, 2.0 and 2.5 for 0.5, 1 and 2μM Pins respectively, Figure 6B). We also observe an increase in the nH,app in our experimental data, but the fitted data have greater nH,app than predicted by modeling (nH,app = 2.6, 3.1 and 3.2 for 0.5, 1 and 2μM Pins respectively, Figure 6C), suggesting that additional activation, likely from GL2, also contributes to the observed steepness. Additionally, the model correctly predicts the effect of in trans GL addition to ΔGL1,2 Pins (Figure 5B, nH,app = 2.2).
Competition mechanisms using decoys can generate thresholding or ultrasensitivity under certain physiological conditions (Gunawardena, 2005; Salazar and Hofer, 2006), depending on the relative affinities of decoys and activating binding sites. We tested the effect of increasing decoy affinity by adding a high affinity GL peptide (Kd approximately 60nM) in trans to the graded ΔGL1,2 Pins protein. Modeling predicts this peptide should build a strong threshold in the response and shift the graded curve to higher G αi concentration (red and black data points, respectively). As seen in Figure 6C, the GL peptide adds a strong threshold to the ΔGL1,2 Pins response curve, but the curve does not have a sigmoidal shape as it is simply an offset hyperbola. This result suggests the affinities of the Pins GL domains are “tuned” in order to shape the response profile from a thresholding to a sigmoidal shape.
Discussion
Pins ultrasensitivity arises from a GoLoco “decoy” mechanism
Complex input/output relationships generated by cell signaling networks allow for a multitude of cellular decision making behaviors, such as bistability or hysteresis, which are necessary to implement diverse physiological processes (Kholodenko, 2006). Ultrasensitivity is a building block for these types of behaviors, yet its molecular origins are poorly understood (Kim and Ferrell, 2007). While cooperativity is a well-described mechanism to generate ultrasensitivity, here we have uncovered a cellular regulatory system that uses another mechanism for obtaining sigmoidal responses with high apparent Hill coefficients. We found activation of the mitotic spindle orientation protein Pins by Gαi is highly ultrasensitive (Figure 1C, nH,app = 3.1 ± 0.1) and this ultrasensitivity arises from a decoy mechanism as binding sites GLs 1 and 2 compete with the activating GL3 for the Gαi input. Cooperativity is commonly thought to be the source for ultrasensitivity in protein -protein interaction networks and protein -DNA interactions (Dueber et al., 2007; Giorgetti et al., 2010). However, our observations of Pins activation are inconsistent with a cooperative mechanism for three reasons. First, activation of ΔGL1,2 occurs at a lower Gαi concentration than WT (Figure 2D). Second, the sigmoidal response can be largely recapitulated through Gαi binding to GLs 1 and 2 in trans (Figure 5C). Lastly, thresholding behavior is entirely dependent on the concentration of Pins present (Figure 6B). These findings are supported by mathematical modeling and suggest that ultrasensitive responses can be generated without cooperativity from binary protein -protein interactions through a simple competition mechanism, similar to the competition that occurs in kinase signaling cascades (Salazar and Hofer, 2006).
Ultrasensitivity generated by competition or cooperative mechanisms
Although competition and cooperativity are both potential origins of ultrasensitive responses, there are inherent differences between curves created by each of these mechanisms. Cooperativity-based ultrasensitivity can dramatically reduce the amount of input necessary to reach maximal output. For example, initial binding events of O2 are of low affinity and, without cooperativity, would require a large change in O2 concentration for saturation. The competition mechanism described here and in kinase cascades generates ultrasensitive responses from a threshold, as activation would occur in a graded fashion at low input concentrations without competition. Therefore, while yielding sigmoidal responses with high apparent Hill coefficients this mechanism may be more important for thresholding than the observed apparent steepness. Modeling studies have shown that multisite phosphorylation builds a good threshold, not necessarily a more switch -like response (although the Hill coefficient is often used as a measure of steepness, this single parameter is also influenced by the threshold; Gunawardena, 2005). However, multisite phosphorylation is required for the bistable signaling nature of Xenopus oocyte maturation (Huang and Ferrell, 1996) and cell cycle progression (Kim and Ferrell, 2007; Nash et al., 2001).
