Abstract
After the first report of induced pluripotent stem cells (iPSCs), considerable efforts have been made to develop more efficient methods for generating iPSCs without foreign gene insertions. Here we show that Sendai virus vector, an RNA virus vector that carries no risk of integrating into the host genome, is a practical solution for the efficient generation of safer iPSCs. We improved the Sendai virus vectors by introducing temperature-sensitive mutations so that the vectors could be easily removed at nonpermissive temperatures. Using these vectors enabled the efficient production of viral/factor-free iPSCs from both human fibroblasts and CD34+ cord blood cells. Temperature-shift treatment was more effective in eliminating remaining viral vector-related genes. The resulting iPSCs expressed human embryonic stem cell markers and exhibited pluripotency. We suggest that generation of transgene-free iPSCs from cord blood cells should be an important step in providing allogeneic iPSC-derived therapy in the future.
Keywords: regenerative medicine, nonintegrating RNA vector
Safer and more efficient reprogramming methods have been explored since the first report of the generation of human induced pluripotent stem cells (iPSCs) (1, 2). Toward this end, several techniques have been used for obtaining integration and/or transgene-free iPSCs, including the use of plasmids (3, 4), the Cre/loxP system (5, 6), adenoviruses (7), piggyBac (8, 9), minicircle vector (10), and proteins (11, 12). However, these methods suffer from low efficiency, require repetitive induction, and/or produce insufficient excision of integrated vectors. Synthetic modified mRNA may solve the problem, but the reagents must be added every day (13). Thus, more efficient and simple methods are needed to generate human iPSCs with no noise of integration or remaining factors for both clinical applications and basic studies.
An alternative technology involves the use of Sendai virus (SeV) vectors. SeV, a member of the Paramyxovirdae family, is an enveloped virus with a single-stranded, negative-sense, nonsegmented RNA genome of ∼15 kb (14). Importantly, recombinant SeV vectors replicate only in the cytoplasm of infected cells and do not go through a DNA phase or integrate into the host genome (15). SeV vectors have proven to be efficient for the introduction of foreign genes in a wide spectrum of host cell species and tissues, and SeV vectors have been studied for applying to clinical studies of gene therapy for cystic fibrosis, critical limb ischemia, vaccines for AIDS, and other areas (reviewed in ref. 16). We previously reported that the SeV vectors efficiently generate human iPSCs from human fibroblasts (17) and human blood cells (18). However, to apply SeV vector technology to the generation of safer iPSCs, the issue of the sustained cytoplasmic replication of viral vectors after the iPSCs have been established had to be overcome, even though viral vectors are slowly diluted during the robust cell division of iPSCs and SeV vector–positive cells can be removed using an anti–SeV-HN antibody (17). In other words, a more efficient shutdown of viral replication is needed to generate human iPSCs. We considered the use of temperature-sensitive (TS) SeV vectors the likeliest best solution.
Here we show that by introducing point mutations in the polymerase-related genes, we obtained new TS vectors and efficiently generated viral/factor-free human iPSCs from fibroblasts using these vectors by two strategies, (i) replacing the MYC vector only and (ii) replacing all reprogramming vectors to the TS vectors. We also applied this method to CD34+ cord blood (CB) cells because SeV vector provides a highly efficient gene transfer into human CB-derived hematopoietic stem cells (19). CD34+ CB cells are the youngest somatic stem cells and are expected to have no postnatal genomic aberration by irritants from the environment or UV rays. They correspond to the hematopoietic stem cells and progenitors with less epigenetic modification related to hematopoietic differentiation. These unique features of CD34+ CB cells suggest that this cell fraction might be an ideal cell source fraction for generating a gold standard iPSC. However, the risk of foreign gene integration (20, 21) needs to be overcome for future clinical applications. We successfully obtained viral/factor-free CB-iPSCs by the TS SeV vectors, and examined the advantages of using the TS SeV vector.
Results
Generation of TS SeV Vectors.
