Abstract
Rett syndrome (RTT) is one of the most prevalent female neurodevelopmental disorders that cause severe mental retardation. Mutations in methyl CpG binding protein 2 (MeCP2) are mainly responsible for RTT. Patients with classical RTT exhibit normal development until age 6–18 mo, at which point they become symptomatic and display loss of language and motor skills, purposeful hand movements, and normal head growth. Murine genetic models and postmortem human brains have been used to study the disease and enable the molecular dissection of RTT. In this work, we applied a recently developed reprogramming approach to generate a novel in vitro human RTT model. Induced pluripotent stem cells (iPSCs) were derived from RTT fibroblasts by overexpressing the reprogramming factors OCT4, SOX2, KLF4, and MYC. Intriguingly, whereas some iPSCs maintained X chromosome inactivation, in others the X chromosome was reactivated. Thus, iPSCs were isolated that retained a single active X chromosome expressing either mutant or WT MeCP2, as well as iPSCs with reactivated X chromosomes expressing both mutant and WT MeCP2. When these cells underwent neuronal differentiation, the mutant monoallelic or biallelelic RTT-iPSCs displayed a defect in neuronal maturation consistent with RTT phenotypes. Our in vitro model of RTT is an important tool allowing the further investigation of the pathophysiology of RTT and the development of the curative therapeutics.
Rett syndrome (RTT) is one of the most common causes of mental retardation in females, with an incidence of 1 in 10,000–15,000 female births (1). Signs and symptoms of RTT appear at 6–18 mo after birth and typically include severe mental retardation, absence of speech, stereotypic hand movements, epileptic seizures, encephalopathy, and respiratory dysfunction (1). Mutations in methyl CpG binding protein 2 (MeCP2) were identified as the primary genetic cause of RTT by Amir et al. (2). Characterization of MeCP2 has revealed three functional domains: a methyl CpG binding domain (MBD), a transcriptional repression domain (TRD), and a carboxyl terminal domain (CTD). The presence of the TRD in MeCP2 suggests a potential role of MeCP2 in transcription repression, and indeed it is involved in gene silencing through binding to methylated CpG and chromatin remodeling (3). In addition, recent molecular and cellular studies in mouse models and cell cultures have shown that MeCP2 also plays roles in transcription activation, long-range chromatin remodeling, and regulation of alternative splicing (4, 5).
More than 300 pathogenic mutations in MeCP2 have been described, most commonly missense or nonsense mutations and most frequently located in the MBD, TRD, or CTD (6). MeCP2 is located on the X chromosome at Xq28. During normal development, one X chromosome in each cell randomly becomes inactivated, and females display a mosaicism of cells with either paternal or maternal X chromosomes. In the case of RTT, variability in the inactivation of the WT or mutant MeCP2 leads to a spectrum of phenotypic severity (7–9). Indeed, females with RTT often exhibit nonrandom activation of X chromosomes favoring the WT allele and resulting in subdued phenotypes (9).
Mice with germline mutations in MeCP2 have provided key models enabling the dissection of the developmental role of MeCP2 in RTT (10). These mice display several phenotypic characteristics mimicking human RTT. Although MeCP2 is broadly expressed in most tissues, neuron-specific deletion of MeCP2 recapitulates the RTT symptoms of the whole-body KO mouse, implicating an essential role for MeCP2 in neurons (11). As neuronal stem cells differentiate into neurons, MeCP2 expression gradually increases. Recently, glial cells were also found to express MeCP2 and to play an important role in pathogenesis of RTT (12–14).
Regardless of the essential information on RTT disease obtained from murine models, the phenotypes of the MeCP2 null KO mouse are not identical to the human phenotypes. Homozygous MeCP2 null female mice are born to term and survive until 4 wk after birth, and heterozygous MeCP2 female mice do not show obvious RTT phenotypes (10). In contrast, only heterozygous human females develop RTT, and homozygotes do not survive. These results suggest that human MeCP2 has a role distinct from murine MeCP2. Likewise, whereas murine and human MeCP2 are essential for neuronal maturation but not for neurogenesis, Xenopus MeCP2 is critical for neurogenesis (15). Thus, developing a human RTT model that more closely recapitulates the human disease is essential for a true understanding of human RTT.
Postmortem human brain tissue samples have provided invaluable insight into RTT (16). Abnormal neuronal development, RTT-specific gene expression including MeCP2, and reliable epigenetic markers have been identified using postmortem brains from patients with RTT. However, the difficulty in obtaining live human brain tissues and cells from either healthy donors or RTT patients necessitates the development of an alternative approach to directly study neurons from human patients with RTT. Human embryonic stem cells (hESCs) are considered an important cellular resource for studying disease in vitro because of their features of self-renewal and pluripotency, which allow differentiation of cells of all three germ layers. The recent successful generation of induced pluripotent stem cells (iPSCs) from patients’ somatic cells by ectopically expressing four transcription factors (Oct4, Sox2, Myc, and Klf4) allows the derivation of pluripotent stem cells from patients with Mendelian genetic or nongenetic diseases. Because human iPSCs acquire similar features as hESCs and can differentiate into cells of all three germ layers, including neuronal and glial cells, iPSCs provide an opportunity to study human neuronal disorders in vitro. iPSCs have been used to model human neuronal diseases, including spinal muscular atrophy, familial dysautonomia, and amyotrophic lateral sclerosis (17–19). iPSCs from patients with spinal muscular atrophy and familial dysautonomia exhibit the phenotypic characteristics of the respective diseases, whereas the in vitro phenotype of amyotrophic lateral sclerosis is yet to be confirmed.