Pins ultrasensitivity is required for robust alignment of the mitotic spindle in vivo
Expressing the non -ultrasensitive ΔGL1,2 Pins failed to fully rescue the spindle positioning defect of the pinsP62 null allele relative to WT Pins (Figure 3A–D), suggesting that ultrasensitive regulation of Pins is important for proper molecular function. The reduced spindle orienting activity of the graded Pins mutant is caused by decreased pathway output because we observed less apical Mud recruitment and spindle pole dynamics relative to NBs expressing WT Pins (Figure 4A–G). The ΔGL1,2 Pins spindle phenotype is similar to loss of Lis1 function, an adaptor protein that physically links the Gαi-Pins-Mud complex at the apical cortex to the Dynein motor protein, generating pulling forces on the spindle (Siller and Doe, 2008). Although ultrasensitivity is important for the robust spindle positioning observed in WT NBs, loss of ultrasensitivity had only a minor effect on spindle orientation, as all spindle angles measured in ΔGL1,2 Pins NBs were within 30° of the apical Pins crescent (Figure 3D). This is likely because of redundant spindle orienting cues in vivo as the mitotic spindle is not completely random in pinsP62 null NBs (Siegrist and Doe, 2005; Johnston et al., 2009; Figure 3D).
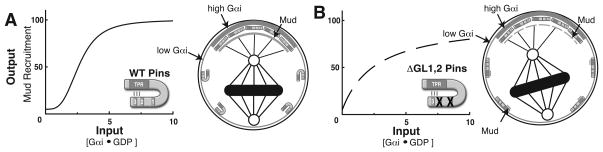
Why might ultrasensitive regulation of Pins be required for robust spindle orienting function? In WT NBs, thresholding limits Pins output to the apical cortex where the Gαi input concentration is high. This Pins is not activated at cortical sites where input concentration is low (Figure 7A). In ΔGL1,2 Pins NBs, thresholding is absent such that Pins output can potentially occur both at the apical cortex and distal cortical regions (Figure 7B). Loss of steepness results in only a slight difference in total Pins output between WT and ΔGL1,2 Pins in vitro (100% vs. 85%, Figure 2B), but transient activation of ΔGL1,2 Pins at cortical sites with low Gαi concentration could magnify this difference in vivo by titrating away Mud from the apical cortex (Figure 7B). Thus, ultrasensitivity may be an important feature of the Gαi-Pins-Mud spindle orientation pathway as it allows for generating maximal pathway output through spatial restriction of Pins activity. In this way, competition -based ultrasensitivity allows for increased pathway output by setting concentration thresholds to restrict signaling protein activity and may be a common theme in other regulatory pathways.
Figure 7. Ultrasensitive regulation of Pins restricts output to the apical cortex.
(A) Thresholding in WT Pins ensures Pins is activated only at the apical cortex where Gαi concentration is above the threshold, allowing for maximal Mud recruitment. Transient interactions of Pins-Gαi at distal sites do not generate output because the decoy GL domains are bound at low Gαi concentration. The steepness in the transition allow for maximal activity at the apical cortex from the present Gαi input.
(B) The graded ΔGL1,2 Pins mutant has lost thresholding, allowing spurious Pins activation at sites distal to the apical cortex where Gαi concentration is low. Combined with loss of steepness, this leads to reduced output at the apical domain as Mud is competed away and causes reduced Mud recruitment relative to WT Pins. Reduced Pins output causes the mitotic spindle to be misaligned relative to the cell polarity axis.
