Abstract
A long-sought milestone in microfluidics research has been the development of integrated technology for scalable analysis of transcription in single cells. Here we present a fully integrated microfluidic device capable of performing high-precision RT-qPCR measurements of gene expression from hundreds of single cells per run. Our device executes all steps of single-cell processing, including cell capture, cell lysis, reverse transcription, and quantitative PCR. In addition to higher throughput and reduced cost, we show that nanoliter volume processing reduced measurement noise, increased sensitivity, and provided single nucleotide specificity. We apply this technology to 3,300 single-cell measurements of (i) miRNA expression in K562 cells, (ii) coregulation of a miRNA and one of its target transcripts during differentiation in embryonic stem cells, and (iii) single nucleotide variant detection in primary lobular breast cancer cells. The core functionality established here provides the foundation from which a variety of on-chip single-cell transcription analyses will be developed.
Keywords: real-time PCR, lab on a chip, transcriptional noise, microRNA, single-cell diagnostics
Single cells represent the fundamental unit of biology; however, the vast majority of biological knowledge has emerged as a consequence of studying cell populations and not individual cells. Inevitably, there are fundamental and applied questions, such as those relating to transcriptional control of stem cell differentiation, intrinsic noise in gene expression, and the origins of disease, that can be addressed only at the single-cell level. For example, single-cell analysis allows for the direct measurement of gene expression kinetics, or for the unambiguous identification of coregulated genes, even in the presence of desynchronization and heterogeneity that could obscure population-averaged measurements. Similarly, single-cell methods are vital in stem cell research and cancer biology, where isolated populations of primary cells are heterogeneous due to limitations in purification protocols, and it is often a minority cell population that is the most relevant. High-throughput single-cell measurement technologies are therefore of intense interest and have broad application in clinical and research settings.
Existing methods for measuring transcript levels in single cells include RT-qPCR (1), single molecule counting using digital PCR (2) or hybridization probes (3, 4), and next generation sequencing (5). Of these, single-cell RT-qPCR provides combined advantages of sensitivity, specificity, and dynamic range, but is limited by low throughput, high reagent cost, and difficulties in accurately measuring low abundance transcripts (6).
Microfluidic systems provide numerous advantages for single-cell analysis: economies of scale, parallelization and automation, and increased sensitivity and precision that comes from small volume reactions. Considerable effort over the last decade has been directed toward developing integrated and scalable single-cell genetic analysis on chip (7, 8). Thus, many of the basic functionalities for microfluidic single-cell gene expression analysis have been demonstrated in isolation, including cell manipulation and trapping (9, 10), RNA purification and cDNA synthesis (11–13), and microfluidic qPCR (14) following off-chip cell isolation, cDNA synthesis, and preamplification. In particular, microfluidic qPCR devices (Biomark Dynamic Array, Fluidigm) have recently been applied to single-cell studies (15, 16). Although these systems provide a high-throughput qPCR readout, they do not address the front end sample preparation and require single-cell isolation by FACS or micropipette followed by off-chip processing and preamplification of starting template prior to analysis. The critical step of integrating all steps of single-cell analysis into a robust system capable of performing measurements on large numbers of cells has yet to be reported. A single demonstration of an integrated device for directly measuring gene expression in single cells was described by Toriello et al., combining all steps of RNA capture, PCR amplification, and end-point detection of amplicons using integrated capillary electrophoresis (17). Despite the engineering complexity of this system, throughput was limited to four cells per run, cell capture required metabolic labeling of the cells, and the analysis was not quantitative. Thus, there remains an unmet need for microfluidic technologies capable of scalable and quantitative single-cell genetic analysis.
Here we describe an integrated microfluidic device for high-throughput RT-qPCR analysis of mRNA and miRNA expression at a throughput of hundreds of single cells per experiment. We show that this technology provides a powerful tool for scalable single-cell gene expression measurements with improved performance, reduced cost, and higher sensitivity as compared to analysis in microliter volumes. This technology represents the implementation of robust and high-throughput single-cell processing and amplification of nucleic acids on a chip, thereby achieving a major milestone in microfluidic biological analysis.
Results and Discussion
Device Design.
An integrated microfluidic device that performs 300 parallel RT-qPCR assays and executes all steps of single-cell capture, lysis, reverse transcription, and qPCR is shown in Fig. 1A. To facilitate the precise comparison of different samples and cell types, our prototype consists of six independent sample-loading lanes, each containing 50 cell-processing units. We resolved previously limiting technical pitfalls by the inclusion of design elements to (i) allow for efficient distribution of single cells without mechanical damage, (ii) minimize background signal arising from free RNA or cell debris in the medium, and (iii) avoid reaction inhibition by cell lysates in nanoliter volumes.