Recently, the Muotri laboratory generated human iPSCs from patients with RTT using retroviral vector expressing the foregoing transcription factors (20). The female RTT-iPSCs thus generated exhibited the reactivation of randomly inactivated fibroblast X chromosome and expressed both WT and mutant MeCP2 from two active X chromosomes. When differentiated into the neuronal lineage, RTT-iPSCs recapitulated the in vivo phenotypes, including synapse defects, smaller soma size, altered calcium signaling, and electrophysiological defects. That study demonstrates the invaluable proof of principle of generating a human RTT syndrome model using iPSCs.
In the present work, we similarly attempted to generate an in vitro RTT model using human iPSCs. Expanding the work of the Muotri laboratory, we found that factor-based reprogramming produces both iPSCs that have the reactivated X chromosomes and iPSCs that retain the randomly inactivated fibroblast X chromosome. which allows the generation of isogenic iPSC clones expressing either WT or mutant MeCP2. These two types of iPSC clones originate from the same fibroblasts and express the same genes except the genes in the X chromosome, including MeCP2. Thus, they are ideal isogenic cell lines for comparing the phenotypes caused by the given mutation. To examine the effect of MeCP2 mutations on neuronal differentiation, we applied monoallelic WT and mutant iPSCs, as well as biallelic iPSCs, to neuronal differentiation. The initial neuronal differentiation of mutant RTT iPSCs was not different from that of WT iPSCs and hESCs; however, during late neuronal maturation, the mutant RTT iPSCs exhibited defects mimicking the RTT syndrome phenotypes. Along with providing an important tool for studying RTT human disease, our results demonstrate the feasibility of generating isogenic iPSCs to study X-linked monogenic disorders.
Results
iPSCs from RTT Fibroblasts.
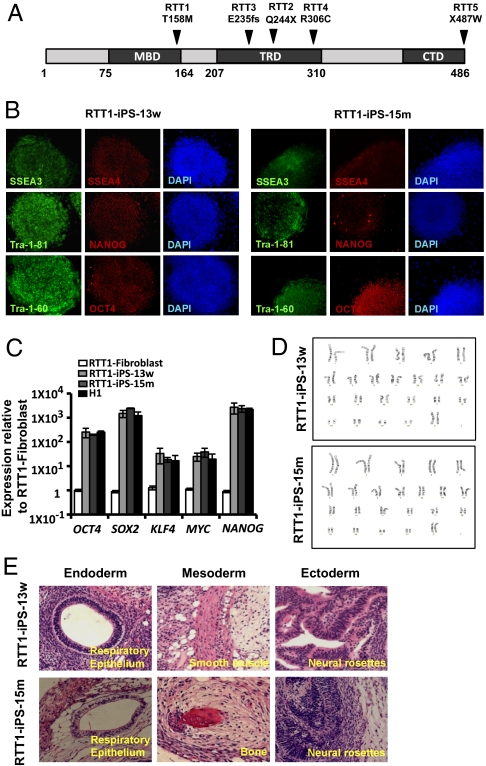
As discussed earlier, MeCP2 has three main domains: MBD, TRD, and CTD (Fig. 1A). Mutations in each of these domains or elsewhere, such as within the two nuclear localization signals, lead to the improper regulation or lack of MeCP2 expression and are a primary cause of RTT. To investigate the impact of mutations in each domain of MeCP2 on the manifestation of the in vitro RTT phenotype, we selected specific fibroblasts from patients with RTT with different mutations to generate the following RTT-iPSCs for the domains of MeCP2: RTT1 (GM17880) for the MBD, RTT2 (GM16548) and RTT3 (GM07982) for the TRD and CTD, RTT4 (GM11270) for the TRD, and RTT5 (GM17567) for the CTD (Table 1).
Fig. 1.
Characterization of RTT- iPSCs derived from RTT patients. (A) Structure of MeCP2 and location (amino acid residues) of mutations. (B) Representative immunofluorescence images of RTT-iPSC-w and RTT-iPSC-m showing expression of pluripotency markers of SSEA3, SSEA4, Tra-1-81, Tra-1-60, NANOG, and OCT4. (C) Relative expression of OCT4, SOX2, KLF4, MYC, and NANOG in monoallelic WT and mutant RTT-iPSCs compared with parental fibroblasts and H1 hESCs by real-time qPCR. qPCR was performed in triplicate, and the relative gene expression was calculated by normalizing against the parental RTT fibroblast cells. Mean ± SEM are shown with error bar. (D) Images for normal karyotype of iPSCs. (E) Representative images of teratomas generated in immunodeficient mice injected with RTT-iPSCs. RTT-iPSCs composed a well-defined cystic teratoma with tissues of all three germ layers including endoderm (respiratory epithelium), mesoderm (smooth muscle and bone), and ectoderm (neuronal rosettes).
Table 1.