Protein modularity can shape cell signaling behavior
The modular architecture of signaling proteins is thought to be a means of coupling different inputs with new output functions, allowing for rapid evolution of new signaling functions (Pawson and Nash, 2003). This feature is also important for creating signaling proteins that integrate multiple inputs to trigger a specific output (Lim, 2002). Protein modularity also can create new input/output relationships such as ultrasensitive responses through cooperative interactions between input domains (Dueber et al., 2007). Our analysis of Pins supports this idea, but adds that modularity can shape pathway responses without cooperativity, simply by including multiple input domains. We have shown in our system that decoys can build either ultrasensitivity or thresholding depending on the affinities of the decoys relative to the activating site for the input. A high affinity decoy sets a strong threshold, but lowering the decoy affinity can change thresholding into a more sigmoidal shaped curve, simply by blending the inflection point between thresholding and activation (Figures 6D, 5B). This type of ultrasensitivity may be a fairly common component of cell signaling pathways because autoinhibition and domain repeats are common features of cell signaling proteins (Dueber et al., 2004; Pufall and Graves, 2002). Thus, incorporating more domain repeats through genetic recombination events can modulate the response profile. The relative affinities of these sites could then be “tuned” through point mutations to build thresholding behavior and/or apparent steepness into the signaling pathway.
Methods
Protein expression and purification
Proteins were expressed in E. coli strain BL21(DE3) using pBH 4 based vectors encoding a 6x histidine tag. H is-tagged proteins were affinity purified on Ni-NTA agarose (Qiagen) and further purified using an AKTA FPLC (GE Healthcare) by anion exchange and/or size exclusion chromatography. Protein stocks were stored in binding buffer (20m M H EPES pH 7.5, 100m M NaCl, 1m M MgCl2 and 1m M DTT). Total protein concentrations were determined by Brad ford assay (Bio-Rad).
In vitro reconstitution of Pins activation
A synthetic peptide containing the minimal Pins TPR binding domain of Mud isoform B residues 1934–1951 was obtained from EZ-Biolabs. Tetramethylrhodamine (TMR) was conjugated to a cysteine residue added to the C-terminus using TMR-maleimide (Life Technologies) according to the manufacturers protocol followed by RP-HPLC and characterization by MALDI-TOF mass spectrometry.
Quantification of the Pins response to Mud as a function of Gαi concentration was conducted under the following conditions: 1μM Pins was incubated with 0.5μM TMR-Mud peptide in binding buffer. Anisotropy was determined by exciting TMR at 555nm and observed emission at 580nm using an ISS-PC1 spectrofluorometer equipped with polarizers and a water bath at 20° C.
The percent of Pins activation was measured relative to the anisotropy of the free TMR-Mud peptide (0%) and the maximal value of WT Pins observed at 20 μM Gαi (100%). Because Pins with inactivating GL mutations had slightly lower final anisotropy, these constructs were normalized relative to the constitutively active Pins 1–610 saturated with one (Pins 1–610 ΔGL1) or two molecules of Gαi (Pins 1–610). For “mini-Pins,” the maximal percent is relative to the maximum anisotropy value in the presence of 20μM Gαi. The apparent Hill coefficient was obtained by fitting the data to the Hill equation.
Construction and analysis of the Pins ΔGL1,2 FRET biosensor
The Pins FRET protein was expressed from a pBH vector encoding an N -terminal YFP (EYFP 1–239) and C-terminal CFP (ECFP 1–239) to create a YFP-ΔGL1,2 Pins-CFP fusion. The protein was expressed and purified as described above with size exclusion chromatography as a final purification step. 100nM FRET protein was incubated in binding buffer in the presence of increasing concentrations of Gαi. FRET was measured by exciting CFP at 433nm and the emissions of CFP at 475nm and YFP at 525nm respectively were measured. The FRET ratio is defined as the ratio of acceptor (YFP) to donor (CFP) emissions. The dissociation constant (Kd) was determined by fitting the data to a standard binding function.
Fly strains and genetics
The transgenic UAS-WT Pins strain was obtained from the Doe lab. The ΔGL1,2 Pins transgenic fly was created by subcloning into the pUAST vector encoding an N -terminal hemagglutinin (HA) epitope. Transgenic UAS-Pins constructs were on the second chromosome and balanced over Cyo, Actin -GFP and were crossed with a stock containing the pinsP62 allele (Yu et al., 2000) balanced over TM3 Ser, Actin -GFP on chromosome three. Flies were crossed at room temperature with the worniu-Gal4; pinsP62/TM3 Ser Actin-GFP driver line (Nipper et al., 2007) for NB specific expression.