Fig. 1.
Design and operation of the microfluidic device for single-cell gene expression analysis. (A) Schematic of microfluidic device. Scale bar: 4 mm. The device features 6 sample input channels, each divided into 50 compound reaction chambers for a total of 300 RT-qPCR reactions using approximately 20 μL of reagents. The rectangular box indicates the region depicted in B. (B) Optical micrograph of array unit. For visualization, the fluid paths and control channels have been loaded with blue and red dyes, respectively. Each unit consists of (i) a reagent injection line, (ii) a 0.6-nL cell capture chamber with integrated cell traps, (iii) a 10-nL reverse transcription (RT) chamber, and (iv) a 50-nL PCR chamber. Scale bar: 400 μm. (C) Optical micrograph of two cell capture chambers with trapped single cells indicated by black arrows. Each trap includes upstream deflectors to direct cells into the capture region. Scale bar: 400 μm. (D–I) Device operation. (D) A single-cell suspension is injected into the device. (E) Cell traps isolate single cells from the fluid stream and permit washing of cells to remove extracellular RNA. (F) Actuation of pneumatic valves results in single-cell isolation prior to heat lysis. (G) Injection of reagent (green) for RT reaction (10 nL). (H) Reagent injection line is flushed with subsequent reagent (blue) for PCR. (I) Reagent for qPCR (blue) is combined with RT product in 50 nL qPCR chamber. Scale bar for D–I: 400 μm.
In order to reduce device complexity and obviate the need for RNA purification, we optimized our device to be compatible with commercially available assays that use “one-pot” RT-qPCR protocols requiring only the sequential addition of reagents into a single reaction vessel. Each cell-processing unit consists of a compound chamber, formed by a cell capture chamber connected sequentially to two larger chambers for RT and qPCR (Fig. 1B). This simple fluidic architecture allows the implementation of either heat lysis followed by two-step RT-qPCR (Fig. 1 D–I), or chemical lysis followed by one-step RT-qPCR. A detailed description of device operation for each of these protocols is provided in SI Materials and Methods. All lanes are connected to a common feed channel that, following the completion of each reaction step, is used to inject the next reaction master mix through the upstream chambers, thereby diluting the intermediate product (cell lysate or cDNA) and assembling the next reaction mixture. This parallelization of reaction assembly in a microfluidic format ensures equal timing of all reaction steps and greatly reduces technical variability associated with pipetting and mixing steps in microliter volumes. Fluorescence measurements were performed to ensure the efficient and reproducible transfer of reactants at each step, showing that losses in sample transfer are below 5%. To minimize device expense and complexity, temperature control and fluorescence detection were performed using peripheral hardware including a CCD detector mounted above a flatbed thermocycler plate.
We designed our chamber volumes to ensure sufficient dilution between each processing step to avoid reaction inhibition while at the same time maintaining high template concentrations and assay sensitivity. Initial attempts to perform RT-qPCR in low nanoliter volumes were found to produce highly variable results, including nonspecific amplification and inconsistent detection of abundant transcripts (18). Cell lysate dilutions showed that reaction inhibition becomes significant at concentrations in excess of 0.2 cells/nL, or 10 cells/50 nL-reaction (Fig. 2D). On the other hand, RT-qPCR measurement noise has been shown to become the dominant source of variability when starting at concentrations below one copy per 100 nL (6), illustrating that minimizing reaction volumes is critical for precise measurements on limited template. Finally, experiments in tubes were performed to determine that a dilution ratio of at least 5∶1 (PCR mix∶RT product) is optimum for PCR efficiency. We therefore designed our combined reactors to have an aggregate total volume of 60.6 nL, consisting of a 0.6-nL cell capture chamber, a 10-nL RT chamber, and a 50-nL qPCR chamber. These volumes allow for the reliable amplification of single molecules (Fig. 2A) and result in a final template concentration of 330 ng/mL when starting from a single-cell equivalent of RNA (20 pg). The use of larger volume RT and PCR chambers has the added advantage of reducing their surface-to-volume ratio, thereby minimizing reagent evaporation through the gas permeable device material (polydimethylsiloxane).
Fig. 2.
