Abstract
The highly oriented filamentous protein network of muscle constantly experiences significant mechanical load during muscle operation. The dimeric protein myomesin has been identified as an important M-band component supporting the mechanical integrity of the entire sarcomere. Recent structural studies have revealed a long α-helical linker between the C-terminal immunoglobulin (Ig) domains My12 and My13 of myomesin. In this paper, we have used single-molecule force spectroscopy in combination with molecular dynamics simulations to characterize the mechanics of the myomesin dimer comprising immunoglobulin domains My12–My13. We find that at forces of approximately 30 pN the α-helical linker reversibly elongates allowing the molecule to extend by more than the folded extension of a full domain. High-resolution measurements directly reveal the equilibrium folding/unfolding kinetics of the individual helix. We show that α-helix unfolding mechanically protects the molecule homodimerization from dissociation at physiologically relevant forces. As fast and reversible molecular springs the myomesin α-helical linkers are an essential component for the structural integrity of the M band.
Keywords: atomic force microscopy, protein folding
Filamentous modular proteins play a key role in the force-bearing structures of the sarcomere (1, 2). The most prominent example is the giant muscle protein titin. For titin, a detailed mechanical hierarchy ranging from entropic stretching of unstructured segments over mechanical kinase activation to unfolding of individual domains has been described (3, 4). Whereas in the sarcomeric I band titin provides the muscle with its passive tension (5), the mechanical properties of the M-band section are less well understood. Here, the 185 kDa protein myomesin (6) as well as other filamentous proteins form a large network constituting, together with metabolic enzymes and kinase domains, a well-organized compartment that has both structural and metabolic properties (7). Myomesin comprises 13 domains, with the first one (My1) being unique and the others (My2–My13) either of the immunoglobulin (Ig) or fibronectin type III fold (8). It is part of a complex network that involves interactions with myosin, titin, obscurin, and obscurin-like 1 (9, 10). Through its N-terminal myosin binding domain (My1) and the ability to form antiparallel homodimers via an interface residing in its C-terminal domain (My13) (11), myomesin acts as a cross-linker of myosin in the M band and its presence is crucial for proper M-band organization (12). Ehler et al. have shown that, together with the C-terminal part of titin, myomesin is a requirement for the integration of myosin into the sarcomere; they further suggest that myomesin in the M band, α-actinin in the Z disk, and titin in between form the basic stabilizing structure of the sarcomere (13). This implicates that myomesin is one of the key factors in maintaining the structural integrity of the M band under load.
During normal muscle operation, the M band and consequently myomesin will constantly be subjected to stress and strain imposed by muscle contraction and relaxation (14, 15). Hence its elastic properties are crucial (15). A special isoform of myomesin predominantly expressed in embryonic heart muscle (EH isoform) contains a long unstructured repeat of amino acids. It has been shown that this insert acts as an entropic spring providing significant elasticity to the EH isoform of myomesin (16, 17). However, the most prevalent isoforms of myomesin lack the specific EH insert. The extensibility of a rigid rod structure consisting of Ig and FnIII domain repeats is limited and therefore stress would be directly transmitted to the dimerization bond of myomesin possibly jeopardizing its structural integrity.
Recent structural studies have revealed a long freestanding α-helical linker connecting the C-terminal Ig domains My12 and My13 of myomesin (18). Such a linker motif is unique in Ig-repeat proteins (18). However, secondary structure prediction suggests that this motif is present between all five C-terminal Ig domains My9–My13 (18). It has therefore been speculated that the α-helical linker segments of myomesin may provide the necessary elasticity for the C-terminal part of this molecule (18). In this study we use high-resolution single-molecule force spectroscopy with the atomic force microscope (AFM) to reveal the unique mechanical characteristics of the My12–My13 linker and its contributions to the elastic properties of the myomesin molecule.
Results
Mechanical Stability of My12 Homooctamers.
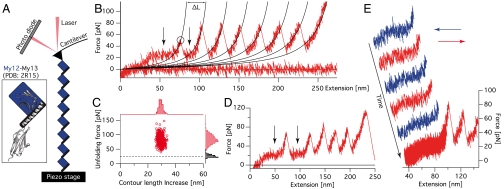
AFM force spectroscopy was performed on a homooctameric construct (My12)8 consisting of a series of eight identical My12 domains, each including the α-helical linker at its C terminus (compare to Fig. 1A). Typical force-extension traces exhibit saw-tooth like patterns as given in Fig. 1B. The contour length increase ΔL = 29.2 ± 0.06 nm (n = 583) is in good agreement with the expected unfolding length of an Ig domain containing 87 residues (Fig. 1C). The force distribution (average unfolding forces of 82.9 ± 0.7 pN, n = 583) is broad and within a typical regime for the unfolding of Ig domains, comparable to the unfolding of Dictyostelium discoideum filamin and about a third of the forces needed to unfold titin Ig domains (19, 20). At the beginning of the unfolding traces, deviating from the typical observed force-extension behavior due to entropic polymer elasticity (19) (black lines in force-extension traces are worm-like chain fits), regions of apparently constant force can be detected (see arrows in Fig. 1B). These plateau regions occur within a relatively narrow force range around 24.0 ± 0.5 pN (n = 72; dashed line in black histogram in Fig. 1C). In contrast to the typical mechanical unfolding of protein domains far from equilibrium, this force plateau reappears between peaks if the force drops below the plateau value after the unfolding of an Ig domain (second arrow in Fig. 1B). This becomes especially clear in the rare cases when two Ig domains unfold coincidentally (arrows in Fig. 1D). To test the reversibility of the plateau behavior, we performed a series of subsequent stretching and relaxation cycles on individual (My12)8 molecules. The plateau exhibits no observable hysteresis between stretch and relaxation cycles up to the highest velocity measured of 1 μm/s (Fig. 1E). From these observations we conclude that the elongation/shortening transitions underlying this force plateau occur fast compared to the experimental time scale.