Fibroblasts from female RTT patients used to derive iPSCs
| Name | Age, years | Nucleotide change | Amino acid change | Affected domain | Representatitve iPSC lines |
| RTT1 | 5 | 473C > T | T158M | MBD | RTT1-iPS-13w |
| GM17880 | RTT1-iPS-15m | ||||
| RTT2 | 5 | 730C > T | Q244X | TRD/CTD | RTT2-iPS-16w |
| GM16548 | RTT2-iPS-12m | ||||
| RTT3 | 25 | 705delG | E235fs | TRD/CTD | RTT3-iPS-42w |
| GM07982 | RTT3-iPS-46m | ||||
| RTT4 | 8 | 916C > T | R306C | TRD | RTT4-iPS-24w |
| GM11270 | RTT4-iPS-19bi | ||||
| RTT5 | 5 | 1461A > G | X487W | CTD | RTT5-iPS-31w |
| GM17567 | RTT5-iPS-32bi |
We have successfully generated iPSCs from RTT fibroblasts by infecting them with retrovirus expressing four reprogramming factors: OCT4, SOX2, KLF4, and MYC (Table 1) (21). Despite the potential involvement of MeCP2 in cell fate maintenance by binding to methylated CpG sequences in fibroblasts, we did not see much difference in the reprogramming efficiency of deriving iPSCs from different RTT fibroblasts compared with normal fibroblasts. We subsequently verified the pluripotency of RTT-iPSCs by immunostaining of the pluripotency markers SSEA3, SSEA4, OCT4, NANOG, Tra-1-81, and Tra-1-60 (Fig. 1B and SI Appendix, Fig. S1). We also confirmed expression of the pluripotency genes OCT4, SOX2, NANOG, KLF4, and MYC by quantitative RT-PCR (qRT-PCR) in iPSCs (Fig. 1C and SI Appendix, Fig. S2). The reactivation of endogenous pluripotency genes and silencing of retroviral ectopic genes confirmed that these were faithfully reprogrammed iPSCs (SI Appendix, Fig. S2E) (22). All of the RTT-iPSCs expressed pluripotency genes at levels comparable to those of hESCs and iPSCs from normal fibroblasts, further confirming that MeCP2 does not play a major role in pluripotency (23). All of the RTT-iPSCs maintained the normal karyotype of 46, XX (Fig. 1D and SI Appendix, Fig. S3). To examine in vivo pluripotency, we injected iPSCs into immune-deficient mice. RTT-iPSCs formed the well-defined cystic teratomas containing tissues of all three embryonic germ layers, confirming their pluripotency (Fig. 1E and SI Appendix, Fig. S4).
Retention and Reactivation of Inactive X Chromosome in RTT-iPSCs.
Reprogramming somatic cells into pluripotent stem cells involves global epigenetic changes, including posttranslational histone modification and DNA methylation (24). iPSCs from murine female fibroblasts display reactivation of the inactive X chromosome, which is one of the features of mouse ESCs derived from the inner cell mass of blastocysts (25). There have been conflicting reports of X chromosome status in female hESCs, however; some cells have both X chromosomes in an active state, whereas others retain random inactivation of X chromosomes (26, 27). Recent successful isolation of murine pluripotent stem cells from epiblasts (EpiESCs) that have an inactive X chromosome suggests that hESCs are equivalent to murine EpiESCs and thus have inactive X chromosomes (28).
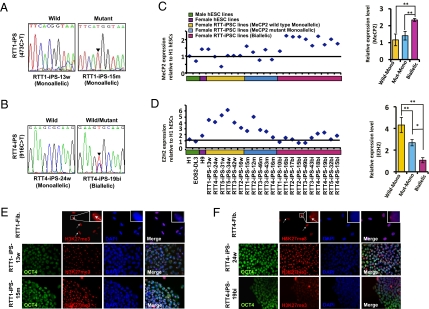
Similar to hESCs, human iPSCs have been shown to have conflicting X chromosome status. The Plath laboratory reported that female iPSCs retain the randomly inactivated X chromosome status of fibroblasts (29), whereas human iPSCs isolated by the Muotri laboratory reactivated the inactive X chromosome (20). Because MeCP2 is present in the X chromosome, the X chromosome status in iPSCs will determine the cells’ normal or pathogenic status. Thus, we examined the X chromosome status in RTT-iPSC lines. We isolated RNA and examined the identity of the MeCP2 transcript expressed by each iPSC line. Expanding the reports of the Plath and Muotri laboratories, we found that some clones expressed MeCP2 of only one allele (e.g., RTT1-iPS-13w, with “w” representing WT; RTT1-iPS-15m, with “m” representing mutant), whereas others expressed MeCP2 of both alleles (RTT4-iPS-19bi, with “bi” representing biallelelic) (Table 1, Fig. 2 A and B, SI Appendix, Table S1 and Fig. S5). Regardless of the types of mutations in MeCP2 of the parental fibroblasts, RTT-iPSCs were isolated that exhibited monoallelic or biallelic expression of MeCP2. Consistent with the allelic specificity, the relative expression of MeCP2 was well correlated among monallelic or biallelic iPSCs (Fig. 2C). MeCP2 expression was approximately twice as great in biallelic iPSCs than in monoallelic iPSCs and male hESCs. Furthermore, MeCP2 expression was inversely correlated with the expression of EZH2, a core component of the polycomb repressive complex, which plays a role in initiation and spreading of X chromosome inactivation (XCI) (Fig. 2D) (30). Interestingly, monoallelic mutant RTT-iPSCs expressed less EZH2 than monoallelic WT-iPSCs, consistent with the recent report that MeCP2 is a positive regulator for EZH2 (31). How MeCP2 regulates EZH2 is not yet fully explored, and will give an important clue to RTT. The formation of foci that represents the inactivated X chromosome was found when monoalellic iPSCs were stained with H3K27me3 antibody, a repressive chromatin mark which accumulates on the inactive X-chromosome (Fig. 2 E and F and SI Appendix, Fig. S6) (30). Male hESCs and biallelic H9 hESCs did not show H3K27me3 foci (SI Appendix, Fig. S7A). Both retention and reactivation of the inactive X chromosome were also found in iPSCs derived from normal fetal female fibroblasts, Detroit 551, and normal adult female fibroblasts, PGP9f (SI Appendix, Fig. S7 B and C). Our results show that human female somatic cell reprogramming in general—not specific to RTT fibroblasts—produce iPSCs that have monoallelic as well as biallelic X chromosomes.