Precision and sensitivity of microfluidic RT-qPCR. (A) Fluorescence image of entire device showing 300 reactions in 6 lanes. Image is taken after 40 cycles of PCR from dilution series of purified total RNA from K562 cells. (Left to Right) The samples are 40 pg/chamber, 5 pg/chamber, 625 fg/chamber, 78 fg/chamber, 10 fg/chamber, and no-template control (NTC). Single molecule amplification at limiting dilution results in a digital amplification pattern for 10- and 78-fg lanes. No amplification is observed in NTC lane (N = 50). (B) Three hundred real-time amplification curves generated from processing sequences of images similar to A. The threshold for determining CT values is indicated by the dashed line. (C) On-chip (black) and off-chip (blue) RT-qPCR for GAPDH from a 8× serial dilution of purified total RNA shows improved sensitivity in nanoliter volume reactions. In the microfluidic system, CT values for the 10-fg sample correspond to single molecule amplifications detected in 19 of 50 chambers. The mean and standard deviation from single-cell measurements is shown in green for both on- and off-chip analysis. CT values obtained on chip correspond to a mean of 20 pg of RNA per cell. Off-chip measurements of single K562 cells washed twice in PBS and isolated by glass capillary exhibit artificially increased levels due to residual signal from debris and free RNA in the supernatant (red). Cells were transferred in approximately 2 μL of supernatant, which was measured to contain approximately 20 pg of extracellular RNA. Error bars represent standard deviation of measured CT values for all amplified reactions. (D) Real-time amplification curves of GAPDH in K562 cell lysate dilutions. Inhibition of RT-PCR occurs at cell lysate concentrations beyond 10 cell equivalents per 50 nL reaction. (E) Measured CT values for GAPDH in dilution series of cell lysate. No inhibition occurs for single-cell lysates.
Another critical step toward integration was to efficiently distribute single cells into each location on the array without mechanical damage. To achieve reproducible and deterministic loading of single cells into each array element, we engineered a hydrodynamic single-cell trap within each capture chamber. Cell traps consisting of a single cup structure (19) were found to be highly inefficient, capturing less than 0.1% of cells passing in close proximity to the center of the channel structure. To improve capture efficiency, we incorporated upstream deflectors, located 22.5 μm from the trap, to focus cells into the central streamlines where capture is most efficient (Fig. 1C). Using these structures we were able to achieve high single-cell occupancy of array locations (Fig. 3 A and B). Over eight separate experiments, a loading protocol of approximately 60 s (106 cells/mL, 20 nL/s per lane) resulted in the successful isolation of single cells in 1,518/1,700 chambers (89.3%), with a cell capture efficiency of 5.0 ± 0.5%. Staining with trypan blue was used to assess the viability of cells after loading and was determined to be equivalent to the viability of the input sample (97.4% viability vs. input 96.8%). Finally, measurements of the distribution of cell diameters prior to and after loading indicated that cell trapping did not introduce significant bias (p = 0.67, two-sample t test) in selecting cells of different sizes (Fig. S1). This cell trap geometry and loading protocol were used in all subsequent qPCR measurements presented below. Further improvement of trap and deflector geometries were found to achieve fill factors of > 99% (100 single cells captured out of 100 traps analyzed) and cell capture efficiencies of 87.0 ± 4.5%, with cell viability again matching the input sample (> 98%) and not significantly biasing cell sizes (p = 0.35, two-sample t test), making this method applicable to the analysis of limited quantity samples such as rare stem cells or clinical samples.
Fig. 3.
Single-cell loading and transcript measurements. (A) The locations of cells in each chamber along all six lanes of a device, as determined by brightfield microscopy, are represented as white circles and overlaid on a heat map of CT values obtained from RT-qPCR measurements of GAPDH in K562 cells. Red circles indicate NTC. (B) Scatter plot showing CT measurements for experiment shown in A. Histogram (Inset) shows 93.2% single-cell occupancy. (C) Distribution of the number of GAPDH transcripts measured in single K562 cells (N = 233).
The immobilization of cells in traps was also used for on-chip washing of cells prior to lysis to remove free RNA, cellular debris, and untrapped cells that would otherwise give rise to background signal or result in low single-cell occupancy (Fig. S2 A and B). The efficiency of chamber washing, determined by loading purified RNA template (36.5 ng/μL), followed by washing and RT-qPCR analysis, was > 99.99% (1.1 × 104 copies measured without wash, 0 copies detected after washing) (Fig. S2C). In addition, RT-qPCR measurements testing different cell loading and washing protocols demonstrated that on-chip washing allows for loading directly from culture medium with low background as compared to off-chip wash steps followed by analysis in microliter volumes (Fig. 2C). Importantly, on-chip washing allows for lysis within seconds of washing, thereby minimizing spurious transcriptional responses that may arise from sequential medium exchange and spin steps.
