Abstract
We used electron cryotomography to study the molecular arrangement of large respiratory chain complexes in mitochondria from bovine heart, potato, and three types of fungi. Long rows of ATP synthase dimers were observed in intact mitochondria and cristae membrane fragments of all species that were examined. The dimer rows were found exclusively on tightly curved cristae edges. The distance between dimers along the rows varied, but within the dimer the distance between F1 heads was constant. The angle between monomers in the dimer was 70° or above. Complex I appeared as L-shaped densities in tomograms of reconstituted proteoliposomes. Similar densities were observed in flat membrane regions of mitochondrial membranes from all species except Saccharomyces cerevisiae and identified as complex I by quantum-dot labeling. The arrangement of respiratory chain proton pumps on flat cristae membranes and ATP synthase dimer rows along cristae edges was conserved in all species investigated. We propose that the supramolecular organization of respiratory chain complexes as proton sources and ATP synthase rows as proton sinks in the mitochondrial cristae ensures optimal conditions for efficient ATP synthesis.
Keywords: cryoelectron tomography, subtomogram averaging, membrane curvature, membrane potential, mitochondrial ultrastructure
Mitochondria, the powerhouses of eukaryotic cells, generate ATP, the universal energy carrier in all life forms. The F1Fo ATP synthase uses the energy stored in the electrochemical proton gradient across the inner mitochondrial membrane to produce ATP from ADP and phosphate. The proton gradient is established by the respiratory chain complexes I, III, and IV, which pump protons out of the mitochondrial matrix into the cristae space while transferring electrons from the electron donors NADH, FADH, or succinate (via complex II) to the final electron acceptor O2. The F1Fo ATP synthase and complex I (NADH dehydrogenase) are the largest membrane protein complexes in mitochondria, composed of more than 20 or 40 individual protein subunits, respectively (1, 2). The 600-kDa ATP synthase consists of the Fo part in the membrane that works like a proton-driven turbine, and the catalytic F1 part on the matrix side. The two parts are held together by a static peripheral stalk and a rotating central stalk that transmits the torque from the rotor unit in the membrane to the catalytic F1 head (3, 4). Complex I is an L-shaped molecule of approximately 1 MDa. Its membrane arm has three or four proton-pumping modules, while the matrix arm catalyzes electron transfer from NADH to the hydrophobic electron acceptor ubiquinol (5). The structures of both complexes have been determined by X-ray crystallography, either partially in the case of the F1Fo ATP synthase (6), or at low resolution in the case of mitochondrial complex I (7, 8), but their relative organization in the mitochondrial inner membrane is largely unknown.
The two large complexes occur at an approximate ratio of one molecule of complex I per 3.5 ATP synthase monomers (9). The ATP synthase is easily identified in mitochondrial membranes by its characteristic 10-nm F1 head connected to the membrane by a 5-nm-long stalk (10–12). Blue-native polyacrylamide gel electrophoresis (BN PAGE) of the mitochondrial F1Fo ATP synthase has shown that the complex forms dimers or larger oligomeric assemblies when solubilized with mild detergents (13). Single-particle electron microscopy indicated angles of 40° (14, 15) or 70–90° (16, 17) between the long axes of monomers in the dimer. Freeze-fracture deep-etch replicas suggested that in Paramecium these dimers form rows along the edge of helical cristae tubes (18). More recently, cryoelectron tomography (cryo-ET) of cristae membranes from bovine heart or rat liver mitochondria has shown that the ATP synthase dimers form long rows along highly curved membrane ridges (12). The dimer rows appear to play a major role in cristae formation and morphology. Deletion of the dimer-specific subunits e, g, or the first helix of subunit b in the peripheral stalk results in the formation of mitochondria with onion-like cristae (19, 20).
BN PAGE has suggested that bovine heart complex I forms supercomplexes with cytochrome c reductase (complex III) and cytochrome c oxidase (complex IV) (21). 3D maps of this supercomplex have been obtained by single-particle electron microscopy of negatively stained samples (22), and, most recently, cryo-EM. The three respiratory chain complexes are oriented in the supercomplex in a way that appears to be optimal for electron transfer and substrate shuttling. Each works as a proton pump, and together they generate the proton motive force (pmf) that drives ATP synthesis. The mutual arrangement of electron transfer complexes as proton sources and ATP synthase complexes as proton sinks in the membrane is therefore of fundamental interest and importance for understanding mitochondrial energy conversion.