The gene trap line G147-GFP, which expresses the GFP-tagged microtubule associated protein Jupiter (Siller et al., 2005) was used for live imaging experiments. A stock with this construct recombined with the pinsP62 allele on chromosome three, was obtained from the Doe lab. This stock was crossed with the worniu-Gal4; pinsP62/TM3 Ser, Actin-GFP driver line to obtain worniu-Gal4; pinsP62, GFP-Jupiter/TM3 Ser, Actin -GFP. The Pins stocks described above were crossed to this new driver line and pinsP62 homozygous larvae were identified from the absence of GFP expression in the gut.
Immunofluorescence
Second to early third instar larval brains were dissected in Schneider’s insect medium (Sigma) and fixed in PBS + 4% paraformaldehyde at room temperature for 20 minutes. Brains were washed in 1x PBS-BT (PBS + 2% BSA, 0.3% Triton-X 100, 0.02% sodium azide) three times and incubated with primary antibodies at 4° C overnight. Brains were washed six times in PBS-BT over 1 hour and incubated with secondary antibodies for 3 hours at 22° C. After washing six times for 1 hour, brains were mounted in Vectashield mounting medium (Vector Labs).
The following primary antibodies and dilutions were used: rat anti-Pins (1:500), mouse anti-tubulin DM1A (1:1500), guinea pig anti-Miranda (1:500), mouse anti-HA (Covance, 1:1000), rat anti-tubulin (abCam, 1:500), rabbit anti-Mud (1:1000), rat anti-Par6 (1:250). Secondary antibodies from Life Technologies or Jackson Immunoresearch were used according to manufacturer specifications.
Acquisition and analysis of images for spindle orientation determination
Fixed NB images were acquired on a Leica SP2 confocal microscope equipped with a 63x 1.4 NA oil immersion objective. The reported spindle angle value is the angle between the spindle vector to the cell center and to the center of the apical Pins or Par -6 crescent, as previously described (Siegrist and Doe, 2005). Spindle angles were measured using ImageJ (NIH ). Only cells in which the apical Pins signal was 1.5x greater than the cytoplasm were scored in our analysis. Figure panels were arranged using ImageJ, Photoshop and Illustrator (Adobe).
Analysis of apical Mud crescent formation
Images of fixed and stained larval brain NBs were acquired as dscribed above. A mud crescent was scored if the pixel intensity at the apical cell cortex was ≥ 2x the signal intensity at the cell center. Cells expressing WT or ΔGL1,2 Pins were scored if the apical Pins intensities were ≥ 1.5x that in the cytoplasm. Signal intensity ratios were determined using Image J.
Live imaging of neuroblasts and quantification of spindle dynamics
Second to early third instar larval brains from animals expressing either WT or ΔGL1,2 Pins and GFP-Jupiter in the pinsP62 genetic null background were dissected in Schneider’s insect medium supplemented with 5% FBS and 0.5μM ascorbic acid (Cabernard and Doe, 2009). Movies were made on a McBain spinning disc confocal microscope equipped with a Hammamatsu CCD camera. Images were acquired at four-second intervals with 2μm z-sections. NBs were identified as large cells in the central brain lobes. Prophase NBs were identified by the presence of two centrosomes that did not radiate microtubules into the cell center. Time zero is the start of prometaphase, when the centrosomes begin to nucleate microtubules form the mitotic spindle. Anaphase onset was determined as the moment when kinetochore microtubules at the center of the spindle began to shorten towards the spindle poles (Siller and Doe, 2008). High velocity spindle movements were scored during the two -minute period immediately prior to anaphase onset. A high velocity spindle movement was scored for either the apical or basal spindle pole if the center of the spindle pole moved ≥ 2 pixels between frames (Siller and Doe, 2008). Movie frames were acquired using Volocity 4 software, processed and analyzed in ImageJ, and movies were compiled in Quicktime.