Fig. 1.
Force spectroscopy on My12 homooctamers. (A) Schematic representation of a My12 homooctamer in the AFM experimental setup (not to scale). The blue squares and the black bars represent the My12 Ig domains and their linker helices, respectively (compare to Inset with cartoon representation of the My12-My13 structure) (B) Typical force-extension trace of (My12)8 unfolding. The circle marks a single unfolding event; black traces represent worm-like chain fits providing the contour length increases ΔL of a single unfolding. The arrows indicate the force plateau. (C) Scatter plot of unfolding forces and corresponding contour length increases with respective distributions (red). The black histogram gives the plateau force distribution and the dashed line its mean value. (D) (My12)8 trace with coincidental double Ig-domain unfolding. The force plateau (arrows) can be observed before and after this event. (E) Force-extension trace containing stretch (red) and relax (blue) cycles to test the reversibility of the plateau. In the final stretching cycle unfoldings of the Ig domains can be observed.
We next estimated the length contribution of an individual unraveling α-helix to the total plateau length. It is important to note that for this estimation only those traces were used where the last peak (reflecting detachment of the protein from cantilever or substrate) was significantly higher than all the preceding peaks (unfolding events); only then it is safe to assume that every single domain has been unfolded resulting in a measurable force peak. Moreover, the length of the plateau can only be measured if nonspecific interactions are absent at the onset of the curves, which further reduces the numbers of usable traces. From the traces fulfilling these requirements (n = 19) the plateau length contribution per domain could be estimated to be 6.1 ± 0.2 nm. This value is in excellent agreement with the calculated contour length increase for the detachment and unfolding of the My12 helix of 6.3 nm (SI Text).
The forces for unfolding the specific α-helical linker are surprisingly high considering that even double-helical coiled-coil structures, triple-helical spectrin repeats, and parallel-stacked α-helical ankyrin repeats exhibit unfolding forces in a similar range (21–23). To investigate the determinants for helix stability in myomesin, we compared stretch-relax cycles at two different extensions in additional experiments. We find that the plateau only appears if the Ig domains are intact and disappears if the Ig domains are unfolded (Fig. S1). This result shows that the ability of an individual helix to fold and unfold against forces as high as 24 pN depends on the presence of its associated Ig-domain interface.
The α-Helix Linker Is an Extensible Element.
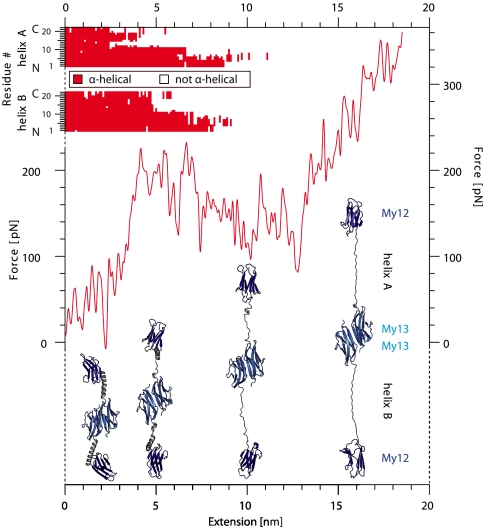
To understand the structural nature of the plateau in the force-extension traces, we performed force-probe molecular dynamics (MD) simulations. A force-extension curve of a myomesin My12–My13 dimer is shown in Fig. 2 (red curve). Beyond extensions of 4 nm a plateau region appears in which the molecule is extended at constant force; beyond 13 nm the force again continues to rise. A series of snapshots of the associated molecular conformations reveals the nature of the force plateau: Whereas all four Ig domains remain intact, the α-helical linkers start to unfold in the plateau region. In our simulations α-helix unfolding starts at those segments of the helix that are solvent exposed (C-terminal part; compare to Fig 2, Top). Part of this region has already unfolded when the plateau force is reached. The other segment of the α-helix forming a hydrophobic interface with the Ig domains is more stable and unfolds later in the plateau. It is important to note that the simulation timescales are much shorter than the experimental timescale and hence, forces from these two methods cannot be directly compared to each other. However, the mechanical hierarchy within the molecule is likely conserved (4, 24), as also suggested by the qualitative agreement between the experimental and calculated force profiles, which both feature a plateau followed by a steep increase in force.