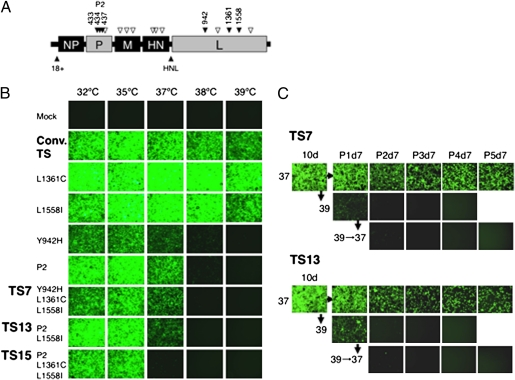
The SeV RNA polymerase comprises the phosphoprotein (P) and the large protein (L), and formation of the P-L complex is required for RNA synthesis (14). Mutations in P or L have been shown to confer temperature sensitivity to the virus (22, 23). Although a conventional nontransmissible F protein-deficient (ΔF)/TS vector was demonstrated to have low cytotoxicity at temperatures above 37 °C (24), it still expressed the GFP gene at 39 °C, albeit at slightly lower levels (Fig. 1B). Thus, we generated a greater number of TS vectors using combination of known point mutations in the P and/or L genes and screened for GFP gene expression in infected cells at various temperatures. The TS vectors obtained were (i) P2 vectors (D433A, R434A, and K437A), which contain a charge-to-alanine mutation in the L-binding domain of the P protein (22); (ii) TS7 vectors (Y942H, L1361C, and L1558I); (iii) TS13 vectors (P2 and L1558I); and (iv) TS15 vectors (P2, L1361C, and L1558I), as indicated in Fig. 1. In the present study, we chose to evaluate these candidate vectors with combined mutations, because a single mutation appeared to be insufficient to confer temperature sensitivity (i.e., the Y942H, L1558I, or L1361C vector). Furthermore, by using TS candidates with combined mutations, the occurrence of WT revertant, as occasionally observed in RNA viruses (14), was less likely. In contrast with the conventional SeV vector, the TS7 and TS13 vectors expressed GFP at 32 °C and 35 °C, and weakly at 37 °C, but not at nonpermissive temperatures of 38 °C or 39 °C. The TS15 vector exhibited greater temperature sensitivity; with this vector, GFP expression was barely detected at 37 °C (Fig. 1B). We confirmed that there was no GFP expression after transfection of cells with the TS7 and TS13 vectors after temperature-shift treatment to 39 °C, even when the infected cells were then cultured at 37 °C for >1 mo with several passages (Fig. 1C). None of the vectors was cytotoxic, and all infected cells were attached and live, with or without temperature-shift treatment. Thus, we used these TS vectors to generate human iPSCs.
Fig. 1.
Generation of TS SeV vectors and inactivation after temperature-shift treatment. (A) Point mutations were introduced into the polymerase-related genes P (P2: 433, 434, and 437) and/or L (942, 1361, and 1558), as indicated in the schematic structure of the ΔF/SeV vector. Open angles indicate conventional mutations in the previous TS vector; closed angles, newly introduced mutations. (B) Confluent LLC-MK2 cells were transduced with each SeV vector carrying GFP at an MOI of 5 and cultured at the indicated temperatures (32, 35, 37, 38, and 39 °C). Green fluorescence was compared at 3 d after infection. (C) To confirm the irreversible inactivation of gene expression by temperature-shift treatment, infected cells were cultured at 37 °C for 10 d and then split into two groups, one group cultured at 37 °C and the other cultured at 39 °C for 28 d, with cells passaged every 7 d. Similarly, cells infected with a TS vector treated at a nonpermissive temperature of 39 °C for 7 d were also cultured for a further 28 d at 37 °C, with cells passaged every 7 d, to evaluate GFP expression.
Generation of Human iPSCs with TS SeV Vectors from Fibroblasts.