Fig. 2.
XCI status of iPSCs. (A and B) Sequences of MeCP2 expressed in RTT-iPSCs. RTT1-iPS-13w, RTT4-iPS-24w, and RTT1-iPS-15m express only WT or mutant MeCP2, whereas RTT4-iPS-19bi expresses both mutant and WT MeCP2. (C and D) Relative expression of MeCP2 (C) and EZH2 (D) in different iPSCs and human ESCs. iPSCs showing the X chromosome reactivation display approximately twice the MeCP2 expression as iPSCs retaining the inactive X chromosome (C). The difference in MeCP2 expression was not observed between WT and mutant monoallelic RTT-iPSCs. EZH2, a polycomb group protein essential for XCI, shows an expression pattern inversely correlated with MeCP2 expression (D). Interestingly, mutant monoallelic RTT-iPSCs has significant less expression of EZH2 than WT monoallelic RTT-iPSCs. Histograms show the average levels of MeCP2 and EZH2 distributions quantified by qPCR and normalized with β-actin. Data are mean ± SEM. *P < 0.01; **P < 0.001. (E and F) Nuclear pattern of H3K27me3 in RTT fibroblasts and iPSCs. RTT1 fibroblast cells and RTT1-derived iPSC lines monoallelically expressing MeCP2 (RTT1-13w and RTT1-15m) exhibit predominant H3K27me3 nuclear foci (E). RTT4 fibroblasts and RTT4-iPS24w cells monoallelically expressing MeCP2 show condensed H3K27me3 staining, whereas RTT4-iPS19bi cells display diffuse staining (F). Nuclei were detected with DAPI, and pluripotency marker (OCT4) was stained. The arrow points to H3K27me3 Xi enrichment in the fibroblast cells. (Insets) Higher-resolution images.
The previous analysis of X chromosome status in hESCs suggested that the culture condition to derive hESCs is a critical determinant of X chromosome status. When hESCs are derived in 5% physiological oxygen concentration, they show two active X chromosomes, whereas those isolated in 20% ambient oxygen concentration have one active X chromosome (32). Cellular stresses, such as oxidative stress, long-term culture, or freezing/thawing of cells, were shown to induce XCI in biallelic hESCs (32). We tested whether cellular stress alters the X chromosome status in female iPSCs. To examine the effect of the long-term culture on the X chromosome status, we cultured RTT-iPSCs over 20 passages and stained them with H3K27me3 antibody. We found that the X chromosome status of iPSCs of early passage was still maintained after 20 passages in both biallelic and monoallelic iPSCs (SI Appendix, Fig. S8A). Whereas biallelic hESCs underwent XCI on freezing and thawing, RTT-iPSCs maintained their biallelic status (SI Appendix, Fig. S8B). This finding suggests that the X chromosome status of iPSCs remains stable even under physiological stress conditions.
Neuronal Maturation Defect in RTT-iPSCs.
RTT is a neurodevelopmental disorder. Brains of patients with RTT and MeCP2 null mice display immature neuronal development (11, 33). Thus, we set out to examine whether the MeCP2 mutations in iPSCs affect neuronal development and recapitulate the phenotypes in vitro. We used iPSCs with monoallelic WT MeCP2 (RTT1-iPS-13w, RTT3-iPS-42w, RTT4-iPS-24w, and RTT5-iPS-31w), those with monoallelic mutant MeCP2 (RTT1-iPS-15m and RTT3-iPS-46m), and those with both WT and mutant MeCP2 (RTT4-iPS-19bi and RTT5-iPS-32bi) (Table 1 and SI Appendix, Fig. S1) for neuronal differentiation (Fig. 3A). iPSCs from normal fibroblasts (normal iPSCs: PGP1-iPS1, PGP9f-iPS1, and 551-iPS-K1) and hESCs (H1 and EOS2) were used as controls.
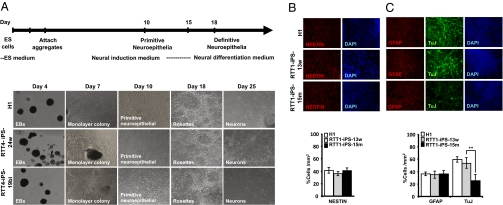
Fig. 3.