To study the macromolecular organization of ATP synthase and complex I in the inner mitochondrial membrane, we imaged mitochondrial membranes and whole mitochondria from mammals, fungi and plants by cryo-ET. Long rows of ATP synthase dimers in cristae membranes were a universal feature of all mitochondria we investigated. The rows were confined to the highly curved edges of lamellar cristae and were formed by dimers with a constant F1 head distance. These dimers corresponded to those with a dimer angle of > 70° observed by single-particle analysis (16, 17). Complex I in the membrane was identified by its shape, or by specific quantum-dot labeling. Unlike the ATP synthase, this complex was irregularly distributed on the mostly flat membrane regions on either side of the ATP synthase dimer rows. The organization of these two large complexes was the same in all species investigated and is thus conserved during evolution. We propose that the conserved organization of respiratory chain complexes into regions that are either rich in ATP synthase or in complex I, and the extended rows of ATP synthase dimers on the cristae edges has important implications for ATP synthesis in mitochondria.
Results and Discussion
Cryo-ET of Whole Mitochondria.
Cryo-ET is the method of choice for visualizing the three-dimensional organization of protein complexes in the membrane. Using this technique, we previously discovered long rows of ATP synthases dimers in membrane fragments of bovine heart and rat liver mitochondria (12). To find out whether this striking arrangement occurs also in intact, functional mitochondria and perhaps extends to other kingdoms, we performed cryo-ET on whole mitochondria from bovine heart (Bos taurus), potato tubers (Solanum tuberosum), and three species of fungi (Saccharomyces cerevisiae, Yarrowia lipolytica, and Podospora anserina).
In tomographic reconstructions, mitochondria appeared as dense, oblate ellipsoids with a diameter around 600–1200 nm (average 900 ± 165 nm) for all species except P. anserina, which were smaller. The mitochondria contained a complete outer membrane with an inner to outer membrane distance of 12 ± 2.5 nm and an intercristae membrane distance of 20 ± 4 nm, consistent with previous studies (11, 23). For potato and the three species of fungi, the cristae consisted of distinct flattened sacs, which merged with the inner boundary membrane in long slit-like junctions (20–300 nm) (Fig. S1A). At their edges in the matrix, cristae often had square corners where the membrane bent abruptly by approximately 90° (Fig. S2). This feature was common in P. anserina and potato but also occurred in S. cerevisiae. In bovine heart, the cristae formed networks of small lamellar patches connected to each other by short tubular segments, as in neuronal mitochondria (24). Cristae were connected to the inner boundary membrane by small circular junctions of 35 ± 5 nm diameter. The tubular cristae segments always originated from the edges of lamellar patches rather than from flat membrane regions (Fig. S1B).
Rows of ATP Synthase Dimers in Whole Mitochondria.
P. anserina mitochondria proved to be the most suitable for examining the arrangement of the ATP synthase in undisrupted organelles. Mitochondria prepared from synchronized, 6-d-old P. anserina cells measured only 300 to 600 nm (average 450 ± 80 nm) in diameter and hence were more transparent compared to those from other species, allowing large protein complexes to be visualized in the tomographic volume. Biochemical assays of similar preparations (25) showed tightly coupled respiration, indicating that the isolated mitochondria were undamaged and active.
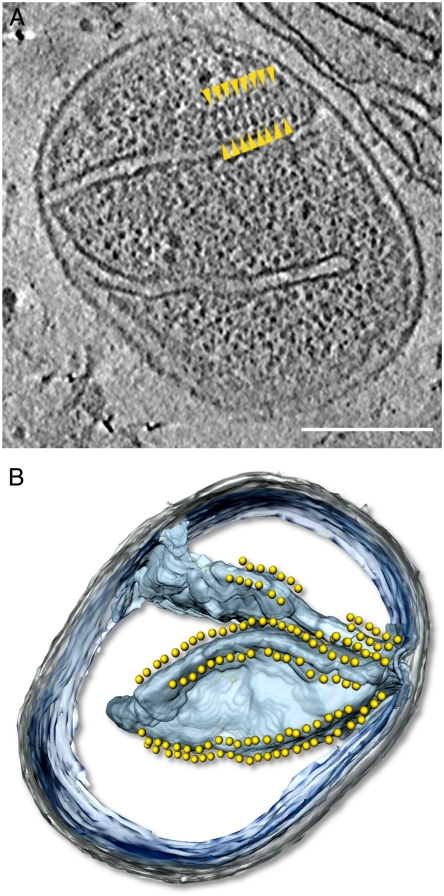
In P. anserina mitochondria we found double rows of 10-nm particles approximately 5 nm above the membrane, along the highly curved cristae edges (Fig. 1 and Movie S1). The particles were connected to the membrane by a stalk of 3–4 nm, giving them the characteristic lollipop shape of the F1Fo ATP synthase (10–12). The center-to-center distance between particles across the dimer rows consistently measured approximately 28 nm, whereas the head-to-head distance along rows varied from 12 nm to greater than 20 nm. We conclude that the double rows are loose arrays of ATP synthase dimers, arranged with their long axis perpendicular to the direction of the row. No ATP synthase dimers or higher oligomers were detected in the flat cristae regions, which suggests that, at least in this species, ATP synthase dimers are confined exclusively to the cristae edges.
