Abstract
Specific regulatory states, i.e., sets of expressed transcription factors, define the gene expression capabilities of cells in animal development. Here we explore the functional significance of an unprecedented example of regulatory state conservation from the cnidarian Nematostella to Drosophila, sea urchin, fish, and mammals. Our probe is a deeply conserved cis-regulatory DNA module of the SRY-box B2 (soxB2), recognizable at the sequence level across many phyla. Transphyletic cis-regulatory DNA transfer experiments reveal that the plesiomorphic control function of this module may have been to respond to a regulatory state associated with neuronal differentiation. By introducing expression constructs driven by this module from any phyletic source into the genomes of diverse developing animals, we discover that the regulatory state to which it responds is used at different levels of the neurogenic developmental process, including patterning and development of the vertebrate forebrain and neurogenesis in the Drosophila optic lobe and brain. The regulatory state recognized by the conserved DNA sequence may have been redeployed to different levels of the developmental regulatory program during evolution of complex central nervous systems.
Keywords: sequence conservation, evolution of development, transgenesis, enhancer, sox21
Comparative evolutionary studies have produced evidence for conservation of genomic regulatory apparatus encoding animal development, sometimes across large phylogenetic distances (for a current review, see ref. 1). Most obvious are examples revealed by genomic sequence comparisons, in which very highly conserved sequences up to several hundred bp long are shown by transgenic reporter assays to possess conserved cis-regulatory function (e.g., refs. 2–7). However, highly conserved cis-regulatory sequences account for only a minor fraction of the large number of genomic cis-regulatory modules likely to exist in animal genomes (8), and cis-regulatory comparisons at significant evolutionary distance often reveal conserved control functions even when sequence conservation is not detectable across the cis-regulatory module. In such examples, where functional assays carried out by insertion of known cis-regulatory modules into the genomes of different species reveal conserved function, it is found that only the specific transcription factor (TF) target sites are retained, spaced in nonconserved DNA sequence, often in diverse site order and number (9–11). Whether the DNA sequence of the cis-regulatory module is largely conserved or not, if the function remains the same, the implication is that the set of TFs to which its target sites respond are largely the same. If the sum of the TFs present in any given development context is considered as the “regulatory state,” a similar cis-regulatory response to a given cis-regulatory construct in phylogenetically diverse systems reveals a regulatory state that has been conserved, at least partially, among these systems. Thus, given cis-regulatory modules can be used as probes for a phylogenetically conserved regulatory state.
The most prominent and concrete examples of phylogenetic regulatory state conservation have been found in cell types that appear across the Bilateria (1, 12). Certain differentiation gene batteries composed of orthologous effector genes and the regulatory state drivers that activate them are known in all bilaterian animals investigated and even in cnidarians, e.g., in muscle cells (5) neurons (13), photoreceptor cells (14–17), and neurosecretory cells (17). In phylogenetically distant animals, however, sometimes only the regulatory state is conserved in cells that perform specifically homologous functions, but the effector genes are all different. For example, the immune cells of sea urchins, lampreys, and amniotes use entirely different receptor proteins, but their deployment is controlled by homologous regulatory states in all three cases (18–20). Similarly, the integument cells that are tasked with wound repair and building the outer epidermis of insects and vertebrates use entirely different effector genes, but these genes are driven by the same regulatory factor (21). Here pan-bilaterian cell-type functions are defined by specific, anciently evolved relations between biological role and conserved regulatory states.
Given regulatory states can be redeployed during evolution for use in developmental contexts other than immediate cell-type specification, this redeployment probably is a major mechanism of evolutionary change in body plan (22). Here, we follow a trail that leads from a very unusually conserved genomic regulatory sequence to conserved cell type-specific regulatory states, but we also show that this regulatory state has been co-opted functionally during evolution, at least in part, for use in different levels of the developmental regulatory process. Although there has been much discussion of the co-option of individual gene regulatory components as a driving mechanism of evolution (e.g., refs. 1, 23), co-option of whole regulatory states has additional implications, as we discuss below.
Results
Transphyletic Conservation of Noncoding Sequences.