Supplementary Material
Acknowledgments
The authors thank C. Doe, K. Guillemin, B. Nolen for comments on the manuscript; members of the Prehoda, Berglund and Nolen labs for helpful discussions; the Doe lab for generously sharing fly stocks and antibodies; K. Tran for help with staining technique and C. Cabernard for help with imaging. This work was supported by NIH grants GM068032 and GM087457 (K.E.P.) and NIH training grant 5-T32-GM007759-30 (N.R.S).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adhikari A, Sprang SR. Thermodynamic characterization of the binding of activator of G protein signaling 3 (AGS3) and peptides derived from AGS3 with G alpha i1. J Biol Chem. 2003;278:51825–32. doi: 10.1074/jbc.M306300200. [DOI] [PubMed] [Google Scholar]
- Albertson R, Doe CQ. Dlg, Scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nat Cell Biol. 2003;5:166–70. doi: 10.1038/ncb922. [DOI] [PubMed] [Google Scholar]
- Atwood SX, Prehoda KE. aPKC Phosphorylates Miranda to Polarize Fate Determinants during Neuroblast Asymmetric Cell Division. Curr Biol. 2009;19:723–729. doi: 10.1016/j.cub.2009.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman SK, Neumuller RA, Novatchkova M, Du Q, Knoblich JA. The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Dev Cell. 2006;10:731–42. doi: 10.1016/j.devcel.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Burrill DR, Silver PA. Making cellular memories. Cell. 140:13–18. doi: 10.1016/j.cell.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabernard C, Doe CQ. Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Dev Cell. 2009;17:134–41. doi: 10.1016/j.devcel.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–87. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- Du Q, Macara IG. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell. 2004;119:503–16. doi: 10.1016/j.cell.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Dueber JE, Mirsky EA, Lim WA. Engineering synthetic signaling proteins with ultrasensitive input/output control. Nat Biotechnol. 2007;25:660–2. doi: 10.1038/nbt1308. [DOI] [PubMed] [Google Scholar]
- Dueber JE, Yeh BJ, Bhattacharyya RP, Lim WA. Rewiring cell signaling: the logic and plasticity of eukaryotic protein circuitry. Curr Opin Struct Biol. 2004;14:690–9. doi: 10.1016/j.sbi.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Ferrell JE., Jr Tripping the switch fantastic: how a protein kinase cascade can convert graded inputs into switch-like outputs. Trends Biochem Sci. 1996;21:460–6. doi: 10.1016/s0968-0004(96)20026-x. [DOI] [PubMed] [Google Scholar]
- Ferrell JE., Jr Building a cellular switch: more lessons from a good egg. Bioessays. 1999;21:866–70. doi: 10.1002/(SICI)1521-1878(199910)21:10<866::AID-BIES9>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Ferrell JE, Jr, Machleder EM. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–8. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- Giorgetti L, Siggers T, Tiana G, Caprara G, Notarbartolo S, Corona T, Pasparakis M, Milani P, Bulyk ML, Natoli G. Noncooperative interactions between transcription factors and clustered DNA binding sites enable graded transcriptional responses to environmental inputs. Mol Cell. 37:418–28. doi: 10.1016/j.molcel.2010.01.016. [DOI] [PubMed] [Google Scholar]
- Goldbeter A, Koshland DE., Jr An amplified sensitivity arising from covalent modification in biological systems. Proc Natl Acad Sci U S A. 1981;78:6840–4. doi: 10.1073/pnas.78.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat Rev Mol Cell Biol. 2008;9:355–66. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- Gunawardena J. Multisite protein phosphorylation makes a good threshold but can be a poor switch. Proc Natl Acad Sci U S A. 2005;102:14617–22. doi: 10.1073/pnas.0507322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Ferrell JE., Jr Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc Natl Acad Sci U S A. 1996;93:10078–83. doi: 10.1073/pnas.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Y, Ohta N, Hisata K, Raabe T, Matsuzaki F. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat Cell Biol. 2006;8:586–93. doi: 10.1038/ncb1409. [DOI] [PubMed] [Google Scholar]