Fig. 1.
Rows of F1-Fo ATP synthase dimers in whole mitochondria. (A) Tomographic slice through a whole mitochondrion of Podospora anserina showing arrays of dimeric ATP synthases (yellow arrowheads). (B) Segmented surface representation of A showing position of ATP synthase dimers (yellow spheres) to cristae membrane (blue). Dimers are confined to the highly curved cristae edges (see Movie S1). The mitochondrion is intact, as shown by the granular appearance of the matrix, in which ribosomes are visible as dark gray, approximately 25-nm particles. Gray, outer membrane; blue, cristae membranes; blue-transparent, inner boundary membrane; yellow, ATP synthase. Scale bar, 200 nm.
In the larger mitochondria from bovine heart, potato, and S. cerevisiae, detailed membrane features were usually obscured by the dense matrix. Occasionally, rows of ATP synthase dimers were visible and could be traced for short distances (Figs. S2 and S3).
The arrangement of ATP synthase in these mitochondria was similar to P. anserina, although the distance between the F1 heads in a dimer from potato was larger. The occurrence of ATP synthase dimer rows in whole mitochondria shows that the linear arrays previously observed in mitochondrial membrane fragments (12) were not due to membrane disruption but are a fundamental feature of intact, active mitochondria.
Subtomogram Averages.
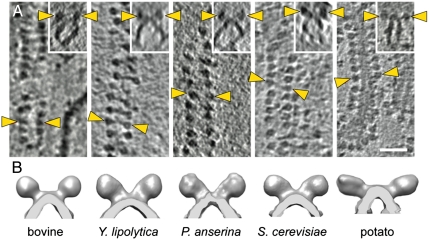
To compare the membrane arrangement of ATP synthase dimers in more detail, we collected tomographic volumes of isolated cristae membranes and calculated subtomogram averages of dimers from all five species (Fig. 2). The signal-to-noise ratio in such preparations was considerably higher than in intact mitochondria, because the membranes were surrounded by dilute aqueous buffer rather than the dense mitochondrial matrix. Cristae membrane fragments were either found amongst intact mitochondria, or created by successive freeze-thaw cycles. Although cristae were predominantly lamellar in whole mitochondria (Fig. 1 and Fig. S1), the isolated membrane fragments were mostly tubular (fungi and potato) or disk-like (bovine heart). As in whole mitochondria, the dimer rows were more or less straight, and followed the undulations of the cristae edges. The distance between F1 heads in a dimer matched those measured in whole mitochondria (Fig. S4). This suggests that the dimer rows are not easily disrupted by mechanical force, and either remain unperturbed or reassemble in isolated cristae membranes. Subtomogram averages of the two yeasts and P. anserina partially resolved the peripheral stalk, which extended from the dimer interface in the membrane toward the F1 heads (Fig. 2B). The angle between monomers in the dimer was approximately 80° for bovine heart and the three species of fungi, and approximately 115° for potato correlating with the difference observed in F1 head distance of the dimers.
Fig. 2.
Rows of F1-Fo ATP synthase dimers from five different species. (A) Tomographic slices showing linear arrays of F1-Fo ATP synthase dimers in mitochondrial membranes from bovine heart, Yarrowia lipolytica, Podospora anserina, Saccharomyces cerevisiae, and potato. (Inset) Side view of each array showing dimers in relation to the membrane. Yellow arrowheads indicate F1 heads of one dimer. Scale bar, 50 nm. (B) Surface representations of subtomogram averages. The number of dimers used in each average was as follows: bovine heart, 84; Y. lipolytica, 136; P. anserina, 24; S. cerevisiae, 138; and potato, 71.