The present study followed from the discovery of a conserved noncoding sequence region (CNR) at corresponding positions in the vicinity of soxB2 class regulatory genes in both vertebrate (human and zebrafish) and invertebrate chordate (Branchiostoma floridae, amphioxus) genomes, in the genomes of a sea urchin (Strongylocentrotus purpuratus) and a hemichordate (Saccoglossus kowalevskii), and, surprisingly, also in the genome of the anthozoan cnidarian Nematostella vectensis (for orthology of the Nematostella gene, see phylogenetic analysis in Fig. S1). Fig. 1 shows the sequence alignments and gene map of the soxB2 CNR for these six genomes. The sequence was found in the course of a computational scan for deeply conserved CNRs across eumetazoan genomes, using as probes a set of chordate CNRs described earlier (ref. 24 and Table S1). We identified seven additional CNRs, 76–217 bp in length, conserved at the sequence level in both chordate and nonchordate deuterostomes (Fig. S2 A–G and Table S1); thus these CNRs must have been present in the last common ancestor of all deuterostomes. One of these CNRs also can be recognized in the Nematostella genome (Fig. S2B; for computational procedures and criterion of conservation see SI Materials and Methods). As with the soxB2 CNR, each of these CNRs occupies a similar syntenic position with respect to the associated gene, which in all cases is a key developmental regulatory gene encoding a TF. We functionally tested all eight CNRs in transgenic systems (SI Materials and Methods and Table S2). In addition to the SoxB2 CNR that is the subject of this paper, we detected positive enhancer activity for a CNR associated with the inhibitor of DNA binding, dominant negative helix-loop-helix (Id) genes across the deuterostomes. Although we focus here only on the analysis and discussion of the SoxB2 CNR, similar results also were obtained for the Id CNR (Fig. S3).
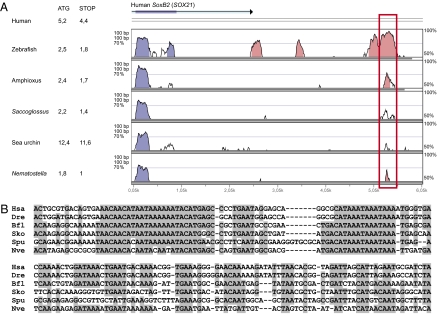
Fig. 1.
The conserved soxB2 CNRs. (A) VISTA plot of soxB2/sox21 loci in human, zebrafish, amphioxus, Saccoglossus, sea urchin, and Nematostella. Blue and pink peaks in VISTA alignments correspond to coding and noncoding regions, respectively. Human SOX21 UTR is depicted in turquoise. The red rectangle indicates the location of the soxB2-CNRs. Distance of soxB2-CNRs to both first (ATG) and last (STOP) soxB2 codons is indicated in kb. (B) Nucleotide alignments of soxB2-CNRs. Shadowed nucleotides in the sequence alignment indicate >65% sequence conservation. Bfl, amphioxus; Dre, zebrafish; Hsa, human; Nve, Nematostella; Sko, Saccoglossus; Spu, sea urchin.
Expression of the soxB2 CNR in Developing Zebrafish Brain.
In vertebrates, sox21 (a soxB2 class gene) is widely expressed in the developing CNS. Previous studies carried out in transgenic mouse embryos showed that a lacZ construct driven by the orthologous human SOX21 CNR is expressed, as is the endogenous mouse gene, in many regions of the brain, spinal column, eye, and otic vesicle. Similar expression patterns were observed in transgenic zebrafish bearing constructs for the orthologous human and pufferfish CNRs (25, 26). These results demonstrate that this vertebrate CNR functions largely as a CNS cis-regulatory module. To compare the in vivo function of the nonvertebrate soxB2 CNR's in the same developmental context, transgenic zebrafish were generated using the various CNRs incorporated in otherwise similar vectors. At 48 h of development, the amphioxus and sea urchin soxB2 CNRs are seen to drive GFP expression in the same gross domains of the developing zebrafish CNS as do the vertebrate CNRs (Fig. 2 A–C; as shown in Fig. S7 A–C, the empty vectors display no activity in the brain). These domains are approximately the same domains in which the endogenous zebrafish sox21b gene is expressed (Fig. S4A), so these CNRs evidently all possess the capacity to recapitulate this overall phase of teleost sox21b expression. Because these images do not afford cellular resolution, this impression pertains only to the general regions marked, i.e., forebrain, eye, midbrain, hindbrain, and spinal cord. At the 48-h stage the hemichordate and Nematostella CNR constructs are expressed less robustly, although all are clearly positive in the forebrains of the transgenic fish (Fig. S4 C–D).
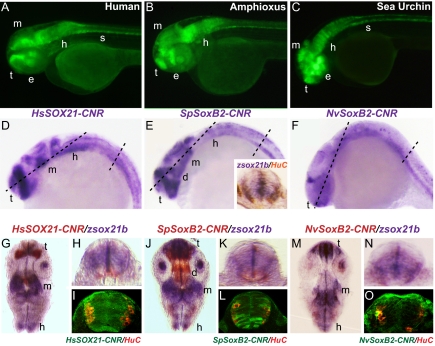
Fig. 2.