Validation of Integrated Single-Cell RT-qPCR.
We first tested the sensitivity and precision of RT-qPCR in our device by performing measurements of GAPDH expression over an 8-fold dilution series of total RNA, ranging from 40 pg (approximately 2 cell equivalents) to 10 fg (approximately 1/2,000 cell equivalents). RNA was purified from K562 cells, a BCR-ABL positive human cell line derived from a patient with chronic myeloid leukemia (20) (Fig. 2 A–C). The efficiency of amplification was determined over the four highest template concentrations (40 pg, 5 pg, 625 fg, and 78.125 fg) as the slope from a linear least squares fit of log 2(C) vs. cycle threshold (CT) and was found to be 0.988 ± 0.055. The standard deviation of CT values was less than 0.15 at the three highest concentrations (SD = 0.08, 0.10, and 0.14 for the 40 pg, 5 pg, and 625 fg samples, respectively), indicating uniform amplification across the array and technical error of less than 10% in absolute concentration, near the limit of qPCR precision. The highest measurement variability was observed in the 78-fg sample, where shot noise (Poisson sampling noise) is most pronounced and accounts for approximately 50% of the measurement variance. Template amounts below 625 fg resulted in a digital pattern characteristic of single molecule amplification (49/50 for 78 fg and 19/50 for 10 fg) and consistent with the expected occupancy of chambers as determined by a binomial distribution (2). Based on the frequency of single molecule detection in the 10-fg sample, we measured the average copy number of GAPDH to be 979 ± 240 transcript copies per single-cell equivalent (20 pg) (Fig. 2). This measurement is comparable to previous reports (11) and is in close agreement with an independent estimate based on normalizing the dilution series to CT values obtained for single molecules (copies/20 pg = 1/2 × copies/40 pg = 1/2 × (1 + efficiency)(CT(40 pg)-CT(single molecule)) = 1,407 ± 153 copies/20 pg). It should be noted that these estimates represent a lower bound because they do not account for RT efficiency; the RT efficiency of GAPDH has been previously estimated to be approximately 50% (13) but is dependent on transcript secondary structure and assay design. A comparison of CT values obtained from on-chip qPCR from cDNA synthesized off-chip demonstrated that on-chip RT efficiency is equal to that obtained off-chip when working from the same RNA concentrations (Fig. S3). Finally, comparison of the same dilution series of RNA, assayed for GAPDH both on-chip and in tubes (20-μL volume) (Fig. 2C), showed that on-chip analysis provides improved sensitivity.
We next evaluated the efficiency and reliability of on-chip cell processing by comparing our GAPDH measurements of purified RNA to measurements performed directly from single K562 cells (Figs. 2C and 3C). K562 cells were loaded directly from culture medium followed by washing and analysis using a chemical lysis and one-step RT-qPCR protocol (Cells Direct™, Invitrogen). Using a CT threshold of 31.5, corresponding to the mean CT of a single molecule of GAPDH (CT = 30.5) plus two standard deviations (SD = 0.5), we observed successful amplification in 100% of single cells (N = 233) (Fig. 3 A and B). Adjacent chambers that did not contain a cell were clearly separated from single-cell measurements with an average Δ CT value of 5.7 (five empty chambers, three of which amplified) (Fig. 3 A and B and Fig. S4). Consistent with previous reports (21), we observed a log-normal distribution of GAPDH in single cells with mean CT values of 20.3 (SD = 0.8) and an average of 1,761 (SD = 648) copies per cell (Fig. 3C). These expression levels are consistent with previous estimates in single cells (11). Additionally, the mean CT of 20.3 observed for single cells matches measurements of single-cell equivalent lysate (CT = 20.2, Fig. 2D). Using digital PCR on cDNA prepared from K562 cell lysate, we measured an average of 1,229 ± 72 GAPDH molecules per single-cell equivalent. We conclude that the relative efficiency of on-chip single-cell lysis and mRNA extraction/accessibility is equal to that achieved when working from RNA purified from large numbers of cells. Finally, as expected, RT-qPCR measurements from chambers loaded with more than one cell show reduced variability and lower CT values (Figs. S2A and S5). Taken together, these results establish the precise measurement of mRNA abundance with single molecule sensitivity and the dynamic range needed for single-cell analysis.