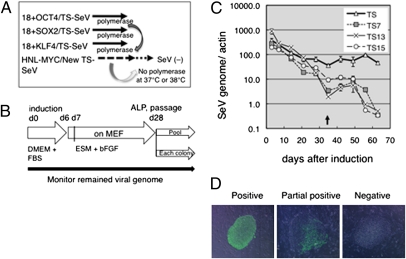
To apply the TS vectors to generate human iPSCs, we adopted two strategies, (i) to replace only c-MYC–carrying vector and (ii) to replace all of the four gene-carrying SeV vector mixtures to the TS vectors. Our previous study showed that the exogenous c-MYC inserted between the HN and L positions in the SeV vector (HNL-MYC) persisted in the infected cells longer than any other vectors carrying OCT3/4, SOX2, or KLF4 at the top of the vectors (18+). When c-MYC was inserted at the 18+ position, such selective retention of the c-MYC–carrying vector was not observed. This was apparently due to the prolonged replication of the HNL-MYC vector (17). Because GFP expression with the TS vectors was relatively weak compared with that obtained using conventional vectors (Fig. 1B), we initially inserted HNL-MYC into the TS vectors so that the initial levels of expression could be restored by the polymerases supplied in trans from the other vectors (OCT3/4, KLF4, and SOX2). In addition, it was hoped that by using this strategy, the viral vectors might easily disappear when the remaining vector was HNL-MYC in the TS vector alone (Fig. 2A). Using these vectors at a multiplicity of infection (MOI) of 3, we obtained colonies from human fibroblast cells that were alkaline phosphatase (ALP)-positive and exhibited human embryonic stem (ES) cell-like morphology ∼28 d after induction (Fig. 2B and Table S1). We then monitored the amount of the SeV genome present during reprogramming and cell expansion using quantitative RT-PCR (qRT-PCR). Surprisingly, replacement of the HNL-MYC vector into only one of the four reprogramming factor mixtures using the TS vectors resulted in a marked decrease in all viral genomes after the appearance of the iPSCs (Fig. 2C). As expected, expression of c-MYC on the TS vectors was correlated with the viral genome (Fig. S1). Then individual colonies were isolated, and the remaining SeV genome in each colony was evaluated. During cell expansion, the vectors were diluted, and most colonies were only partially positive for SeV (Fig. 2D, Middle). At passage 4, 80% of the colonies were negative for the viral genome using the TS13-HNL-MYC and TS15-HNL-MYC vectors, and by passage 10, all colonies were negative for the vector by qRT-PCR (Fig. 3A, Left). Because the number of SeV-negative colonies was not increased using the TS7 vector at passage 10, although the partial negative colonies were increased, the iPSCs were subjected to a temperature shift to 38 °C (Fig. 3A, Right). Incubation of cells at 38 °C for 3–5 d was sufficient to obtain SeV-negative iPSCs (Figs. 3A and 4B) with no changes in the expression of human ES cell (hESC) marker genes, NANOG (Fig. 4B), or the other related marker genes. Quantitative RT-PCR analysis and Western blot analysis of these iPSCs obtained by the first strategy revealed no detectable viral genome or protein expression in the established iPSCs at the late passage numbers, with slightly detectable viral genome or protein expression in the pooled colonies at passage 8 (1/1,000–1/10,000 compared with day 3; Fig. 3 B and C and Table S3). TS15 showed higher dilution than other vectors, comparable to their temperature sensitivities. More importantly, copy numbers of OCT3/4, SOX2, KLF4, and c-MYC genes in SeV-generated iPSCs were the same as in parental cells, in contrast to retroviral-generated iPSCs, in which copy numbers were verified because of the vector integration (Fig. 3D). The condition and the efficiencies of these experiments and characteristics of obtained iPS clones are summarized in Tables S1 and S3.
Fig. 2.