Similar, although not identical, transcriptional regulatory activities of orthologous soxB2 CNRs. A–C show GFP expression controlled by the indicated soxB2 CNRs in lateral views of transgenic 48-h zebrafish embryos. (D–O) Expression of indicated constructs in 24-h transgenic zebrafish, (D–F) Whole-mount in situ hybridization (WMISH) to detect GFP expression. Inset in E shows a transverse section of the spinal cord double stained for zsox21b (purple) and the neuronal marker HuC (brown). (G, J, M) Coronal sections through brain as indicated, respectively, in D–F. (H, I, K, L, N, O) Cross-sections through spinal cord. (H, K, N) Transgenic embryos were double stained for zsox21b (purple) and GFP (red); reporter constructs are identified at the tops of panels. (I, L, O) Confocal sections showing GFP (green) and HuC (red) protein localization in the respective transgenic embryonic spinal cords. d, diencephalon; e, eye; h, hindbrain; m, midbrain; s, spinal cord; t, telencephalon.
Embryos at 24 h bearing the human, sea urchin, and Nematostella soxB2 CNR constructs were sectioned as indicated by the dashed lines in Fig. 2 D–F, and expression of the endogenous sox21b gene as well as of construct-driven GFP were displayed by double in situ hybridization to provide higher resolution spatial information (Fig. 2 G–O). Coronal sections through the telencephalon and diencephalon plus midbrain and hindbrain, and transversely through the spinal column, are provided for each construct, and in the spinal sections an additional marker, HuC, which identifies mature neurons (27), also has been included. In the brain sections, both similarities and differences between the domains of expression driven by the three constructs are apparent (Fig. 2 G, J, M). All, including the Nematostella construct, express clearly in the anterior forebrain (Fig. 2 D–F), as does the endogenous sox21b gene (Fig. 2 G, J, M). However, the human and sea urchin constructs express in midbrain as well (Fig. 2 D and E), but the Nematostella construct does not (Fig. 2F). On the other hand, the sea urchin construct displays a strong additional domain of expression in the diencephalon (Fig. 2 E and J). In the spinal cord sections it can be seen that sox21 is expressed through almost all the cross-sectional area (Fig. 2 E Inset, H, K), as are the human CNR construct (Fig. 2I) and, to a lesser extent, the sea urchin construct (Fig. 2L). Although at least some of the small patches of mature neurons denoted by HuC expression overlap with GFP expression domains, most of the spinal cord sectional area where the CNRs are active lacks mature neurons. Instead we see regionalized expression throughout all or large parts of the structure. Similarly, in the brain, the lack of diencephalon expression of the human CNR construct, in accord with the endogenous gene expression pattern, cannot be caused by the absence of neuronal tissue there but rather implies a regional repression function to which the sea urchin CNR is insensitive, evidently lacking the requisite target sites (Fig. 2 G and J). We see that in the 24-h embryo the regulatory states to which the CNRs respond are deployed according to future domains of the CNS.
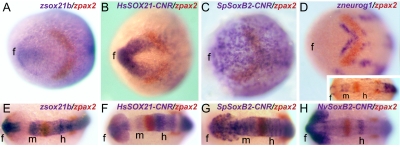
Moving to earlier stages of CNS development, regional expression in the anterior neuroectoderm in advance of neurogenesis is seen for both the endogenous sox21b gene and the human and sea urchin CNR-driven constructs, although again the patterns are not identical. At tailbud stage the endogenous sox21b is expressed in an anchor-like pattern at the forward end of the neural plate far anterior to the paired box gene 2 (pax2) which is an integral component of the gene network establishing the midbrain/hindbrain boundary (28) (Fig. 3A). At this stage the only region undergoing neurogenesis, as marked by neurogenin expression, is immediately adjacent to this boundary (Fig. 3D). The human SOX21 CNR construct is expressed like the endogenous zebrafish sox21b, whereas the sea urchin construct is expressed over all the anterior neuroepithelium (Fig. 3 B and C). At the 5–15 somite stage, neurogenesis has commenced in the forebrain (Fig. 3D Inset), but the human, sea urchin, and Nematostella soxB2 CNR constructs are expressed much more widely, including regions where there still is no neurogenin expression posterior to the pax2 stripe (Fig. 3 E–H and Fig. S5). Again, both the regional expression seen in these figures and the gaps in the expression pattern at the future diencephalon domain (except for the sea urchin construct; Fig. 3G and Fig. S5J) indicate expression that foreshadows the parts of the brain, rather than simply reflecting neurogenesis or the presence of differentiated neurons. As in the later stages, the most general and uniform expression domain generated by all of the CNRs as well the endogenous sox21b control system is throughout the future forebrain (Fig. 3 E–H and Fig. S5).