Application to Measurement of Single-Cell miRNA Expression.
We next applied our technology to the study of single-cell miRNA expression. miRNAs are thought to provide a unique signature of cellular state and are central players in orchestrating development and oncogenesis, making them a promising class of biomarker for single-cell analysis (15, 22, 23). Importantly, the short length of miRNAs (approximately 22 nucleotides) makes them difficult to detect by hybridization approaches, so that RT-qPCR is the dominant quantification strategy. To demonstrate the robustness and throughput of our technology, we performed a total of 1,672 single-cell measurements to examine single-cell variability in the expression of nine miRNAs spanning a wide range of abundance (> 16,000 copies per cell to < 0.2 average copies per cell). K562 cells were again chosen as a heterogeneous population for this study because they are known to exhibit mixed characteristics of erythrocytes, granulocytes, and monocytes (20, 24). We first measured the expression of miR-16, a highly expressed microRNA that is found in many tissue types (25) and has been suggested as a suitable internal standard for normalization (26). We found that miR-16 was log-normally distributed across K562 cells, but with slightly lower expression and notably tighter regulation than GAPDH, having an average of 804 (SD = 261) copies per cell and a standard deviation of 30% (mean CT = 21.4, SD = 0.4). This strikingly low variability is within our estimates of cell volume differences (Fig. S1). Matched experiments on single cells, isolated by micropipette into 20-μL volume tubes displayed an increase in measurement variability to approximately 90% (mean CT = 29.5, SD = 0.9), demonstrating the improved precision of parallel microfluidic cell processing in nanoliter volumes (Fig. 4A). Microliter volume experiments also showed a pronounced increase in measured CT values that results from the low concentration of template and the large number of required PCR cycles.
Fig. 4.
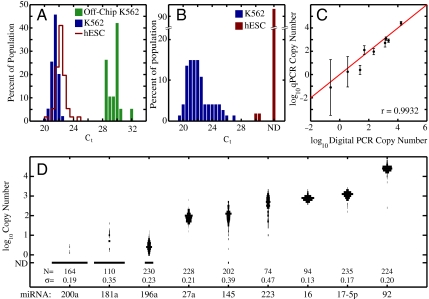
Single-cell miRNA measurements. (A) Single-cell measurements of miR-16 expression in K562 cells and hESCs. Measurements of single K562 cells isolated using a microcapillary and assayed in 20-μL volumes are shown for comparison of technical variability. The observed shift in mean CT values between on- and off-chip measurements is due to lower template concentrations, and hence increased required PCR cycles, in the off-chip samples. (B) Differential expression of miR-223 between K562 cells and hESCs. Right-most bar indicates cells for which miR-223 was not detected (ND). (C) Mean single-cell miRNA copy numbers measured by RT-qPCR in the microfluidic device compared to digital PCR measurements from bulk cell lysate. Error bars represent standard deviation of single-cell measurements for each miRNA. (D) One thousand five hundred and sixty-one single-cell measurements of the expression of 9 miRNA in K562 cells. Reflected histograms represent the expression distributions for each miRNA.
To demonstrate the utility of our device for measuring differential expression in single cells, we next measured the expression of miR-223, a miRNA implicated in myeloid differentiation (24, 27). In contrast to miR-16, K562 cell miR-223 expression was found to be highly variable (mean CT = 22.2, SD = 1.6, copy number = 513, SD = 406) and was not log-normally distributed (Fig. 4B), consistent with the known functional heterogeneity of K562 cells. These measurements highlight the utility of single-cell miRNA expression analysis for assessing the heterogeneity of cell populations and for identifying miRNAs that are useful biomarkers of cellular state. To further explore this possibility, we measured the expression of an additional seven miRNAs (nine total) and plotted the patterns of single-cell expression in K562 populations (Fig. 4C). Following the procedure described above, we used single molecule CT values, obtained by digital PCR, to translate measured CT values to absolute copy number. Assuming 100% efficient amplification, we observed that the copy number, calculated as 2(CT(single cell)-CT(single molecule)), was well correlated (coefficient of 0.9932) with the average copy number obtained by digital PCR of cDNA prepared from bulk lysates (Fig. 4D). Single-cell measurements revealed distinct patterns of miRNA expression, with miR-16, miR-92, and miR-17-5p each exhibiting unimodal and tightly regulated distributions, whereas miR-223, miR-196a, and miR-145 showed multimodal distributions and a high level of cellular heterogeneity. Notably, for the lowest abundance miRNA, miR-200a, we detected expression in only a small fraction of cells and at levels below approximately five copies per cell. The average miR-200a copy number over all cells was within a factor of two of that obtained by digital PCR (0.2 copies per cell). In contrast, miR-92 was found to be the most abundant miRNA and was present at approximately 16,000 copies per cell. These measurements established miRNA quantification in single cells with a dynamic range of greater than 104 and at single molecule sensitivity.