(A) Strategy used to obtain vector/factor-free iPSCs with an SeV vector mixture. Human fibroblasts were infected with the SeV vector mixture containing four factors. During reprogramming, polymerases may be supplied to the TS vector (which has c-MYC at the HNL position) by OCT3/4-, SOX2-, and KLF4-carrying conventional SeV vectors. Then the SeV vector may disappear when there is TS vector alone at nonpermissive temperature. (B) Procedure for reprogramming using SeV vectors. (C) qRT-PCR of existing SeV genomes in induced cells. RNA was extracted from cells between 3 d and 2 mo after infection, and the amount of SeV genome was analyzed by qRT-PCR. iPSC colonies were passaged every 7 d after day 35 (arrow), and whole colonies on the culture dish were analyzed. (D) Typical staining of iPSC colonies with anti-SeV antibodies. (Left) At passage (P) 1, many colonies were positive for SeV. (Middle) After several passages (P4), many colonies were partially positive. (Right) Colonies were found to be negative for SeV at P10. (Scale bar: 200 μm.)
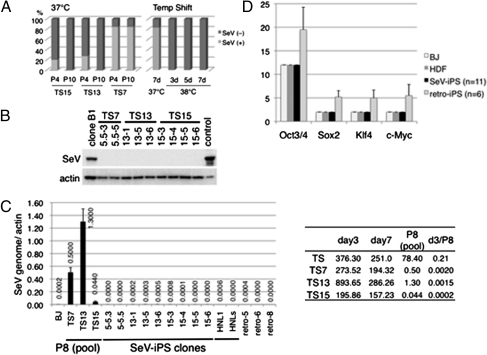
Fig. 3.
(A) Ratio of SeV-positive colonies. P, passage number. (Left) Randomly chosen colonies were expanded independently, and the existence of SeV vectors was evaluated by qRT-PCR at P4 and P10. The number of positive and negative colonies was counted and is expressed for each as a ratio of all colonies chosen (n = 12 per each TS vector). (Right) Temperature shift to a nonpermissive temperature of 38 °C effectively removed SeV vectors from the iPSC colonies generated using TS7 vectors. The culture dishes at P4 were split and transferred to culture at 37 or 38 °C for the number of days indicated. The ratio was calculated as left panel. (B) SeV proteins were not detected in iPSC colonies by Western blot analysis with anti-SeV antibodies. Clone B1, positive control for iPSCs in which the SeV persisted (17); control, LLC-MK2 cells transfected with plasmids encoding SeV vectors. 5.5-3 and 5.5-5 were generated with MYC/TS7; 13-1, 13-5, and 13-6 were generated with MYC/TS13; and 15-3, 15-4, 15-5, and 15-6 were generated with MYC/TS15. (C) qRT-PCR of viral genome in SeV-generated iPSCs (viral genome/actin). BJ, parental fibroblasts; day 3 and day 7, BJ cells infected with SeV vectors after 3 and 7 d; HNL1 and HNLs, iPSCs established using conventional SeV (17). P8, pooled colonies at passage 8. Values <0.001 were backgrounds under the calculation curve as detected in parental cells. (D) Copy numbers of OCT3/4, SOX2, KLF4, and c-MYC in parental cells [BJ and human dermal fibroblast (HDF) cells] and iPSCs generated by SeV (SeV-iPS; n = 11) or retrovirus (retro-iPS; n = 6), as determined by qRT-PCR of genomic DNA. The passage numbers of tested clones are listed in Table S3.
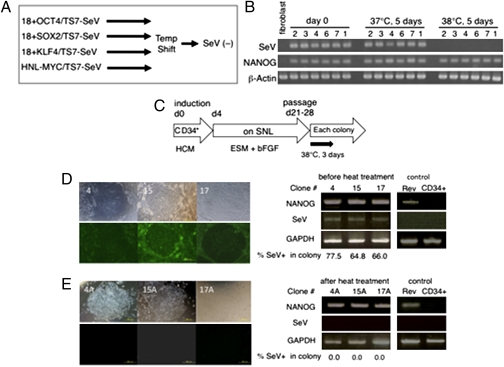
Fig. 4.