Defect in neuronal maturation of mutant RTT-iPSCs. (A) Schematic depicting the neural differentiation procedure. See Materials and Methods for a detailed description. Representative images showing morphological changes during neural differentiation of hESCs (H1), and RTT-iPSCs (RTT4-iPS-24w, RTT4-iPS-19bi). (B and C) H1 hESCs and RTT-iPSCs were induced for neuronal differentiation and stained with neuronal stem cell marker (NESTIN), mature neuronal marker (TuJ), and astrocyte marker (GFAP) during the differentiation. H1 hESCs and WT and mutant RTT-iPSCs all show a similar neuronal commitment, represented by NESTIN staining after 11 d of neural differentiation (B). H1 hESCs and WT RTT1-iPS-13w cells are fully differentiated into neuronal differentiation for 25 d and display a robust expression of the mature neuronal marker TuJ, whereas mutant RTT1-iPS-15m cells demonstrate less neural maturation. In contrast, glial differentiation (GFAP+ cells) is similar in all cell lines (C). **P < 0.01. Bar graphs show the percentages of TuJ+ and GFAP+ cells. The densities of TuJ+ and GFAP+ cells vs. DAPI per square mm were analyzed using Image J analysis software. Data are mean ± SEM of one experiment performed in triplicate.
Neural differentiation was initiated by scraping the hESCs or iPSCs and creating embryoid bodies (EBs) in hES medium. hESCs and normal and RTT-iPSCs exhibited no noticeable difference in the ability to form EBs, consistent with the fact that mutations in MeCP2 do not affect the global developmental potential of pluripotent stem cells (23). After 4 d of EB formation, EBs were incubated in neural induction medium to initiate neuronal differentiation. At 2 d after incubation in neural induction medium, cells were dissociated and plated on laminin-coated wells in neural induction medium. Cells migrated outward from the spherical center within 1 d and formed neural rosettes after 5 d, suggesting the successful initiation of neural differentiation (Fig. 3A) (34). Whereas the ability to form rosettes varied between hESCs and iPSCs, all of the RTT-iPSCs as well as the normal iPSCs and hESCs showed robust neuronal differentiation. Previous analyses revealed differences in the proliferation rate and differentiation potential even among normal hESC and iPSC clones (35, 36), Thus, the differences in rosette-forming ability among WT and mutant RTT-iPSC clones is not unexpected. However, more detailed analysis is needed to determine whether the mutations in MeCP2 have any effect on early neuronal differentiation.
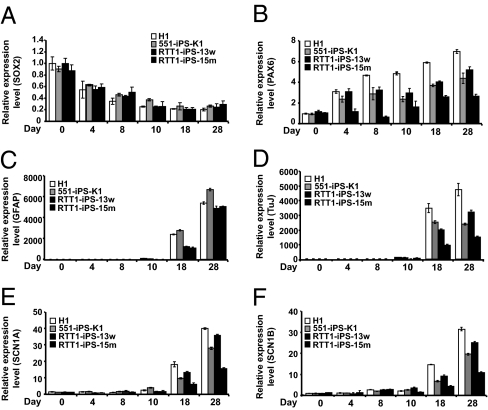
To examine whether MeCP2 mutations affect neural maturation, we further differentiated rosettes in neural differentiation medium, and performed immunostaining at various stages of differentiation (Fig. 3 B and C). We also examined the change in neuronal gene expression with qRT-PCR (Fig. 4 A–D and SI Appendix, Fig. S12). At 5 d after plating of EBs on laminin, the majority of cells expressed NESTIN, a specific marker for the neuronal stem cells, with no significant differences among hESCs, normal iPSCs, WT RTT-iPSCs, and monoallelic or biallelic mutant RTT-iPSCs (Fig. 3B and SI Appendix, Fig. S9). Expression of SOX2, a marker for neuronal stem cells, was not significantly different among hESCs, normal iPSCs, and RTT-iPSCs (Fig. 4A and SI Appendix, Fig. S12), whereas expression of PAX6 was lower in mutant RTT-iPSCs (Fig. 4B and SI Appendix, Fig. S12), suggesting a late maturation defect in mutant RTT-iPSCs. When stained for GFAP, a marker for glial cells, mutant RTT-iPSCs did not differ significantly from hESCs, normal iPSCs, and WT RTT-iPSCs (Figs. 3C and 4C and SI Appendix, Fig. S9) (1). However, mutant RTT-iPSCs, both monoallelic and bialleleic, formed significant fewer TuJ+ cells than hESCs (H1), normal iPSCs (PGP1-iPS1, PGP9f-iPS1, and 551-iPS-K1), and WT RTT-iPSCs (Fig. 3C and SI Appendix, Figs. S9–S11). In agreement with the immunostaining data, mutant RTT-iPSCs had lower expression of TuJ and the sodium channels, markers of the mature neuron (Fig. 4 D–F and SI Appendix, Fig. S12) (37). A recent report by the Muotri laboratory provided a detailed analysis of neuronal maturation defects caused by MeCP2 mutations (20), noting the lower neurite aborization and electrophysiological defects in neurons differentiated from biallelic RTT-iPSCs. We found similarly lower neuronal maturation in monoallelic mutant RTT-iPSCs as in biallelic RTT-iPSCs.
Fig. 4.