Finally, to illustrate the utility of single-cell measurements in precisely assessing differences in both the average expression and the heterogeneity between two different cell populations, the expression levels of miR-16 and miR-223 in K562 cells were compared to those in CA1S cells (28, 29), a human embryonic stem cell line (hESC). Although miR-16 was found to be expressed in hESC at similar levels to K562 (ΔCT = 0.6), we observed approximately a twofold greater variability in expression (mean CT = 22.0, SD = 0.7) (Fig. 4A). In contrast, when compared to K562, single CA1S cell measurements of miR-223 showed strong down-regulation, with miR-223 detected in only 3.6% of cells. The absence of significant miR-223 expression in hESC is expected due to the role of miR-223 as a differentiation-specific miRNA (24, 27).
Coregulation of miR-145 and OCT4 in Single Cells.
The measurement of multiple transcripts in single cells allows for quantitative measurements of gene coregulation that would otherwise be masked by cellular heterogeneity (14). To demonstrate this capability we designed an optically multiplexed assay to study the coregulation of miR-145 and OCT4, a known target of miR-145 (30), during the differentiation of hESCs (Fig. 5 A–C). A total of 1,094 single-cell measurements were performed at 0, 4, 6, and 8 d of differentiation. Cell distributions at each time point were used to map out the evolution of these transcripts and showed that average miR-145 levels increased approximately 20-fold (copy numbers: D0: mean = 18.9, SD = 25.5, D8: mean = 380.3, SD = 259.4) over 8 d. Increases in miR-145 were accompanied by progressive down-regulation of OCT4, ultimately reaching an average of 30-fold suppression (copy numbers: D0: mean = 755.7, SD = 306.4, D8: mean = 27.8, SD = 124.5) after 8 d (independently verified by mRNA-FISH) (Fig. S6 and Table S1). Notably, single-cell analysis at day 6 showed a bimodal distribution in both OCT4 and miR-145, revealing a transition of cellular state (30) that likely reflects the spontaneous differentiation of a subpopulation of cells. The observed single-cell dynamics of miR-145 and OCT4 coregulation are not apparent in population measurements, highlighting the use of scalable single-cell transcriptional analysis in correlating molecular signatures to cellular decision making (14).
Fig. 5.
Optical multiplexing of single-cell RT-qPCR. (A) Multiplexed analysis of the coexpression of OCT4 and miR-145 in differentiating hESC. Points are color-coded to represent single-cell measurements (N = 547) for each time point. Crosses represent population mean copy number. (B and C) Histograms showing the distribution of each transcript are projected on the axes with the mean copy number indicated by a dashed line. (D) Coexpression measurements of SP1 wild-type and SNV mutant transcripts in primary cells isolated from a lobular breast cancer sample. Mutant SP1 is detected in 23 of 117 primary cells, and undetected in K562 cells (N = 37).
Single Nucleotide Variant (SNV) Detection in Primary Cells.
Finally, to establish the specificity of our method we used multiplexed measurements of mRNA SNVs to assess the genomic heterogeneity within a primary tumor sample. A total of 117 single cells isolated from a plural effusion of a metastatic breast cancer were assayed for the expression of a SNV mutant of the transcription factor SP1, previously identified by deep sequencing (31) (Fig. 5D). Primers were designed using sequences flanking the SNV location and do not discriminate between the genomic DNA and mRNA transcript. Of the 117 primary cells analyzed, 22 (18.8%) were heterozygous for the mutant and wild-type allele, 85 (72.6%) were homozygous wild type, 1 (0.9%) was homozygous mutant, and the transcripts were undetected in 9 (7.7%). We did not detect the SP1 mutation in 37 control K562 cells and failed to detect the wild-type transcript in only 2 of these cells. In the absence of copy number alterations in the primary sample, these observed frequencies would suggest a mutant to wild-type SP1 ratio of 11.2% (18.8 × 1 + 0.9 × 2 = 20.6 mutant to 18.8 × 1 + 72.6 × 2 = 164 wild type). However, using digital PCR on purified DNA from the primary sample, we found the ratio of mutant to wild-type SP1 alleles to be 18.7 ± 2.3%, in agreement with the previously reported ratio of 21.9%, obtained by deep sequencing (31). The lower frequency of cells expressing the mutant SP1 allele may be due to allelic expression bias or an amplification of the SP1 mutant allele, both of which are supported by Shah et al. (31). Regardless, given that the frequency of tumor cells within the original sample was approximately 89% (31), both DNA molecule counting and single-cell RNA expression measurements show that the metastasis of this tumor is derived from multiple cancer cell lineages.