Fig. 3.
Early expression promoted by orthologous SoxB2-CNRs. (A–D) Dorsal views of tailbud embryos, double WMISH as indicated in each panel. (E–H and Inset in D) Dorsal views of embryos at the 5–15 somite stage, double WMISH as indicated. f, forebrain; h, hindbrain; m, midbrain.
A trivial explanation for those aspects of the CNR expression patterns that spatially overlap endogenous sox21b expression is that the CNRs simply are responding to autoregulation by the Sox21 factors themselves. This explanation is unlikely, however, because Sox21 is known to act in a repressive manner (29, 30). Nonetheless, we tested this possibility by knocking down zebrafish sox21a and sox21b genes, either individually or in combination, using specific morpholinos (MOs). These MOs were introduced into transgenic embryos that expressed GFP under the control of different soxB2 CNRs. For zsox21a, we used a previously reported MO (29); for zsox21b, the MO was shown to block translation of a zsox21b-GFP fusion mRNA (Fig. S6). Injection of 15 ng of individual MOs or 7.5 ng of each MO injected together had no effect on the GFP levels in transgenic embryos expressing human or sea urchin soxB2 CNR constructs. Thus positive autoregulation of the CNRs by endogenous Sox21 is ruled out.
Function of the soxB2 CNRs in Transgenic Sea Urchins.
These results, taken together with the striking sequence homology displayed in Fig. 1, raise an immediate paradox: Sea urchins have no forebrains, midbrains, spinal columns, eyes, or otic vesicles. Where in such an organism would the extremely similar cis-regulatory sequences isolated from the soxB2 genes drive expression? The endogenous soxB2 of the sea urchin is expressed in feeding larvae in the heavily innervated ciliary band, as well as in cells of the midgut wall, as shown by whole-mount FISH (Fig. 4A). When the sea urchin, human, or Nematostella soxB2 CNRs are inserted in expression vectors and injected into fertilized eggs, they also are expressed in individual ciliary band neurons of the feeding larva and can be identified by the GFP-stained axons (Fig. 4 B–D; for empty vector controls, see Fig. S7 K–M). In this organism, injected DNA is incorporated stably and randomly in all cell lineages, but in a mosaic fashion (31). If the expression in sea urchin and some vertebrate neurons is indicative of ancestral function, and because soxB2 is a regulatory gene, it may be assumed that at least one plesiomorphic function of the CNR is to contribute [likely positively (30)] to the execution of neuronal differentiation by causing the expression of SoxB2. In turn, the sequences of this CNR plesiomorphically recognize a regulatory state associated with neuronal differentiation.
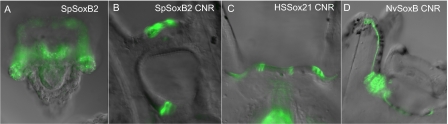
Fig. 4.
SoxB2-CNRs generate neuron-specific expression in sea urchin larvae. Transgenic sea urchin larvae carrying reporter genes under the control of SoxB2-CNRs from the indicated species (upper right corners). (A) Endogenous expression of sea urchin soxB2, fluorescent WMISH. (B and C) Expression of sea urchin (B) and human (C) soxB2 CNR vectors. (F) Nematostella CNR vector.
Functions of the soxB2 CNRs in Drosophila.
The conserved SoxB2 CNRs cannot be detected in the genomes of a mollusk (Lottia), an annelid (Capitella), a crustacean (Daphnia), or in the Drosophila genome, and perhaps it is not conserved at the gross sequence level, as in Fig. 1, in any protostome genome. However, as noted above, cis-regulatory function often is preserved when essential TF target sites remain present, even though no overall sequence similarity can be recognized. As a direct test of functionality across a huge phylogenetic gulf, human and sea urchin soxB2 CNR expression constructs using GFP reporters were introduced into Drosophila.