- Johnston CA, Hirono K, Prehoda KE, Doe CQ. Identification of an Aurora-A/PinsLINKER/Dlg spindle orientation pathway using induced cell polarity in S2 cells. Cell. 2009;138:1150–63. doi: 10.1016/j.cell.2009.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland DE, Jr, Némethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–85. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- Kholodenko BN. Cell-signalling dynamics in time and space. Nat Rev Mol Cell Biol. 2006;7:165–76. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Ferrell JE., Jr Substrate competition as a source of ultrasensitivity in the inactivation of Wee1. Cell. 2007;128:1133–45. doi: 10.1016/j.cell.2007.01.039. [DOI] [PubMed] [Google Scholar]
- Kimple RJ, Kimple ME, Betts L, Sondek J, Siderovski DP. Structural determinants for GoLoco-induced inhibition of nucleotide release by Galpha subunits. Nature. 2002;416:878–81. doi: 10.1038/416878a. [DOI] [PubMed] [Google Scholar]
- Lechler T, Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437:275–80. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim WA. The modular logic of signaling proteins: building allosteric switches from simple binding domains. Curr Opin Cell Biol. 2002;14:149–54. doi: 10.1016/s0959-440x(02)00290-7. [DOI] [PubMed] [Google Scholar]
- Morin X, Jaouen F, Durbec P. Control of planar divisions by the G-protein regulator LGN maintains progenitors in the chick neuroepithelium. Nat Neurosci. 2007;10:1440–8. doi: 10.1038/nn1984. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–74. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–21. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- Nipper RW, Siller KH, Smith NR, Doe CQ, Prehoda KE. Galphai generates multiple Pins activation states to link cortical polarity and spindle orientation in Drosophila neuroblasts. Proc Natl Acad Sci U S A. 2007;104:14306–11. doi: 10.1073/pnas.0701812104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–52. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- Pufall MA, Graves BJ. Autoinhibitory domains: modular effectors of cellular regulation. Annu Rev Cell Dev Biol. 2002;18:421–62. doi: 10.1146/annurev.cellbio.18.031502.133614. [DOI] [PubMed] [Google Scholar]
- Salazar C, Hofer T. Competition effects shape the response sensitivity and kinetics of phosphorylation cycles in cell signaling. Ann N Y Acad Sci. 2006;1091:517–30. doi: 10.1196/annals.1378.093. [DOI] [PubMed] [Google Scholar]
- Siegrist SE, Doe CQ. Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell. 2005;123:1323–35. doi: 10.1016/j.cell.2005.09.043. [DOI] [PubMed] [Google Scholar]
- Siller KH, Cabernard C, Doe CQ. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat Cell Biol. 2006;8:594–600. doi: 10.1038/ncb1412. [DOI] [PubMed] [Google Scholar]
- Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–74. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- Siller KH, Serr M, Steward R, Hays TS, Doe CQ. Live imaging of Drosophila brain neuroblasts reveals a role for Lis1/dynactin in spindle assembly and mitotic checkpoint control. Mol Biol Cell. 2005;16:5127–40. doi: 10.1091/mbc.E05-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan DG, Fisk RM, Xu H, van den Heuvel S. A complex of LIN-5 and GPR proteins regulates G protein signaling and spindle function in C elegans. Genes Dev. 2003;17:1225–39. doi: 10.1101/gad.1081203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson JJ, Chen KC, Novak B. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr Opin Cell Biol. 2003;15:221–31. doi: 10.1016/s0955-0674(03)00017-6. [DOI] [PubMed] [Google Scholar]
- Yu F, Cai Y, Kaushik R, Yang X, Chia W. Distinct roles of Galphai and Gbeta13F subunits of the heterotrimeric G protein complex in the mediation of Drosophila neuroblast asymmetric divisions. J Cell Biol. 2003;162:623–33. doi: 10.1083/jcb.200303174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Kuo CT, Jan YN. Drosophila neuroblast asymmetric cell division: recent advances and implications for stem cell biology. Neuron. 2006;51:13–20. doi: 10.1016/j.neuron.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Yu F, Morin X, Cai Y, Yang X, Chia W. Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell. 2000;100:399–409. doi: 10.1016/s0092-8674(00)80676-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.