Our second strategy with temperature-shift treatment. (A) Schemes to remove viral genome. (B) qRT-PCR of the SeV genome and hESC markers before and after temperature-shift treatment. iPSCs generated from HDF cells using TS7 vectors were treated with a temperature shift to 38 °C for 5 d. The SeV genome was not detected after this temperature-shift treatment, whereas expression of NANOG was not affected. (C) Schemes for generation of virus vector-free iPSCs from CD34+ CB cells with SeV vectors. The ESC-like colonies 4, 15, and 17 that emerged were subcloned after heat treatment for 3 d at 38 °C. (D and E) The remaining SeV construct in SeV iPSC 4, SeV iPSC 15, and SeV iPSC 17 (D; passage 2, before heat treatment) and heat-treated subclone SeV iPSC 4A (from 4), SeV iPSC 15A (from 15), and SeV iPSC 17A (from 17) (E; passage 4) were determined by immunostaining with anti-HN antibody (Left) and by qRT-PCR against endogenous NANOG transcript and SeV RNA construct (Right). Retrovirally generated iPSCs from CB cells (Rev) and CD34+ CB cells (CD34) were used as controls. The percentage of SeV-positive cells in the respective colony was determined using a two-value recognition function and is given below the qRT-PCR image.
Expression of Human ES Cell Markers and Epigenetics.
All iPSCs examined expressed hESC markers, as determined by qRT-PCR, and were positive for stage-specific embryonic antigen (SSEA)-4, TRA-1-60, TRA-1-81, NANOG, and OCT4, as demonstrated by immunostaining (Fig. S2 A and B). Furthermore, CpG dinucleotides at the OCT3/4 promoter region in the iPSCs were demethylated (Fig. S2C). Global gene expression in the iPSCs was similar to that of hESCs (Fig. S3A). Clustering analysis revealed a high degree of similarity among the reprogrammed iPSCs obtained using the TS7, TS13, or TS15 vectors that clustered together with hESCs and was distant from that of the parental somatic cells. These iPSCs had a normal 46 XY or 46 XX karyotype, even after temperature-shift treatment, and could be maintained for more than 20 passages (Fig. S4A and Table S3). DNA fingerprinting analysis verified that these iPSCs were indeed derived from the parental fibroblast cells (Fig. S4B).
Established iPSCs Show Potentiality of Differentiation to Three Germ Layers.
The ability of hESCs/iPSCs to differentiate into all cell types provides the basis for their potential in regenerative medicine. Thus, we investigated the differentiating potential of SeV-generated iPSCs by evaluating teratoma formation. In these experiments, the virus-negative iPSCs were injected s.c. or i.m. into NOD/SCID mice. Histological examination of the teratomas revealed that the tissues had originated from the three embryonic germ layers and included neural and epithelial tissues, muscle, cartilage, bone, gut-like structures, and various glandular structures (Fig. S3B). We further tested the pluripotency of SeV-generated iPSCs in vitro. Like hESCs, these iPSCs formed embryoid bodies in suspension culture (Fig. S4C). For mesoderm-derived differentiation, the embryoid bodies were grown in adherent culture with 0.1 mM ascorbic acid and 20% FBS to enhance cardiomyocyte differentiation (25), whereas an established protocol for hESCs involving activin A treatment (26) was used for the induction of definitive endoderm cells. As a result, SeV-generated iPSCs differentiated into beating cardiomyocytes and endoderm-derived pancreatic cells that stained positive for pancreatic and duodenal homeobox 1 (PDX1; Fig. S4C). Coculture of these iPSCs with PA6 feeder cells resulted in the generation of dopaminergic neurons (ectoderm derivatives) that were positive for βIII tubulin and tyrosine hydroxylase (27) (Fig. S4C). These data indicate that the SeV-generated iPSCs are pluripotent, like hESCs, and respond to different differentiation stimuli. Based on these results, we conclude that the viral/factor-free iPSCs generated using TS SeV vectors meet the criteria of hESCs and could serve as a clinically important source of stem cells without the danger of integration of any foreign genes.
Generation of Human iPSCs from CB Cells.