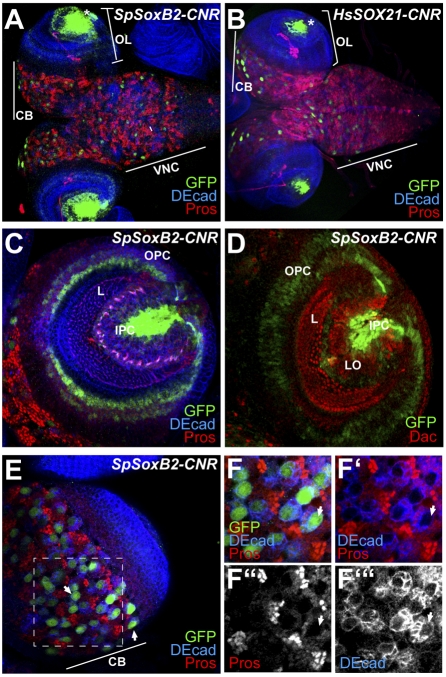
Both constructs are expressed strongly and specifically in the late embryonic and the third-instar larval CNS. In transgenic Drosophila embryos SpSoxB2-CNR and HsSOX21-CNR both drive GFP in cephalic neuroblasts, and, in addition, the human construct is expressed in a small subset of neuroblasts of the ventral nerve cord and, in later stages, in the peripheral nervous system chordotonal organs and in dorsal and ventral epidermis (Fig. S8; empty vector controls are shown in Fig. S7 D–I). In the larval CNS, SpSoxB2-CNR and HsSOX21-CNR both drive expression in neuroblasts of the central brain and ventral nerve cord as well as in the optic lobes (Fig. 5 A and B). A high-magnification analysis using markers to identify the cell types expressing these constructs is shown in Fig. 5 C–F. Here we see that the sea urchin CNR is active in the inner proliferative center of the optic lobes, which will give rise to inner medulla and lobulla neurons, and also in the outer proliferative region, which develops into lamina and outer medulla neurons (32). No expression is detected in differentiated neurons, which are marked by dachshund (dac) expression (Fig. 5D) (32). As in zebrafish, some additional expression is seen in a few neurons (i.e., axon-bearing cells) of the lobulla. In the central brain (CB, Fig. 5A and E), the sea urchin soxB2 and the human sox21 CNR constructs also are expressed in neuroblasts but not in the adjacent ganglion mother cells marked by prospero expression (Fig. 5 E and F and not shown). The expression pattern driven by these CNRs is reminiscent of that of Drosophila sox neuro (33, 34), a SoxB1 class gene. The main conclusion from Fig. 5 is that at least key components of the regulatory state used plesiomorphically in differentiated sea urchin and vertebrate neurons also are expressed in the Drosophila brain and optic lobes, even though no sequence grossly similar to a soxB2 CNR exists in the Drosophila genome. However, the cellular resolution of these studies also shows that in Drosophila this regulatory state is used in neuroblasts and proliferating precursors of neurons, rather than in differentiated mature neurons or ganglion mother cells.
Fig. 5.
Enhancer activity of SoxB2-CNRs in larval Drosophila CNS. Confocal immunofluorescence images of third-stage larval CNSs are shown. GFP expression driven by (A) SpSoxB2 and (B) HsSox21 CNRs is detected in the optic lobes (OL), central brain (CB) and ventral nerve cord (VNC). (C and D) High-magnification views of the optic lobes of SpSoxB2-CNR larvae. DE-cadherin (DEcad) staining outlines cell shapes and allows defining of specific regions within the optic lobes. IPC, inner proliferative center, L, lamina; OPC, outer proliferative center. In D, dac expression marks differentiated lamina neurons. LO, lobulla. (E) Expression of SpSoxB2-CNR in central brain (CB). Inset in D is enlarged in F–F′′′. Large GFP+ neuroblasts (marked with arrows) are surrounded by ganglion mother cells expressing nuclear prospero (Pros).
Discussion
The soxB2 CNRs that are the subject of this study are unique in two respects of apparently opposing import. On the one hand, they are the most deeply conserved enhancers described to date, and their level of sequence conservation, extending through three deuterostome phyla all of the way to an anthozoan cnidarian, is most unusual. On the other hand, when looked at closely, their developmental functions in the different phyla tested are not the same. The conserved sequence elements seen in Fig. 1B obviously suffice to interact with a number of different TFs, and together these factors constitute the regulatory state to which they respond. This regulatory state may include TFs from Sox, Fox, and Pou families (Fig. S9A). In fact, Sox2 has been found bound at the Sox21 CNR in human stem cells (Fig. S9B) (35). The basis of the unusual degree of sequence conservation could be extremely close packing of target sites so that the structure differs from most cis-regulatory modules in lacking divergent intersite sequence. With a couple of exceptions, in each transphyletic context the CNRs from diverse sources generate a very similar regulatory output. Thus, whether of human, sea urchin, or anthozoan origin, they express in differentiated neurons in transgenic sea urchin larvae (Fig. 2); whether of sea urchin or human origin, they drive expression in the same neuroblasts in Drosophila larvae (Fig. 3); whether of anthozoan, sea urchin, or human origin, they activate transcription in developing fish forebrain; and whether of sea urchin or human origin, they express in the anterior neural plate in advance of neurogenesis per se. Therefore, the different CNRs “see” largely the same regulatory states in each of these contexts. It also is true that the sea urchin CNR evidently lacks a site for a diencephalon repressor that the other sequences retain, and the Nematostella CNR evidently also lacks sites for some regional brain activators. However, if in general the CNRs see the same regulatory sequence in each context but operate in different phases of nervous system development, then two revealing issues arise. First, what does it mean in terms of evolutionary mechanism that the same (or similar and overlapping) regulatory states are used in different phases of nervous system development? Second, what is implied by the confinement of this regulatory state to development of structures that have in common only that they all are aspects of the respective CNSs?