Fig. 2.
Force-probe molecular dynamics simulation of My12-My13 dimer unfolding. The red force-extension trace clearly exhibits a plateau. Corresponding structural snapshots at 2, 5, 10, and 16 nm are given below in cartoon representation and show that the plateau corresponds to α-helix unfolding. The individual conformational state of both helices is mapped for each residue against extension (Top).
Reversible Single Helix Unfolding Dynamics Under Load.
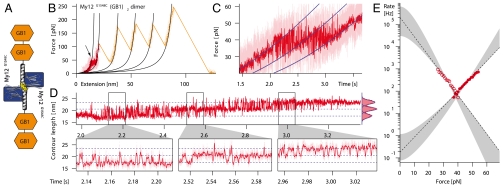
To measure the dynamics of folding and unfolding of individual helices we designed a molecular construct that ensures that force is only applied to the α-helical linker. In this construct the associated Ig domain is not exposed to load and can continuously provide the interface required for the folding/unfolding transition of the helix. To this end we introduced a cysteine residue at the N-terminal end of the α-helix (G1548C), which, at the same time, marks the C terminus of the associated Ig-domain My12. To provide attachment sites to tip and substrate, we fused two immunoglobulin-binding domains B1 of streptococcal protein G (GB1) next to the C terminus of the helix. The mechanics of GB1 domains have been characterized in detail (25). The respective construct My12G1548C(GB1)2 can form a dimer through the cysteine at position 1548. A schematic of the construct is shown in Fig. 3A. Thus, by stretching the dimer, force is applied only to the two α-helices and the GB1-handles, but not to My12 (see Fig 3A).
Fig. 3.
Fast and reversible unfolding transitions of the My12 α-helical linker. (A) Schematics of the construct. The cysteine mutation is colored yellow. (B) Typical force-extension trace of My12G1548C(GB1)2 dimer unfolding. The light trace is the original data, the dark trace is filtered. In the red part the pulling velocity was 5 nm/s, in the orange part 100 nm/s. The first four peaks correspond to GB1 unfolding, the last one to detachment of the sample. Black lines are worm-like chain fits. The arrow marks the region of fast transitions. (C) Zoom into transitions region. In a representation where the data are plotted as force vs. time rapid transitions between folded and unfolded states are clearly visible. Blue lines follow worm-like chain elasticity. (D) (Upper) The transitions region plotted as (filtered) contour length against time. Blue dashed lines correspond to worm-like chain traces in C. The corresponding contour length histogram (Right) is well described by three Gaussians (blue lines). (Lower) Three zooms into the indicated regions of the trace above. (E) Force-dependent folding (open circles) and unfolding (filled circles) rates as obtained from the analysis of the transition traces (compare to SI Materials and Methods). The gray areas illustrate the variability of meaningful extrapolations to zero-force following the model of Schlierf et al. (32, 33), the dashed lines are linear fits according to the Bell model (30, 31).
A typical force-extension trace is given in Fig. 3B. The major unfolding peaks correspond to the GB1 domains with expected values for unfolding force and contour length increase (25). Because the stability of GB1 domains exceeds helix stability, we expect unfolding of the two cross-linked helices to occur at the onset of the pulling curve. Indeed, a region of fast transitions at about 40 pN before the first unfolding peak can be observed (see arrow in Fig. 3B). In this extension range, we performed measurements at very low pulling velocities of 5–10 nm/s to increase the amount of data in the relevant force window. A zoom into the plateau region of the force curve is provided in Fig. 3C. For clarity, the data in this graph are plotted as force versus time and show rapid equilibrium transitions between three states with distinct contour lengths (blue lines). The different states can be interpreted as both helices folded, one helix unfolded, or both helices unfolded, respectively.
To visualize the transitions between the three states in a more direct way, we chose to plot the time series data as contour length vs. time (Fig. 3D; for additional traces, see Fig. S2). This procedure has been suggested by Puchner et al. (26) and transforms each force-extension data point into contour length space by solving the worm-like chain formula for the contour length. Instead of the tilted blue levels in Fig. 3C, we now obtain vertical transitions between strictly horizontal levels (Fig. 3D; note that an unfolding event leads to a decrease in force, but to an increase in contour length). During pulling, a slow force ramp is applied to the myomesin chimera (compare to Fig. 3C) and hence the population probability shifts over time from the fully folded initial state to the fully unfolded final state. The three zooms at the bottom show different regions of the transition zone (Fig. 3D, Bottom). The contour lengths of the three states are well fit by Gaussian distributions (Fig. 3D, Top Right); the mean contour length increase for one unfolding transition is 3.05 ± 0.06 nm (n = 32). This number is significantly shorter than the expected contour length increase for complete helix unfolding of 4.7 nm. Note that the expected contour length increase is different from the earlier value because of the change in the direction of force application. Instead of pulling the helix off the folded Ig domain, we now apply force along the axis of the helix. A contour length change of 3.05 nm, however, would correspond to unfolding/refolding transitions of only the N-terminal two thirds of the helix (G1548–K1562), which is just the part of the helix that has a hydrophobic interface with My12 and that is terminated by a hydrogen bond between K1562 and E1520. The rest of the helix protrudes from the domain and is entirely solvent exposed, which apparently results in a lower mechanical stability. This result is in full agreement with our force-probe MD simulations (Fig. 2). The difference in plateau force we observe between this construct (approximately 40 pN) and the homooctameric protein (24 pN; Fig. 1C) can also be explained by the differences in pulling geometry in those two experiments. In equilibrium, the smaller length gain in the pulling geometry along the helix axis requires a higher average unfolding force, because the free energy difference between the folded and the unfolded state likely remains the same.