Expression of neuronal markers during differentiation of hESCs and iPSCs. (A–F) RNA was isolated from each time point during neuronal differentiation, and real-time qPCR was performed with primers to detect SOX2, PAX6, GFAP, TuJ, SCN1A, and SCN1B. hES cells (H1), normal iPSCs (551-iPS-K1), WT RTT1-iPS-13w cells, and mutant RTT1-iPS-15m iPSCs were used. Gene expression was normalized against β-actin and calculated against H1 at day 0. Data are mean ± SEM in one experiment performed in triplicate.
We further tested the neuronal maturation defect in RTT-iPSCs using a different neuronal differentiation protocol based on inhibition of BMP signaling by noggin and isolating neural progenitor cells (NPCs) from rosettes (38). NPCs isolated from hESCs, normal iPSCs, WT RTT-iPSCs, and mutant RTT-iPSCs demonstrated no differences in morphology and expression of the early neuronal marker NESTIN (SI Appendix, Fig. S11A). NPCs were further differentiated and examined for their glial and neuronal differentiation. Like the neuronal cells directly differentiated from rosettes, differentiated NPCs from mutant RTT-iPSCs exhibited fewer TuJ1+ cells than hESCs, normal iPSCs, or WT RTT-iPSCs (SI Appendix, Fig. S11B), but no differences in the number of GFAP+ cells differentiated from these cells. Moreover, we exclude the possibility of apoptosis as a cause of manifestation of neuronal maturation defect in mutant RTT-iPS cells, because WT and mutant RTT-iPS cells showed no differences in apoptosis (SI Appendix, Fig. S13). These results support our conclusion that MeCP2 contributes to the maturation of neuronal cells.
Discussion
In this study, we isolated pluripotent stem cells from patients with RTT. Although some iPSC clones retain the inactive X chromosome of fibroblasts, others reactivate the inactive X chromosome, thereby allowing the generation of mutant, WT, or biallelic RTT-iPSCs. Although the factors that direct the activation or inactivation of X chromosomes in human iPSCs are not clear, RTT-iPSCs demonstrated pluripotency regardless of X chromosome status. Female fibroblast cell lines with no MeCP2 mutations (Detroit 551 or PGP9f) were also shown to become both monoallelic and biallelic iPSCs, suggesting that both the retention and reactivation of X chromosome status during reprogramming are not restricted to MeCP2 mutant cell lines. Recently, the Ellis group generated iPSCs from patients with RTT and similarly observed retention of the inactive somatic X chromosome in iPSCs (39), independently confirming our findings. Pluripotent ESCs derived from inner cell mass are considered to be in the “naïve” state and have two active X chromosomes, whereas pluripotent EpiESCs derived from epiblasts have undergone random XCI and considered to be in the “primed” state (28). hESCs cultured in serum-free conditions with bFGF have similar features as EpiESCs; however, when hESCs are cultured in the presence of LIF, ERK1/2 inhibitor (PD0325901), and GSK3 inhibitor (CHIR99021), the ectopic expression of OCT4 and KLF4 or of KLF4 and KLF2 reactivates the X chromosomes in female hESCs and iPSCs (40), suggesting that the X chromosomes are in a highly reversible state. The continuous expression of OCT4 and KLF4 is required to maintain them in this naïve state (40). In the present study, the formation of biallelic iPSCs without ERK1/2 inhibitor and GSK3 inhibitor treatment might be related to the high ectopic expression of OCT4 and KLF4. However, the relative expression of total OCT4 and KLF4 does not differ in monoallelic and biallelic iPSCs (SI Appendix, Fig. S2); thus, it seems likely that intrinsic or environmental factors other than the level of ectopic gene expression influence X chromosome status.
A recent meta-analysis of entire genes in the X chromosome in hESCs and iPSCs by the Benvenisty laboratory showed the presence of both monoallelic and biallelic hESCs and iPSCs (41), consistent with our findings. Depending on the expression of genes on the X chromosome, the Benvenisty laboratory grouped hESCs and iPSCs into three categories: cell lines with no XCI, those with partial XCI, and those with complete XCI. A total of 22 iPSC lines derived via retroviral or lentiviral infection of reprogramming factors, direct delivery of reprogramming proteins, and minicircle derivation were included in the analysis and categorized into the three groups. Interestingly, iPSC lines generated by nonviral methods tend to have biallelic X chromosome status, while those by lentivirus seems to have monoallelic X chromosome status (41). Retrovirus-mediated reprogramming generated iPSCs that showed both biallelic and monoallic expression of X chromosome genes (41). However, more female iPSCs derived from various methods need to be evaluated to investigate the effect of reprogramming methods on X chromosome status in iPSCs.
Monoallelic iPSCs with WT MeCP2 in active X chromosomes and those with mutant MeCP2 in active X chromosomes yielded the unexpected but important isogenic cell lines. These cells have exactly the same genetic composition with the exception of one X chromosome and MeCP2 status, allowing for direct comparison between the lines. This finding will be important in the development of in vitro genetic models and cell therapies for X-linked diseases, such as Duchenne muscular dystrophy, fragile X syndrome, Wiskott–Aldrich syndrome, Aicardi syndrome, and α-thalassemia (29). Female iPSCs with either WT or mutant alleles can be derived from patients of these diseases. Following the direct differentiation of lineages that are affected in the disease, molecular signaling pathways can be investigated to study the disease, or cells from iPSCs with WT allele can be transplanted to cure the disease.