Evolutionary Redeployment of a Regulatory State.
Specific regulatory states are generated by specific gene regulatory network (GRN) subcircuits. In animal development these GRNs are hierarchical in structure, such that the earliest-acting subcircuits at the highest levels control the activation of following subcircuits that execute the immediately succeeding jobs of a developmental process, and these subcircuits, together with external inputs, in turn control the next, and so forth, until the terminal state reached (1, 36, 37). The last stage consists of the expression of differentiation gene batteries. A developmental process, such as the regional development of an element of the nervous system, can be considered as the outcome of a given hierarchical GRN (1). Thus, the general answer to the questions above is that the regulatory state recognized by the soxB2 CNRs always has been generated by developmental GRNs of the nervous system, but during evolution this regulatory state has been used at altered hierarchical positions in the GRN.
The plesiomorphic role could have been the terminal one: triggering neuronal differentiation (or maintenance thereof) (30), as seen in the sea urchin feeding larva and in some vertebrate cell populations. In the development of the adult sea urchin body plan, a fivefold radial CNS develops. It will be interesting to see whether the CNR and the soxB2 gene that it controls operate in this later process of CNS development, and, if so, at what level. However, in addition to the ancestral role, in both Drosophila and vertebrates the CNRs are active in brain patterning and regionalization. These animals may have inherited from their last Precambrian common ancestor GRN kernels that encode the fundamental anterior/posterior organization of the CNS (17, 35, 36, 38–40), but it is interesting that in the Drosophila brain and optic lobes the soxB2 CNRs are active specifically in proliferating precursors and neuroblasts (Fig. 3). In zebrafish, however, the soxB2 CNRs become active before neurogenesis, perhaps in forebrain pattern formation, and their function then extends to some mature neurons in the spinal cord (Figs. 4 and 5). Thus, the regulatory states to which the CNRs respond were co-opted for different sets of related functions in these branches of evolution.
Structural Implications of Evolutionary Regulatory State Redeployment.
The idea of “intercalary evolution” has been adduced to explain the utilization of given regulatory genes in both terminal differentiation gene batteries and upstream developmental GRNs (8, 41). Here the plesiomorphic role of the regulatory gene is to operate a differentiation gene battery, requiring it to be activated at a given developmental address. As regulatory circuitry controlling elaboration of the domain in which the battery is expressed builds up, the same gene is reused at higher levels of the GRN at the same address. This useful concept could be extended from individual gene to regulatory states, i.e., to the GRN subcircuits generating given regulatory states. The examples cited at the beginning of this article in which the regulators controlling differentiation of a given cell type remain unchanged in evolution, although controlling different sets of effector genes, show that the regulatory state can be disassociated from its downstream targets, and new downstream targets can be inserted. In considering vertical changes in regulatory state deployment in a developmental GRN, the regulatory state is disassociated from its upstream drivers and is expressed instead at a different level of the GRN. A key aspect of developmental regulatory evolution could be the redeployment of regulatory states from plesiomorphic roles as drivers of differentiation gene batteries to functions that in modern GRNs control earlier developmental processes.
Materials and Methods
Exact procedures are detailed in SI Materials and Methods, where the computational procedures used for the identification of the soxB2 CNRs and the others denoted in Fig. S2 are outlined. The problem of defining the orthology of the soxB2 genes of Nematostella is nontrivial, and the mode of solution also is given in SI Materials and Methods and Fig. S1. The remainder of SI Materials and Methods and Tables S1 and S2 contains the detailed procedures by which the CNRs were cloned and built into expression vectors. For insertion into sea urchin eggs, the vectors were injected into egg cytoplasm; for insertion into zebrafish, the Tol2 system was used as described in SI Materials and Methods and the references therein.