To quantify the force-dependent helix dynamics, a Hidden Markov Model (HMM) (27, 28) with force-dependent transition rates was applied to the contour length traces (Fig. S2, top trace) to objectively attribute every data point to a unique state. Using a method described by Oberbarnscheidt et al. (29), the force-dependent transition rates for unfolding and refolding of a single helix in presence of its folded domain were extracted from the HMM reconstructions. The rates are given in a Chevron plot like graph in Fig. 3E, with the force acting as denaturant. The open and closed circles are shifted with respect to each other because unfolding and refolding proceed from different starting forces determined by the cantilever spring constant. The force dependency of the rates is linear to good approximation over the experimentally accessible range. Several models can be used to extrapolate this data over the whole range of forces. The roughest approximation is the Bell model (30, 31), which is fully linear (dashed lines in Fig. 3E). In addition (gray shaded regions), we used a model introduced earlier (32, 33) that takes into account the finite extension of the folded structure as well as the energy contribution of the whole system including cantilever bending and entropic polymer spacer elasticity. The outer boundaries of the shaded regions are fits requiring the zero force rates to be consistent with the equilibrium free energy (16 kBT) as obtained from integrating the area under force vs. distance curve. The inner boundaries are a best fit with no additional thermodynamic constraint.
Mechanical Stability of the My13 Dimer Interface.
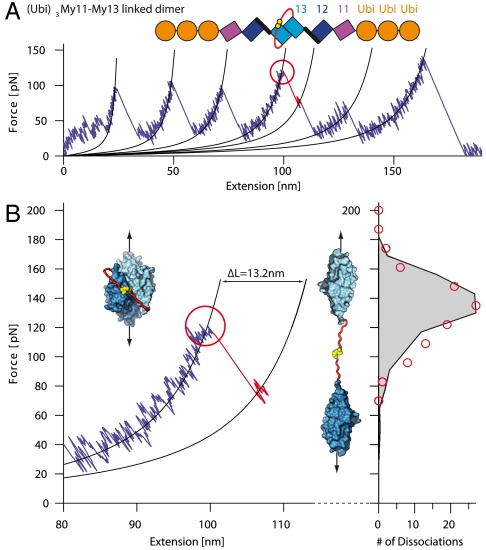
To assess the overall mechanical properties of myomesin, it is essential to evaluate also the stability of the myomesin dimerization. A quantitative measurement of the dissociation force of the dimer, however, requires a special experimental approach, because dissociation of the dimer is indistinguishable from the detachment of the protein from cantilever or substrate, with both appearing as the last peak in a force trace (34). To overcome this problem, we fused an unstructured linker consisting of nine residues (red in Fig. 4A, Top) with an additional C-terminal cysteine (yellow) to the My13 C terminus of a (Ubi)3My11–My13 monomer (see schematics in Fig. 4A; human ubiquitin domains (Ubi) serve as handles for attachment to the cantilever and the substrate). The cysteines at the termini of the linkers form a disulfide bridge that connects the two polypeptide chains even after the My13 dimer has been dissociated by force. Dissociation of the dimer stretches the flexible polypeptide linker, which, together with the reorientation of the separated My13 domains, will lead to an expected length increase of 13.2 nm. A similar strategy has been successfully employed to measure unbinding forces of the titin-obscurin interaction (35). This approach permits the clear distinction between desorption and dimer dissociation events.
Fig. 4.
Forced dissociation of linked My13 dimer domains. (A) Typical force-extension trace of (Ubi)3My11-My13 linked dimers. A schematic representation of the construct is given on top. Each monomer consists of three ubiquitins (orange) and the myomesin domains My11-My13 (purple, blue, and cyan). The two monomers are linked via an unstructured amino acid sequence (red) with C-terminal cysteines (yellow) that form a covalent disulfide bond. Dimer dissociation (red circle) is identified by a shorter contour length increase than for domain unfolding. (B) Zoom into part of (A) where dimer dissociation occurs. Insets show structure of the My13 domains (cyan) with linker (red) and bonded cysteines (yellow) in the dimeric (Left) and dissociated state (Right). The dissociation force distribution to the right (red circles) is well reproduced by a Monte Carlo simulation (black trace).