Conclusion
Here we have demonstrated the implementation of scalable and quantitative single-cell gene expression measurements on an integrated microfluidic system. The presented device performs 300 high-precision single-cell RT-qPCR measurements per run, surpassing previous microfluidic systems by a factor of approximately 100 in throughput. Further scaling the throughput to over 1,000 measurements on a device with an area of one square inch is straightforward as each array element occupies an area of 0.6 mm2. In terms of performance, we have established a dynamic range of at least 104, measurement precision of better than 10%, single molecule sensitivity, and specificity capable of discriminating the relative abundance of alleles differing by a single nucleotide. Compared to tube-based single-cell RT-qPCR, microfluidic processing provides improved reproducibility, precision, and sensitivity, all of which may be critical in identifying subtle differences in cell populations. Nanoliter volume also results in a 1,000-fold reduction in reagent consumption, thereby enabling cost-effective analysis of large numbers of single cells.
In over 3,300 single-cell experiments, using adherent and suspension cell lines as well as clinical samples, we have shown that microfluidic RT-qPCR is well-suited to the quantitative analysis of miRNA expression and SNV detection, both of which are difficult or inaccessible by alternative hybridization methods. Notably, our device allowed for precise comparison of the distributions of GAPDH and miR-16 expression. miR-16 was found to be exquisitely regulated in K562 cells, a finding that is striking given the known functional heterogeneity of this population and the high variability in the expression of other measured miRNAs. We postulate that higher variability of GAPDH expression reflects the fundamentally stochastic process of transcriptional bursts followed by mRNA degradation. Incorporation of miRNA into the RNA-induced silencing complex (RISC) is known to provide enhanced stability so that miRNA are inherently less subject to temporal fluctuations; miRNA are thus particularly suited as biomarkers for assessing single-cell state and population heterogeneity. We anticipate that scalable and precise single-cell miRNA analysis will become an invaluable tool in stratifying populations of mixed differentiation state (15).
Here we have established the critical element of combining all single-cell-processing steps into an integrated platform. This functionality provides a solid foundation upon which increasingly advanced microfluidic single-cell transcription analysis may be built. We anticipate that more complex fluid routing (32), to distribute cell contents across multiple chambers, will allow for the multiplexed measurements of tens of targets across hundreds of cells, and for combining this technology with single molecule detection by digital PCR. Alternatively, the microfluidic system described here could be used for single-cell processing and preamplification, with recovered reaction products analyzed by high-throughput microfluidic qPCR or sequencing. We contend that the simplicity of device operation will soon allow for the robust and automated implementation of single-cell RT-qPCR, leading to its widespread adoption in research applications and opening the prospect of diagnostic tests based on single-cell analysis.
Materials and Methods
Experimental details and protocols for device fabrication, device operation, on-chip and off-chip RT-qPCR, cell culture, hESC differentiation, digital PCR, mRNA-FISH, image analysis, transfer efficiency measurements, and cell trapping measurements are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments.
This research was funded by Genome BC, Genome Canada, Western Diversification, Canadian Institutes of Health Research (CIHR), Terry Fox Foundation, and the Natural Sciences and Engineering Research Council (NSERC). Salary support was provided by the Michael Smith Foundation for Health Research (to A.K.W. and C.L.H.), NSERC (to A.K.W., M.V., M.H., and D.S.), and CIHR (to C.L.H.).
Footnotes
Conflict of interest statement: C.L.H. has a financial interest in Fluidigm Corporation, which has products related to the subject matter of this study.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019446108/-/DCSupplemental.