Supplementary Material
Acknowledgments
We thank Billie J. Swalla, Paola Oliveri, Pedro Martínez, and F. Rentzsch for providing genomic DNAs and José Luis Ferrán for helpful comments. We also thank A. Cameron for technical assistance with sea urchins and J. Barsi from the E.H.D. laboratory for protocols. F.G. was supported by National Science Foundation Grant IOS-0641398 (to E.H.D.) and by the Camilla Chandler Frost Fellowship. I.S.P. was supported by National Institutes of Health Grant HD037105 (to E.H.D.). M.I., I.M., and J.G.-F. were funded by Grants BFU2005-00252 and BMC2008-03776 from the Spanish Ministry of Science and Innovation. M.I. and I.M. hold Formación de Personal Investigador and Formación de Personal Universitario grants, respectively. J.G.-F. was funded by Institució Catalana de Recerca i Estudis Avançats Academia, Generalitat de Catalunya, Spain. F.C. was supported by Grants BFU2009-07044 from the Spanish Ministry of Science and Innovation and CVI 2658 (Junta de Andalucía); and J.L.G.-S. was supported by Grants BFU2010-14839, CSD2007-00008 (Ministerio de Ciencia e Innovación), and Proyecto de Excelencia CVI 3488 (Junta de Andalucía).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109037108/-/DCSupplemental.
References
- 1.Peter IS, Davidson EH. Evolution of gene regulatory networks controlling body plan development. Cell. 2011;144:970–985. doi: 10.1016/j.cell.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elgar G, Vavouri T. Tuning in to the signals: Noncoding sequence conservation in vertebrate genomes. Trends Genet. 2008;24:344–352. doi: 10.1016/j.tig.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Pennacchio LA, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 4.Rastegar S, et al. The words of the regulatory code are arranged in a variable manner in highly conserved enhancers. Dev Biol. 2008;318:366–377. doi: 10.1016/j.ydbio.2008.03.034. [DOI] [PubMed] [Google Scholar]
- 5.Seipel K, Schmid V. Evolution of striated muscle: Jellyfish and the origin of triploblasty. Dev Biol. 2005;282:14–26. doi: 10.1016/j.ydbio.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 6.Vavouri T, Walter K, Gilks WR, Lehner B, Elgar G. Parallel evolution of conserved non-coding elements that target a common set of developmental regulatory genes from worms to humans. Genome Biol. 2007;8:R15. doi: 10.1186/gb-2007-8-2-r15. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Lee AP, Kodzius R, Brenner S, Venkatesh B. Large number of ultraconserved elements were already present in the jawed vertebrate ancestor. Mol Biol Evol. 2009;26:487–490. doi: 10.1093/molbev/msn278. [DOI] [PubMed] [Google Scholar]
- 8.Davidson EH. The Regulatory Genome. Gene Regulatory Networks in Development and Evolution. San Diego: Academic/Elsevier; 2006. [Google Scholar]
- 9.Oda-Ishii I, Bertrand V, Matsuo I, Lemaire P, Saiga H. Making very similar embryos with divergent genomes: Conservation of regulatory mechanisms of Otx between the ascidians Halocynthia roretzi and Ciona intestinalis. Development. 2005;132:1663–1674. doi: 10.1242/dev.01707. [DOI] [PubMed] [Google Scholar]
- 10.Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet. 2008;4:e1000106. doi: 10.1371/journal.pgen.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinman VF, Nguyen A, Davidson EH. Caught in the evolutionary act: Precise cis-regulatory basis of difference in the organization of gene networks of sea stars and sea urchins. Dev Biol. 2007;312:584–595. doi: 10.1016/j.ydbio.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Davidson EH, Erwin DH. An integrated view of Precambrian eumetazoan evolution. Cold Spring Harb Symp Quant Biol. 2009;74:65–80. doi: 10.1101/sqb.2009.74.042. [DOI] [PubMed] [Google Scholar]
- 13.Hayakawa E, Fujisawa C, Fujisawa T. Involvement of Hydra achaete-scute gene CnASH in the differentiation pathway of sensory neurons in the tentacles. Dev Genes Evol. 2004;214:486–492. doi: 10.1007/s00427-004-0430-4. [DOI] [PubMed] [Google Scholar]
- 14.Hennig AK, Peng GH, Chen S. Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res. 2008;1192:114–133. doi: 10.1016/j.brainres.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ranade SS, et al. Analysis of the Otd-dependent transcriptome supports the evolutionary conservation of CRX/OTX/OTD functions in flies and vertebrates. Dev Biol. 2008;315:521–534. doi: 10.1016/j.ydbio.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swaroop A, Kim D, Forrest D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci. 2010;11:563–576. doi: 10.1038/nrn2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tessmar-Raible K, et al. Conserved sensory-neurosecretory cell types in annelid and fish forebrain: Insights into hypothalamus evolution. Cell. 2007;129:1389–1400. doi: 10.1016/j.cell.2007.04.041. [DOI] [PubMed] [Google Scholar]