A typical force trace of the specific protein construct is shown in Fig. 4A. Unfolding events of My11, My12, and ubiquitin could be identified by their unfolding forces and by their contour length increases (note that in the example trace in Fig. 4A detachment from the cantilever occurs before ubiquitin could be unfolded; for additional traces, see Fig. S3). A unique event (red circle), characterized by a contour length increase of ca. 13 nm and a short-lived subsequent dwell can be observed in the unfolding traces (see zoom in Fig. 4B). The contour length increase (13.1 ± 0.2 nm (n = 97); see Fig. S4) is identical with the expected increase upon dimer dissociation within resolution of our instrument. Dimer dissociation is always followed by two unfolding events of ca. 29 nm contour length increase that occur at significantly lower force than the other domain unfolding forces of similar length increase (see also Fig. S3). We attribute these events to the unfolding of the separated My13 monomers. The distribution of dissociation forces (red circles, Fig. 4B, to the right) is well reproduced by a Monte Carlo simulation with a dissociation rate of koff = 0.01 s-1 and a barrier width of Δx = 2.9 Å (Fig. 4B, Right, black line). The mean dimer dissociation force of 137 ± 2 pN (n = 97) is significantly higher than the most probable unfolding force of My12 of 83 pN (see Fig. 1C).
Discussion
For its role as a cross-linker of myosin filaments, the elastic properties of myomesin as well as the stability of its dimerization interface are important. In the absence of detailed structural insight, early mechanical measurements of myomesin have naturally focused on the stability of the fold of Ig and Fn domains within the myomesin rod (16, 17). In those earlier studies, in addition to the domain unfolding forces, the entropic elasticity of the EH segment, a putatively unstructured element containing ca. 100 amino acid residues, was investigated. However this segment only occurs in an isoform predominantly expressed in embryonic heart muscle. The sources of elasticity for the most commonly expressed myomesin isoforms lacking the unstructured segment have so far remained unclear. The recent discovery of a long α-helical linker segment in between My12 and My13 and putatively also in between the other C-terminal Ig domains as well as the detailed structure of the dimerization complex have raised the possibility of a unique elastic element within myomesin (18).
The data presented in this paper show that the C terminus of myomesin comprises three distinct mechanical elements that govern the overall elastic behavior of the protein: At low stretching forces, an interdomain α-helical linker unfolds and refolds rapidly and reversibly. When the force increases, Ig domains of the myomesin rod unfold followed by the dissociation of the dimerization interface.
The reversibility of the α-helical linker as elastic segment ensuring rapid contraction of the overstretched myomesin dimer becomes directly apparent in Fig. 3C. Already at the midpoint forces of 40 pN, folding/unfolding kinetics occurs at 100 s-1 (Fig. 3E). Extrapolating to zero force values, we arrive at refolding rates of the α-helix in the order of 105–106 s-1. Even though this extrapolation is not unambiguous due to the reasons mentioned in the results section, this number is consistent with values determined for α-helix formation in solution (36, 37). It is interesting to note that in the Ig domain I27 from titin, Marszalek et al. reported a short β-strand segment that detaches from the domain core in a close to equilibrium transition prior to its complete unfolding (24). This equilibrium transition has been shown to act as a force buffer minimizing the probability of titin Ig unfolding at low loading rates (38). Whereas this β-strand interacts with the rest of the Ig domain through hydrogen bonds, in myomesin, the nature of the interactions between helix and domain core is a mixture between hydrophobic and hydrogen bond interactions (18).
The equilibrium force-extension curves (Fig. 3 B–D) allow us to determine the gain in free energy associated with the formation of the α-helix by integrating force vs. extension under the curve. The obtained value of 16 kBT is substantial even compared to values for folding of much larger domains. Typical folding free energies for Ig domains are in range of 4–15 kBT (39, 40). Hence, nature uses a substantial amount of interaction free energy to design a rapidly folding element that can reversibly extend under load.
Small force imbalances due to statistical variations in the number of force-generating actin-bindings on either half of a myosin filament have been suggested to result in axial displacement of myosin filaments (15, 41, 42). Imbalances of four to eight myosin motor domains lead up to forces of a few tens of piconewtons and displacements in the order of 10 nm (4). This results in axial shear strain on the M band and its components like myomesin. Intriguingly, according to current models of the M-line structure, myomesin is oriented along the longitudinal axis of the sarcomere filaments only with its C-terminal part (My7–My13) (15). This C-terminal portion of the myomesin dimer then spans the region between the M4 and M4′ line being a constituent of the M filament as seen in EM micrographs (43–45). Toward the N terminus, myomesin is oriented rather perpendicular to the sarcomere long axis and features binding sites for titin and/or obscurin (9, 10, 15). Hence, longitudinal mechanical stress acting on the C-terminal repeats of myomesin as well as on the dimer interface will likely be distributed over a complex network of filament interactions involving titin, obscurin, myosin, and the N-terminal part of myomesin (9, 10, 46). Even though the details of the mechanical interactions among those filaments are just emerging (35), they likely provide an overall stable anchoring of the myomesin filament also at its N terminus.