References
- 1.Taniguchi K, Kajiyama T, Kambara H. Quantitative analysis of gene expression in a single cell by qPCR. Nat Methods. 2009;6:503–506. doi: 10.1038/nmeth.1338. [DOI] [PubMed] [Google Scholar]
- 2.Warren L, Bryder D, Weissman IL, Quake SR. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc Natl Acad Sci USA. 2006;103:17807–17812. doi: 10.1073/pnas.0608512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larsson C, Grundberg I, Soderberg O, Nilsson M. In situ detection and genotyping of individual mRNA molecules. Nat Methods. 2010;7:395–397. doi: 10.1038/nmeth.1448. [DOI] [PubMed] [Google Scholar]
- 5.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 6.Bengtsson M, Hemberg M, Rorsman P, Stahlberg A. Quantification of mRNA in single cells and modelling of RT-qPCR induced noise. BMC Mol Biol. 2008;9:63–74. doi: 10.1186/1471-2199-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zare RN, Kim S. Microfluidic platforms for single-cell analysis. Annu Rev Biomed Eng. 2010;12:187–201. doi: 10.1146/annurev-bioeng-070909-105238. [DOI] [PubMed] [Google Scholar]
- 8.Sims CE, Allbritton NL. Analysis of single mammalian cells on-chip. Lab Chip. 2007;7:423–440. doi: 10.1039/b615235j. [DOI] [PubMed] [Google Scholar]
- 9.Wheeler AR, et al. Microfluidic device for single-cell analysis. Anal Chem. 2003;75:3581–3586. doi: 10.1021/ac0340758. [DOI] [PubMed] [Google Scholar]
- 10.Skelley AM, Kirak O, Suh H, Jaenisch R, Voldman J. Microfluidic control of cell pairing and fusion. Nat Methods. 2009;6:147–152. doi: 10.1038/nmeth.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcus JS, Anderson WF, Quake SR. Microfluidic single-cell mRNA isolation and analysis. Anal Chem. 2006;78:3084–3089. doi: 10.1021/ac0519460. [DOI] [PubMed] [Google Scholar]
- 12.Bontoux N, et al. Integrating whole transcriptome assays on a lab-on-a-chip for single cell gene profiling. Lab Chip. 2008;8:443–450. doi: 10.1039/b716543a. [DOI] [PubMed] [Google Scholar]
- 13.Zhong JF, et al. A microfluidic processor for gene expression profiling of single human embryonic stem cells. Lab Chip. 2008;8:68–74. doi: 10.1039/b712116d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo GJ, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 15.Petriv OI, et al. Comprehensive microRNA expression profiling of the hematopoietic hierarchy. Proc Natl Acad Sci USA. 2010;107:15443–15448. doi: 10.1073/pnas.1009320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diehn M, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toriello NM, et al. Integrated microfluidic bioprocessor for single-cell gene expression analysis. Proc Natl Acad Sci USA. 2008;105:20173–20178. doi: 10.1073/pnas.0806355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong Y, Ogunniyi AO, Love JC. Massively parallel detection of gene expression in single cells using subnanolitre wells. Lab Chip. 2010;10:2334–2337. doi: 10.1039/c004847j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Carlo D, Aghdam N, Lee LP. Single-cell enzyme concentrations, kinetics, and inhibition analysis using high-density hydrodynamic cell isolation arrays. Anal Chem. 2006;78:4925–4930. doi: 10.1021/ac060541s. [DOI] [PubMed] [Google Scholar]
- 20.Lozzio BB, Lozzio CB, Bamberger EG, Feliu AS. A multipotential leukemia cell line (K-562) of human origin. Proc Soc Exp Biol Med. 1981;166:546–550. doi: 10.3181/00379727-166-41106. [DOI] [PubMed] [Google Scholar]
- 21.Bengtsson M, Stahlberg A, Rorsman P, Kubista M. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res. 2005;15:1388–1392. doi: 10.1101/gr.3820805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 23.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 24.Yuan JY, et al. MicroRNA-223 reversibly regulates erythroid and megakaryocytic differentiation of K562 cells. J Cell Mol Med. 2009;13:4551–4559. doi: 10.1111/j.1582-4934.2008.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang F, Hajkova P, Barton SC, Lao K, Surani MA. MicroRNA expression profiling of single whole embryonic stem cells. Nucleic Acids Res. 2006;34:e9. doi: 10.1093/nar/gnj009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 28.Schulze HG, et al. Assessing differentiation status of human embryonic stem cells noninvasively using Raman microspectroscopy. Anal Chem. 2010;82:5020–5027. doi: 10.1021/ac902697q. [DOI] [PubMed] [Google Scholar]
- 29.Adewumi O, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- 30.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 31.Shah SP, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 32.Huft J, Da Costa D, Walker D, Hansen CL. Three-dimensional large-scale microfluidic integration by laser ablation of interlayer connections. Lab Chip. 2010;10:2358–2365. doi: 10.1039/c004051g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.