Consistent with previous findings in postmortem brains of patients with RTT and MeCP2 mutant mice, we found that mutant RTT-iPSCs showed a defect in neuronal maturation and, interestingly, biallelic RTT-iPSCs and monoallelic mutant RTT-iPSCs had similar defects in neuronal maturation. Whereas homozygous MeCP2 null mice had a more severe neuronal and behavioral phenotype than heterozygous MeCP2 mice (11, 23), we found no phenotypic difference between biallelic and monalllelic mutant RTT-iPSCs. This might be due to the limited resolution in our in vitro neuronal differentiation assay, or, althernatively, the phenotypic manifestation in the whole organism might be due to a systemic effect of MeCP2 mutations. If the in vitro neuronal maturation is solely regulated cell-autonomously, then biallelic iPSCs should exhibit fewer defects than monoallelic mutant RTT-iPSCs. Thus, our results suggest a noncell autonomous effect of MeCP2 mutations on neuronal maturation. Likewise, MeCP2 mutant glia has been shown to play an important role in the pathogenesis of RTT (12, 14). Monoallelic WT and mutant RTT-iPSCs generated in the current research will be useful resources for investigating their reciprocal effect from neurons and glia differentiated from WT or mutant RTT-iPSCs.
Here we have compared neuronal differentiation in four different MeCP2 mutant iPSCs: RTT1, RTT3, RTT4, and RTT5 (SI Appendix, Table S1). All four of these mutant iPSCs exhibited similar defects in neuronal maturation. Recently, the Muotri laboratory independently reported these neuronal maturation defects in biallelic iPSCs isolated from the same parental fibroblasts (RTT1 and RTT2) (20). Whether the different mutations of MeCP2 cause the same phenotypes via common molecular signaling pathways or different pathways remains unclear. RTT1 has a mutation in MBD and presumably has lost its interaction with targets, whereas RTT4 has a mutation in the transcription repression domain and may still play a role in long-range gene regulation despite the loss of its role in direct transcription regulation (4). Future in-depth analysis of how the different mutants of MeCP2 lead to RTT phenotypes will be very informative in developing the therapeutics specific for RTT caused by distinct mutations. Overall, our RTT in vitro model is an important tool for investigating the pathophysiology of RTT and identifying new therapies to cure the disease.
Materials and Methods
Cell Culture and Reprogramming.
Fibroblast cell lines from patients with RTT (RTT1: GM17880; RTT2: GM16548; RTT3: GM07982; RTT4: GM11270; RTT5: GM17567) (Table 1) were obtained from the Coriell Institute for Medical Research. Normal fetal lung fibroblast Detroit 551 was purchased from American Type Culture Collection (CCL-110). Fibroblasts were infected with pMIG retrovirus expressing four reprogramming factors and selected for iPSCs as described previously (42). Normal adult male iPSCs (PGP1-iPS1) and female iPSCs from PGP9f fibroblasts were derived previously (43). Human ES cells (H1, ES02) and iPSCs were maintained and passaged according to standard protocol (42).
Neuronal Differentiation, Gene Expression Analysis, and Immunostaining.
hESCs and iPSCs were differentiated into neural rosettes and neurons as described previously (34). NPCs and neurons were isolated from these cells using a method based on blocking the BMP pathway (38). RNA was isolated and used for gene expression analysis using primers listed in SI Appendix, Table S2. The allele-specific expression of MeCP2 was analyzed using primers that specifically bind the known mutated nucleotides of MeCP2 (SI Appendix, Table S3). For immunostaining, undifferentiated or differentiated hESCs and iPSCs were fixed with 4% paraformaldehyde and stained using the antibodies listed in SI Appendix, Table S4. Detailed descriptions of these procedures are provided in SI Appendix, Materials and Methods.
Teratoma Formation Assay, Karyotypes, and Determination of Apoptotic Cells.
Detailed descriptions of these procedures are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Flora Vaccarino for a critical reading of the manuscript and helpful comments, Dr. Jeonghee Kim and Dr. Hyesook Ahn for advice on cellular quantification, and Dr. Yinghong Ma for advice on neuronal differentiation. I.-H.P.’s work was supported by the Yale School of Medicine and the Charles Hood Foundation.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018979108/-/DCSupplemental.