The dimer unbinding forces of 137 pN set a clear limit for the force range a myomesin molecule can bear in vivo. It is important to note that whereas this value lies in the upper range of My12 Ig domain unfolding forces, the two force distributions still overlap due to the stochastical nature of mechanical protein unfolding (see Fig. 4B, in comparison with Fig. 1C). This can be observed in the sample traces in Fig. S3 where dimer dissociation often occurs before all Ig domains are unfolded. For the I-band part of titin, unfolding of Ig domains under load has been postulated as a mechanism to compensate extreme stresses in the I band (47). Whereas the much higher anchoring forces of titin in the Z disk are consistent with such a scenario (48), it appears highly unlikely that a single myomesin molecule will routinely experience forces high enough for Ig-domain unfolding because the risk of dimer breakage would be too large.
Generally, an elastic mechanism based on nonequilibrium processes such as Ig-domain unfolding exhibits broad force distributions and can therefore not supply sharply defined force values. Equilibrium unfolding/refolding of the α-helical linker combines two essential features for a reliable elastic mechanism: the possibility for considerable elongation at force values far below dimer dissociation at the level of 30 pN whereas still providing stability and rigidity at forces below 20 pN (from the parameters obtained in the Monte Carlo simulation (Fig. 4B, Right) we estimate a lifetime of 12 s for the dimer interface at the plateau force). At first sight, the possible elongation upon single α-helix unfolding may appear small. However, secondary structure prediction suggests that α-helices are present between all five C-terminal Ig domains My9–My13 (18). Hence, full extension of the supposed eight helices in a dimer can elongate the molecule by approximately 50 nm, which corresponds to 50% of the total folded length of the myomesin dimer as roughly estimated by assuming a diameter of 4 nm per domain. This provides myomesin with enough adaptability to react to misalignment or changes in spacing of thick filaments (15). When tension subsides after muscle contraction, in contrast to the titin PEVK or myomesin EH segments, the fast reversibility of My12 α-helix unfolding maintains a restoring force at the plateau level until the helix has refolded, thus actively contributing to the recovery of the highly organized M-band alignment.
In summary, we show that the unique α-helical linker found between the C-terminal Ig domains My12 and My13 in myomesin has mechanical properties that make it ideally suited to act as a strain absorber in the M band. The fast and reversible two-state-folding kinetics protect the stability of the myomesin dimer under load up to strains of presumably 150%. The mechanical properties of myomesin elasticity presented here form an important building block for the emerging mechanical and structural understanding of the M band.
Materials and Methods
All constructs were cloned and expressed by standard molecular biology techniques. Single-molecule force spectroscopy was performed on a custom-built high-resolution AFM setup using procedures described before (49, 50). For MD simulations the structure was solvated with water in a rectangular box; force-probe MD simulations (51) were executed using GROMACS 3.3.1 and the OPLS-AA force field (52, 53). Analysis of the force-dependent transition rate follows a protocol where the force-extension data are transformed to contour length vs. time (26) and filtered using a customized HMM (27, 28); the resulting conformational state sequence is then analyzed following a method introduced by Oberbarnscheidt et al. (29).
Complete descriptions of the methods used are given in SI Materials and Methods.
Supplementary Material
Acknowledgments.
This work was supported by a Deutsche Forschungsgemeinschaft FOR1352 (P8) Grant (M.R.). F.B. was supported by CompInt in the framework of Elitenetzwerk Bayern.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105734108/-/DCSupplemental.
References
- 1.Au Y. The muscle ultrastructure: A structural perspective of the sarcomere. Cell Mol Life Sci. 2004;61:3016–3033. doi: 10.1007/s00018-004-4282-x. [DOI] [PubMed] [Google Scholar]
- 2.Gregorio CC, Granzier H, Sorimachi H, Labeit S. Muscle assembly: A titanic achievement? Curr Opin Cell Biol. 1999;11:18–25. doi: 10.1016/s0955-0674(99)80003-9. [DOI] [PubMed] [Google Scholar]
- 3.Li H, et al. Reverse engineering of the giant muscle protein titin. Nature. 2002;418:998–1002. doi: 10.1038/nature00938. [DOI] [PubMed] [Google Scholar]
- 4.Puchner EM, et al. Mechanoenzymatics of titin kinase. Proc Natl Acad Sci USA. 2008;105:13385–13390. doi: 10.1073/pnas.0805034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Granzier HL, Labeit S. Titin and its associated proteins: The third myofilament system of the sarcomere. Adv Protein Chem. 2005;71:89–119. doi: 10.1016/S0065-3233(04)71003-7. [DOI] [PubMed] [Google Scholar]