In the second strategy, we obtained viral-free iPSCs from human fibroblasts using TS13 or TS7 vector mixtures consisting of four reprogramming factors (i.e., 4F/TS13 and 4F/TS7, respectively) at a higher MOI of 30 at 37 °C, with (TS7) or without (TS13) temperature-shift treatment (Fig. 4 A and B and Table S1). In this case, we used a higher MOI because we could not obtain iPSCs at a lower MOI, likely due to the weak expression of TS vectors at 37 °C, lacking any supplemental polymerases from mixed conventional vectors. However, an MOI of only 2 was sufficient to generate iPSCs from the CB cells described below, possibly because of the highly efficient gene transfer into human CB stem cells (19). We infected freshly isolated mononuclear cells with the GFP construct at an MOI of 2 and found that GFP expression was limited to the CD34+ fraction (43% of the CD34+ cells were GFP+ cells), whereas no GFP+ cells were found in the CD34− fraction at 2 d after SeV infection (19). The rate of SeV infection determined by GFP expression in CD34+ cells was increased up to 100% if we used SeV vectors at an MOI of 20. This fraction corresponds to hematopoietic stem cells or progenitors, as reported elsewhere (28). Then freshly isolated purified CD34+ CB cells (purity of CD34+, 96–99%) or the frozen CD34+ CB cells (RIKEN BioResource Center; purity of CD34+, 97–99%), thawed 1 d before infection, were used for SeV infection. These cells were treated with a SeV vector mixture consisting of SeV TS7-OCT3/4, -SOX2, -KLF4, and -c-MYC. Infected cells were incubated under hematopoietic cell culture conditions, followed by cocultivation on mitomycin C (MMC)-treated SNL cells on day 4 or day 10 (Fig. 4C). hESC-like colonies expressing SSEA-4 appeared after 14 d of cocultivation with SNL (18 d after SeV infection), whereas no colonies were obtained after transfer onto SNL on day 10 (Table S2). The efficiency with which iPSC colonies emerged on SNL feeder cells using SeV TS7 vectors is also shown in Table S2. To eliminate remaining SeV virus constructs, ESC-like colonies were subjected to heat treatment at 38 °C for 3 d and recloned. SeV-positive cells constituted 60–80% of total cells within hESC-like colonies before heat treatment (clones SeV iPSC 4, 15, and 17; passage 2). This was reduced to 0% after recloning (clones SeV iPSC 4A, 15A, and 17A; passage 4), followed by the heat treatment. The percentages of SeV-positive cells were determined by the proportion of sum of the area positively detected with anti-SeV HN (envelope) antibody against total colony area. In the agreement with this staining result, SeV constructs were not detected by qRT-PCR at passage 4 after heat treatment (Fig. 4E). The virus-negative ESC-like clones showed the expression of hESC markers (Fig. S5 A and B), global gene expression profile similar to that of hESCs (Fig. S5 C and E), and normal karyotype (Fig. S5D). Seven established cell clones were tested for embryoid body–mediated in vitro differentiation potential, and three out of the seven clones were found to have in vivo differentiation potential in a teratoma formation assay with SCID mice. All three of these clones were able to give rise to cells of all three germ layers as detected by immunocytochemistry and cell morphology studies (Fig. S5F). These three lines formed the teratomas with a cystic mass containing differentiated tissues morphologically corresponding to all three germ layers (Fig. S5F). The characterization of established clones from fibroblasts and CB cells is summarized in Table S3.
Discussion
The present study has demonstrated that the established TS SeV vectors TS15, TS13, and TS7 are highly effective tools with which we were able to obtain transgene-free human iPSCs. These iPSCs were generated efficiently by robust gene replication, with a subsequent rapid decrease in the level of factor-carrying SeV vectors during cell expansion (Fig. 2C). Almost all of the colonies picked up were virus-negative at late passages, and even if vectors were present, they could be easily removed using the temperature-shift protocol (Figs. 3A and 4). Indeed, the iPSCs generated using TS SeV vectors were free of any integrated viral factors, in contrast to retroviral-generated iPSCs, which express variable copy numbers of OCT3/4, SOX2, KLF4, and c-MYC (Fig. 3D). We also demonstrated that using TS SeV vectors, virus-negative iPSCs were efficiently generated from CD34+ human CB cells (Fig. 4).