- 18.Herrin BR, Cooper MD. Alternative adaptive immunity in jawless vertebrates. J Immunol. 2010;185:1367–1374. doi: 10.4049/jimmunol.0903128. [DOI] [PubMed] [Google Scholar]
- 19.Hibino T, et al. The immune gene repertoire encoded in the purple sea urchin genome. Dev Biol. 2006;300:349–365. doi: 10.1016/j.ydbio.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 20.Messier-Solek C, Buckley KM, Rast JP. Highly diversified innate receptor systems and new forms of animal immunity. Semin Immunol. 2010;22:39–47. doi: 10.1016/j.smim.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 21.Pearson JC, Juarez MT, Kim M, Drivenes O, McGinnis W. Multiple transcription factor codes activate epidermal wound-response genes in Drosophila. Proc Natl Acad Sci USA. 2009;106:2224–2229. doi: 10.1073/pnas.0810219106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao F, Davidson EH. Transfer of a large gene regulatory apparatus to a new developmental address in echinoid evolution. Proc Natl Acad Sci USA. 2008;105:6091–6096. doi: 10.1073/pnas.0801201105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davidson EH, Erwin DH. Evolutionary innovation and stability in animal gene networks. J Exp Zoolog B Mol Dev Evol. 2010;314:182–186. doi: 10.1002/jez.b.21329. [DOI] [PubMed] [Google Scholar]
- 24.Putnam NH, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 25.Woolfe A, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holland LZ, et al. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 2008;18:1100–1111. doi: 10.1101/gr.073676.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park HC, et al. Structural comparison of zebrafish Elav/Hu and their differential expressions during neurogenesis. Neurosci Lett. 2000;279:81–84. doi: 10.1016/s0304-3940(99)00940-4. [DOI] [PubMed] [Google Scholar]
- 28.Raible F, Brand M. Divide et Impera—the midbrain-hindbrain boundary and its organizer. Trends Neurosci. 2004;27:727–734. doi: 10.1016/j.tins.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Argenton F, et al. Ectopic expression and knockdown of a zebrafish sox21 reveal its role as a transcriptional repressor in early development. Mech Dev. 2004;121:131–142. doi: 10.1016/j.mod.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Sandberg M, Källström M, Muhr J. Sox21 promotes the progression of vertebrate neurogenesis. Nat Neurosci. 2005;8:995–1001. doi: 10.1038/nn1493. [DOI] [PubMed] [Google Scholar]
- 31.Livant DL, Hough-Evans BR, Moore JG, Britten RJ, Davidson EH. Differential stability of expression of similarly specified endogenous and exogenous genes in the sea urchin embryo. Development. 1991;113:385–398. doi: 10.1242/dev.113.2.385. [DOI] [PubMed] [Google Scholar]
- 32.Meinertzhagen IA, Hanson JA. The development of the optic lobe. The Development of Drosophila melanogaster. In: Bate M, Martinez-Arias A, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 1363–1491. [Google Scholar]
- 33.Crémazy F, Berta P, Girard F. Sox neuro, a new Drosophila Sox gene expressed in the developing central nervous system. Mech Dev. 2000;93:215–219. doi: 10.1016/s0925-4773(00)00268-9. [DOI] [PubMed] [Google Scholar]
- 34.Buescher M, Hing FS, Chia W. Formation of neuroblasts in the embryonic central nervous system of Drosophila melanogaster is controlled by SoxNeuro. Development. 2002;129:4193–4203. doi: 10.1242/dev.129.18.4193. [DOI] [PubMed] [Google Scholar]
- 35.Hawkins RD, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 37.Erwin DH, Davidson EH. The evolution of hierarchical gene regulatory networks. Nat Rev Genet. 2009;10:141–148. doi: 10.1038/nrg2499. [DOI] [PubMed] [Google Scholar]
- 38.Denes AS, et al. Molecular architecture of annelid nerve cord supports common origin of nervous system centralization in Bilateria. Cell. 2007;129:277–288. doi: 10.1016/j.cell.2007.02.040. [DOI] [PubMed] [Google Scholar]
- 39.Lowe CJ, et al. Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell. 2003;113:853–865. doi: 10.1016/s0092-8674(03)00469-0. [DOI] [PubMed] [Google Scholar]
- 40.Seibert J, Urbach R. Role of en and novel interactions between msh, ind, and vnd in dorsoventral patterning of the Drosophila brain and ventral nerve cord. Dev Biol. 2010;346:332–345. doi: 10.1016/j.ydbio.2010.07.024. [DOI] [PubMed] [Google Scholar]
- 41.Gehring WJ, Ikeo K. Pax 6: Mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15:371–377. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.