- 6.Grove BK, et al. A new 185,000-dalton skeletal muscle protein detected by monoclonal antibodies. J Cell Biol. 1984;98:518–524. doi: 10.1083/jcb.98.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Squire JM, Al-Khayat HA, Knupp C, Luther PK. Molecular architecture in muscle contractile assemblies. Adv Protein Chem. 2005;71:17–87. doi: 10.1016/S0065-3233(04)71002-5. [DOI] [PubMed] [Google Scholar]
- 8.Obermann WM, Plessmann U, Weber K, Fürst DO. Purification and biochemical characterization of myomesin, a myosin-binding and titin-binding protein, from bovine skeletal muscle. Eur J Biochem. 1995;233:110–115. doi: 10.1111/j.1432-1033.1995.110_1.x. [DOI] [PubMed] [Google Scholar]
- 9.Obermann WM, Gautel M, Weber K, Fürst DO. Molecular structure of the sarcomeric M band: Mapping of titin and myosin binding domains in myomesin and the identification of a potential regulatory phosphorylation site in myomesin. EMBO J. 1997;16:211–220. doi: 10.1093/emboj/16.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukuzawa A, et al. Interactions with titin and myomesin target obscurin and obscurin-like 1 to the M-band: implications for hereditary myopathies. J Cell Sci. 2008;121:1841–1851. doi: 10.1242/jcs.028019. [DOI] [PubMed] [Google Scholar]
- 11.Lange S, et al. Dimerisation of myomesin: Implications for the structure of the sarcomeric M-band. J Mol Biol. 2005;345:289–298. doi: 10.1016/j.jmb.2004.10.040. [DOI] [PubMed] [Google Scholar]
- 12.Potthoff MJ, et al. Regulation of skeletal muscle sarcomere integrity and postnatal muscle function by Mef2c. Mol Cell Biol. 2007;27:8143–8151. doi: 10.1128/MCB.01187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehler E, Rothen BM, Hämmerle SP, Komiyama M, Perriard JC. Myofibrillogenesis in the developing chicken heart: Assembly of Z-disk, M-line, and the thick filaments. J Cell Sci. 1999;112:1529–1539. doi: 10.1242/jcs.112.10.1529. [DOI] [PubMed] [Google Scholar]
- 14.Agarkova I, Ehler E, Lange S, Schoenauer R, Perriard J-C. M-band: A safeguard for sarcomere stability? J Muscle Res Cell Motil. 2003;24:191–203. doi: 10.1023/a:1026094924677. [DOI] [PubMed] [Google Scholar]
- 15.Agarkova I, Perriard J-C. The M-band: An elastic web that crosslinks thick filaments in the center of the sarcomere. Trends Cell Biol. 2005;15:477–485. doi: 10.1016/j.tcb.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Bertoncini P, et al. Study of the mechanical properties of myomesin proteins using dynamic force spectroscopy. J Mol Biol. 2005;348:1127–1137. doi: 10.1016/j.jmb.2005.03.040. [DOI] [PubMed] [Google Scholar]
- 17.Schoenauer R, et al. Myomesin is a molecular spring with adaptable elasticity. J Mol Biol. 2005;349:367–379. doi: 10.1016/j.jmb.2005.03.055. [DOI] [PubMed] [Google Scholar]
- 18.Pinotsis N, Lange S, Perriard J-C, Svergun DI, Wilmanns M. Molecular basis of the C-terminal tail-to-tail assembly of the sarcomeric filament protein myomesin. EMBO J. 2008;27:253–264. doi: 10.1038/sj.emboj.7601944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 20.Schwaiger I, Schleicher M, Noegel AA, Rief M. The folding pathway of a fast-folding immunoglobulin domain revealed by single-molecule mechanical experiments. EMBO Rep. 2005;6:46–51. doi: 10.1038/sj.embor.7400317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwaiger I, Sattler C, Hostetter DR, Rief M. The myosin coiled-coil is a truly elastic protein structure. Nat Mater. 2002;1:232–235. doi: 10.1038/nmat776. [DOI] [PubMed] [Google Scholar]
- 22.Rief M, Pascual J, Saraste M, Gaub HE. Single molecule force spectroscopy of spectrin repeats: Low unfolding forces in helix bundles. J Mol Biol. 1999;286:553–561. doi: 10.1006/jmbi.1998.2466. [DOI] [PubMed] [Google Scholar]
- 23.Li L, Wetzel S, Plückthun A, Fernandez JM. Stepwise unfolding of ankyrin repeats in a single protein revealed by atomic force microscopy. Biophys J. 2006;90:L30–32. doi: 10.1529/biophysj.105.078436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marszalek PE, et al. Mechanical unfolding intermediates in titin modules. Nature. 1999;402:100–103. doi: 10.1038/47083. [DOI] [PubMed] [Google Scholar]
- 25.Cao Y, Lam C, Wang M, Li H. Nonmechanical protein can have significant mechanical stability. Angew Chem Int Ed Engl. 2006;45:642–645. doi: 10.1002/anie.200502623. [DOI] [PubMed] [Google Scholar]
- 26.Puchner EM, Franzen G, Gautel M, Gaub HE. Comparing proteins by their unfolding pattern. Biophys J. 2008;95:426–434. doi: 10.1529/biophysj.108.129999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabiner L, Juang B. An introduction to hidden Markov models. IEEE ASSP Mag. 1986;3:4–16. [Google Scholar]
- 28.Eddy SR. What is a hidden Markov model? Nat Biotechnol. 2004;22:1315–1316. doi: 10.1038/nbt1004-1315. [DOI] [PubMed] [Google Scholar]