Although the use of synthetic modified mRNA to generate iPSCs has been reported, this is highly dependent on the gene delivery system, because it requires repetitive transfection for 16 d (13). Therefore, it might not be applicable to difficult-to-transfect cells, such as primary peripheral blood cells. The addition of decoy receptor to prevent IFN production by host cells is also needed. In contrast, SeV vectors require no transfection reagents or decoy receptors. We suggest that this is a considerable advantage to using nonintegrating SeV; the iPSCs thus generated have a homogeneous genetic background. Several studies on the generation of iPSCs from blood cells and CB cells with integration-free episomal vectors have been published recently (29–31). The efficiency of generating iPSCs is lower with episomal vectors than with SeV (>0.1% at an MOI of 2) and varies depending on the construction of the vectors and the nature of the transfection medium used. The major advantage of using nonintegrated SeV vectors is a potent and robust protein-expressing property that does not require optimization of the transfection medium. The volume of collected CB is 80–120 mL and may contain 2–4 × 105 CD34+ cells on average. Our results show that only 1 × 104 CD34+ cells, corresponding to ∼5 mL of CB, are needed to obtain 10 independent iPSC clones with SeV. The rest of the CB can be sorted and used for the regular bone marrow transplantation therapy. In addition, there have been no reports of pathogenicity associated with SeV in primates, and the safety of the SeV vector is further enhanced by the F-deficiency that makes the vector nontransmissible (16).
In the present study, we have confirmed that the SeV vectors were not reactivated or detected in iPSCs at late passages or after temperature-shift treatment (Figs. 1C and 3). It has been suggested that TS mutations in P and L affect polymerase activity and promote degradation of the virus vector after treatment at nonpermissive temperatures (23). Based on our findings, we believe the TS SeV vector system that allows the elimination of remaining SeV construct from reprogrammed cells by temperature-shift treatment could accelerate future clinical application of iPSCs generated from cells obtained by less invasive or even noninvasive methods.
Materials and Methods
The induction of human iPSCs was done as reported previously (1, 17). Unless indicated otherwise, SeV vectors (OCT3/4, SOX2, KLF4, and c-MYC) were used at an MOI of 3 or 30 for 1 × 106 fibroblasts. For retroviral induction, fibroblasts were infected with mCAT containing simian immunodeficiency virus (SIV) vector at an MOI of 50, and then expanded by induction with retrovirus vectors containing reprogramming factors. Infected cells were transferred onto MMC-treated mouse embryonic fibroblast (MEF) feeder cells on day 6 after induction. Details of the experimental procedures are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Satomi Nishikawa and K. Kobayashi for useful discussions; T. Aoi for RNA from hESCs; A. Iida for a critical reading of the manuscript; T. Kitamura for pMX vector; RIKEN BioResource Center for CD34+ CB cells; S. Seino, T. Kanaya, K. Washizawa, T. Fujikawa, E. Suzuki, and T. Yamamoto for technical assistance; H. Iwasaki for help with the histological studies; and T. Ofuji and M. Miyako for help with manuscript editing. This work was supported in part by the Japan Science and Technology Agency's Precursory Research for Embryonic Science and Technology and S-Innovation Programs, Regional Consortium Project Grant 19K5510 from the Ministry of Industry and Economy (2007–2009), Grant-in-Aid 2210853 from the Japan Society for the Promotion of Science Fellows, and Japan Science and Technology Agency “Tests for Safety Issue of Pluripotent Stem Cell” (2010-14).
Footnotes
Conflict of interest statement: H.B., T.T., K.S., M.I., and N.F. are employees of DNAVEC Corporation. M.H. is a founder of DNAVEC Corporation.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, ww.ncbi.nlm.nih.gov/geo (accession nos. GSE24240 and GSE25090).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103509108/-/DCSupplemental.