References
- 1.Chahrour M, Zoghbi HY. The story of Rett syndrome: From clinic to neurobiology. Neuron. 2007;56:422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG–binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 3.Nan X, et al. Transcriptional repression by the methyl-CpG–binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 4.Chahrour M, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yasui DH, et al. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc Natl Acad Sci USA. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christodoulou J, Grimm A, Maher T, Bennetts B. RettBASE: The IRSA MECP2 variation database—a new mutation database in evolution. Hum Mutat. 2003;21:466–472. doi: 10.1002/humu.10194. [DOI] [PubMed] [Google Scholar]
- 7.Ishii T, et al. The role of different X-inactivation pattern on the variable clinical phenotype with Rett syndrome. Brain Dev. 2001;23(Suppl 1):S161–S164. doi: 10.1016/s0387-7604(01)00344-8. [DOI] [PubMed] [Google Scholar]
- 8.Amir RE, et al. Influence of mutation type and X chromosome inactivation on Rett syndrome phenotypes. Ann Neurol. 2000;47:670–679. [PubMed] [Google Scholar]
- 9.Young JI, Zoghbi HY. X-chromosome inactivation patterns are unbalanced and affect the phenotypic outcome in a mouse model of Rett syndrome. Am J Hum Genet. 2004;74:511–520. doi: 10.1086/382228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stearns NA, et al. Behavioral and anatomical abnormalities in Mecp2 mutant mice: A model for Rett syndrome. Neuroscience. 2007;146:907–921. doi: 10.1016/j.neuroscience.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 11.Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- 12.Ballas N, Lioy DT, Grunseich C, Mandel G. Non-cell autonomous influence of MeCP2-deficient glia on neuronal dendritic morphology. Nat Neurosci. 2009;12:311–317. doi: 10.1038/nn.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maezawa I, Jin LW. Rett syndrome microglia damage dendrites and synapses by the elevated release of glutamate. J Neurosci. 2010;30:5346–5356. doi: 10.1523/JNEUROSCI.5966-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maezawa I, Swanberg S, Harvey D, LaSalle JM, Jin LW. Rett syndrome astrocytes are abnormal and spread MeCP2 deficiency through gap junctions. J Neurosci. 2009;29:5051–5061. doi: 10.1523/JNEUROSCI.0324-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stancheva I, Collins AL, Van den Veyver IB, Zoghbi H, Meehan RR. A mutant form of MeCP2 protein associated with human Rett syndrome cannot be displaced from methylated DNA by notch in Xenopus embryos. Mol Cell. 2003;12:425–435. doi: 10.1016/s1097-2765(03)00276-4. [DOI] [PubMed] [Google Scholar]
- 16.Samaco RC, Nagarajan RP, Braunschweig D, LaSalle JM. Multiple pathways regulate MeCP2 expression in normal brain development and exhibit defects in autism-spectrum disorders. Hum Mol Genet. 2004;13:629–639. doi: 10.1093/hmg/ddh063. [DOI] [PubMed] [Google Scholar]
- 17.Lee G, et al. Modeling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ebert AD, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dimos JT, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 20.Marchetto MC, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 22.Chan EM, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 23.Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 24.Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silva SS, Rowntree RK, Mekhoubad S, Lee JT. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc Natl Acad Sci USA. 2008;105:4820–4825. doi: 10.1073/pnas.0712136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dvash T, Lavon N, Fan G. Variations of X chromosome inactivation occur in early passages of female human embryonic stem cells. PLoS ONE. 2010;5:e11330. doi: 10.1371/journal.pone.0011330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: Facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tchieu J, et al. Female human iPSCs retain an inactive X chromosome. Cell Stem Cell. 2010;7:329–342. doi: 10.1016/j.stem.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 31.Mann J, et al. MeCP2 controls an epigenetic pathway that promotes myofibroblast transdifferentiation and fibrosis. Gastroenterology. 2010;138:705–714. doi: 10.1053/j.gastro.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lengner CJ, et al. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141:872–883. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 33.Francke U. Mechanisms of disease: Neurogenetics of MeCP2 deficiency. Nat Clin Pract Neurol. 2006;2:212–221. doi: 10.1038/ncpneuro0148. [DOI] [PubMed] [Google Scholar]
- 34.Li XJ, Zhang SC. In vitro differentiation of neural precursors from human embryonic stem cells. Methods Mol Biol. 2006;331:169–177. doi: 10.1385/1-59745-046-4:168. [DOI] [PubMed] [Google Scholar]
- 35.Osafune K, et al. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- 36.Hu BY, et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci USA. 2010;107:4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitaker WR, et al. Distribution of voltage-gated sodium channel α-subunit and β-subunit mRNAs in human hippocampal formation, cortex, and cerebellum. J Comp Neurol. 2000;422:123–139. doi: 10.1002/(sici)1096-9861(20000619)422:1<123::aid-cne8>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 38.Gerrard L, Rodgers L, Cui W. Differentiation of human embryonic stem cells to neural lineages in adherent culture by blocking bone morphogenetic protein signaling. Stem Cells. 2005;23:1234–1241. doi: 10.1634/stemcells.2005-0110. [DOI] [PubMed] [Google Scholar]
- 39.Cheung AY, et al. Isolation of MECP2-null Rett syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation. Hum Mol Genet. 2011;20:2103–2115. doi: 10.1093/hmg/ddr093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanna J, et al. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci USA. 2010;107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bruck T, Benvenisty N. Meta-analysis of the heterogeneity of X chromosome inactivation in human pluripotent stem cells. Stem Cell Res (Amst) 2011;6:187–193. doi: 10.1016/j.scr.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 42.Park IH, Lerou PH, Zhao R, Huo H, Daley GQ. Generation of human induced pluripotent stem cells. Nat Protoc. 2008;3:1180–1186. doi: 10.1038/nprot.2008.92. [DOI] [PubMed] [Google Scholar]
- 43.Lee JH, et al. A robust approach to identifying tissue-specific gene expression regulatory variants using personalized human induced pluripotent stem cells. PLoS Genet. 2009;5:e1000718. doi: 10.1371/journal.pgen.1000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.