- 29.Oberbarnscheidt L, Janissen R, Oesterhelt F. Direct and model free calculation of force-dependent dissociation rates from force spectroscopic data. Biophys J. 2009;97:L19–21. doi: 10.1016/j.bpj.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 31.Evans E, Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlierf M, Berkemeier F, Rief M. Direct observation of active protein folding using lock-in force spectroscopy. Biophys J. 2007;93:3989–3998. doi: 10.1529/biophysj.107.114397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gebhardt JCM, Bornschlögl T, Rief M. Full distance-resolved folding energy landscape of one single protein molecule. Proc Natl Acad Sci USA. 2010;107:2013–2018. doi: 10.1073/pnas.0909854107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertz M, et al. Structural and mechanical hierarchies in the alpha-crystallin domain dimer of the hyperthermophilic small heat shock protein Hsp16.5. J Mol Biol. 2010;400:1046–1056. doi: 10.1016/j.jmb.2010.05.065. [DOI] [PubMed] [Google Scholar]
- 35.Pernigo S, et al. Structural insight into M-band assembly and mechanics from the titin-obscurin-like-1 complex. Proc Natl Acad Sci USA. 2010;107:2908–2913. doi: 10.1073/pnas.0913736107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eaton WA, Muñoz V, Thompson PA, Chan CK, Hofrichter J. Submillisecond kinetics of protein folding. Curr Opin Struct Biol. 1997;7:10–14. doi: 10.1016/s0959-440x(97)80003-6. [DOI] [PubMed] [Google Scholar]
- 37.Kubelka J, Hofrichter J, Eaton WA. The protein folding “speed limit”. Curr Opin Struct Biol. 2004;14:76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 38.Nunes JM, et al. A “force buffer” protecting immunoglobulin titin. Angew Chem Int Ed Engl. 2010;49:3528–3531. doi: 10.1002/anie.200906388. [DOI] [PubMed] [Google Scholar]
- 39.Politou AS, Thomas DJ, Pastore A. The folding and stability of titin immunoglobulin-like modules, with implications for the mechanism of elasticity. Biophys J. 1995;69:2601–2610. doi: 10.1016/S0006-3495(95)80131-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carrion-Vazquez M, et al. Mechanical and chemical unfolding of a single protein: A comparison. Proc Natl Acad Sci USA. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huxley HE, Faruqi AR, Kress M, Bordas J, Koch MH. Time-resolved X-ray diffraction studies of the myosin layer-line reflections during muscle contraction. J Mol Biol. 1982;158:637–684. doi: 10.1016/0022-2836(82)90253-4. [DOI] [PubMed] [Google Scholar]
- 42.Linari M, et al. Interference fine structure and sarcomere length dependence of the axial X-ray pattern from active single muscle fibers. Proc Natl Acad Sci USA. 2000;97:7226–7231. doi: 10.1073/pnas.97.13.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knappeis GG, Carlsen F. The ultrastructure of the M line in skeletal muscle. J Cell Biol. 1968;38:202–211. doi: 10.1083/jcb.38.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luther P, Squire J. Three-dimensional structure of the vertebrate muscle M-region. J Mol Biol. 1978;125:313–324. doi: 10.1016/0022-2836(78)90405-9. [DOI] [PubMed] [Google Scholar]
- 45.Obermann WM, et al. The structure of the sarcomeric M band: localization of defined domains of myomesin, M-protein, and the 250-kD carboxy-terminal region of titin by immunoelectron microscopy. J Cell Biol. 1996;134:1441–1453. doi: 10.1083/jcb.134.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gautel M. The sarcomeric cytoskeleton: Who picks up the strain? Curr Opin Cell Biol. 2011;23:39–46. doi: 10.1016/j.ceb.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 47.Linke WA, Fernandez JM. Cardiac titin: molecular basis of elasticity and cellular contribution to elastic and viscous stiffness components in myocardium. J Muscle Res Cell Motil. 2002;23:483–497. doi: 10.1023/a:1023462507254. [DOI] [PubMed] [Google Scholar]
- 48.Bertz M, Wilmanns M, Rief M. The titin-telethonin complex is a directed, superstable molecular bond in the muscle Z-disk. Proc Natl Acad Sci USA. 2009;106:13307–13310. doi: 10.1073/pnas.0902312106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwaiger I, Kardinal A, Schleicher M, Noegel AA, Rief M. A mechanical unfolding intermediate in an actin-cross-linking protein. Nat Struct Mol Biol. 2004;11:81–85. doi: 10.1038/nsmb705. [DOI] [PubMed] [Google Scholar]
- 50.Junker JP, Ziegler F, Rief M. Ligand-dependent equilibrium fluctuations of single calmodulin molecules. Science. 2009;323:633–637. doi: 10.1126/science.1166191. [DOI] [PubMed] [Google Scholar]
- 51.Grubmüller H, Heymann B, Tavan P. Ligand binding: Molecular mechanics calculation of the streptavidin-biotin rupture force. Science. 1996;271:997–999. doi: 10.1126/science.271.5251.997. [DOI] [PubMed] [Google Scholar]
- 52.Kutzner C, et al. Speeding up parallel GROMACS on high-latency networks. J Comput Chem. 2007;28:2075–2084. doi: 10.1002/jcc.20703. [DOI] [PubMed] [Google Scholar]
- 53.Jorgensen WL, Tirado-Rives J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.