References
- 1.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 3.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse-induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 4.Yu J, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soldner F, et al. Parkinson's disease patient–derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sommer CA, et al. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woltjen K, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaji K, et al. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia F, et al. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7:197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou H, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim D, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warren L, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamb RA, Kolakofsky D. Paramyxoviridae: The viruses and their replication. Fields Virology. In: Fields BN, Knipe DM, Howley PM, editors. 3rd Ed. Philadelphia: Lippincott-Raven; 1996. pp. 1177–1204. [Google Scholar]
- 15.Li HO, et al. A cytoplasmic RNA vector derived from nontransmissible Sendai virus with efficient gene transfer and expression. J Virol. 2000;74:6564–6569. doi: 10.1128/jvi.74.14.6564-6569.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagai Y, Takakura A, Irie T, Yonemitsu M, Gotoh B. Evolution of Sendai virus: The journey from mouse pathogen to a state-of-the-art tool in virus research and biotechnology. The Paramyxoviruses. In: Samal SK, editor. Norwich, UK: Horizon Scientific Press; 2011. [Google Scholar]
- 17.Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad, Ser B, Phys Biol Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seki T, et al. Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell. 2010;7:11–14. doi: 10.1016/j.stem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Jin CH, et al. Recombinant Sendai virus provides a highly efficient gene transfer into human cord blood–derived hematopoietic stem cells. Gene Ther. 2003;10:272–277. doi: 10.1038/sj.gt.3301877. [DOI] [PubMed] [Google Scholar]
- 20.Giorgetti A, et al. Generation of induced pluripotent stem cells from human cord blood using OCT4 and SOX2. Cell Stem Cell. 2009;5:353–357. doi: 10.1016/j.stem.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takenaka C, Nishishita N, Takada N, Jakt LM, Kawamata S. Effective generation of iPS cells from CD34+ cord blood cells by inhibition of p53. Exp Hematol. 2010;38:154–162. doi: 10.1016/j.exphem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Bowman MC, Smallwood S, Moyer SA. Dissection of individual functions of the Sendai virus phosphoprotein in transcription. J Virol. 1999;73:6474–6483. doi: 10.1128/jvi.73.8.6474-6483.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feller JA, Smallwood S, Skiadopoulos MH, Murphy BR, Moyer SA. Comparison of identical temperature-sensitive mutations in the L polymerase proteins of Sendai and parainfluenza3 viruses. Virology. 2000;276:190–201. doi: 10.1006/viro.2000.0535. [DOI] [PubMed] [Google Scholar]
- 24.Inoue M, et al. Nontransmissible virus-like particle formation by F-deficient Sendai virus is temperature-sensitive and reduced by mutations in M and HN proteins. J Virol. 2003;77:3238–3246. doi: 10.1128/JVI.77.5.3238-3246.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo XM, et al. Creation of engineered cardiac tissue in vitro from mouse embryonic stem cells. Circulation. 2006;113:2229–2237. doi: 10.1161/CIRCULATIONAHA.105.583039. [DOI] [PubMed] [Google Scholar]
- 26.D'Amour KA, et al. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 27.Kawasaki H, et al. Induction of midbrain dopaminergic neurons from ES cells by stromal cell–derived inducing activity. Neuron. 2000;28:31–40. doi: 10.1016/s0896-6273(00)00083-0. [DOI] [PubMed] [Google Scholar]
- 28.Majeti R, Park CY, Weissman IL. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell. 2007;1:635–645. doi: 10.1016/j.stem.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chou BK, et al. Efficient human iPS cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures. Cell Res. 2011;21:518–529. doi: 10.1038/cr.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu K, et al. Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood. 2011;117:e109–e119. doi: 10.1182/blood-2010-07-298331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu J, Chau KF, Vodyanik MA, Jiang J, Jiang Y. Efficient feeder-free episomal reprogramming with small molecules. PLoS ONE. 2011;6:e17557. doi: 10.1371/journal.pone.0017557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.