Single-particle analyses of detergent-solublized ATP synthase dimers from S. cerevisiae have indicated two different conformations: one with an angle > 70° between the long axis of the monomers, and another with a smaller angle of approximately 40° (14–17). Clearly, all dimers we found in whole mitochondria and cristae fragments were of the former class with the wider dimer angle. Dimers with the smaller angle have been proposed to reflect the interaction of monomers along the rows (17). However, a dimer angle of 40° in this direction would result in an approximately 20-nm radius of curvature along the row, which we did not observe.
Alternatively, the 40° dimers might be due to an interaction with the IF1 factor that binds to the F1 heads and inhibits ATP hydrolysis. S. cerevisiae dimers with the 40° angle were isolated at pH 7.0. It is known that mammalian IF1 is mainly dimeric at pH 6.5 (26). It is thus conceivable that the 40° dimers formed by association with dimeric IF1 during isolation. Indeed, single-particle analysis of bovine heart ATP synthase revealed a similar 40° dimer with a bridging density between the F1 heads (15), which was attributed to the IF1 protein. Accordingly, the dimers in the linear arrays we found in whole mitochondria and isolated cristae membranes would be the uninhibited dimer, whereas the 40° dimers seem to be the inhibited IF1 complex.
Dimer Rows are Ubiquitous in All Mitochondria.
The linear rows of ATP synthase dimers contrast with the helical rows observed in the two protist species Paramecium and Polytomella (18, 27), which have been proposed to induce the formation of helical tubular cristae from flat membranes (28). The dimer rows in these species appear to be more regular, such that each dimer interacts specifically with its neighbors, each offset by a small angle [9° in Polytomella (27)]. This gives rise to a helical arrangement on the outer edge of spiralling tubular cristae (18). In the six species we investigated [including rat liver, (12)], no offset was observed between dimers, and thus the dimer arrays were never helical. Therefore, although details of molecular interaction within the dimer or between dimers along rows may vary, the linear arrays of ATP synthase dimers are a ubiquitous, fundamental feature in mitochondria of all eukaryotes, including protists (29).
Linear dimer rows induce tight bends or ridges in the inner membrane, often extending for several hundred nanometers. Our observation that the tightly bent membrane regions persist after disruption of the mitochondria supports our earlier conclusion that the dimers shape the membrane, rather than the other way round. The dimer rows thus exert a bending force on the lipid bilayer, and any newly added dimers converge at the point of highest membrane curvature, where the bending energy exerted by each dimer would be minimal. This suggests that the linear rows of ATP synthase dimers are sufficient to create cristae ridges, whereas other factors are probably required to generate or maintain cristae junctions.
Complex I in Cristae Membranes.
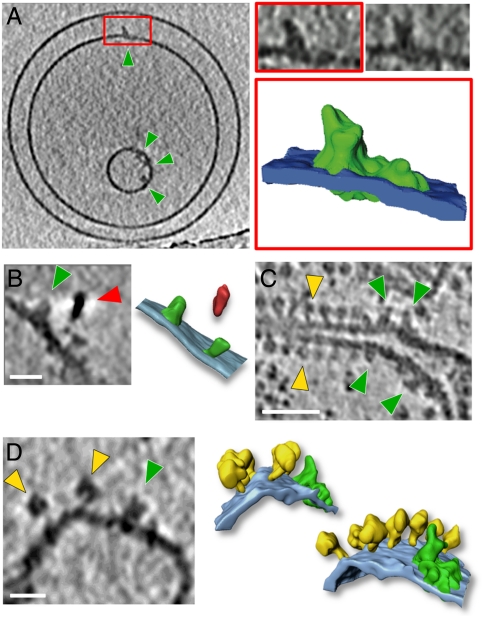
Rectangular particles rising approximately 15 nm above the inner membrane surface were often seen in tomographic volumes of isolated cristae membranes of all species investigated except S. cerevisiae (Fig. 3). Comparison with tomograms of complex I reconstituted into proteoliposomes suggested that these densities were most probably complex I (Fig. 3A). To confirm this, cristae membranes from Y. lipolytica were labeled with quantum dots conjugated to antibodies against complex I (Fig. 3B). Although the labeling efficiency was low, quantum dots were found only on one side of the membrane, and always within 25 nm of a complex I-like density, corresponding to the length of two IgG molecules. Apart from the ATP synthase, complex I is the only other membrane protein in the inner mitochondrial membrane large enough to be easily visible by cryo-ET. Complex I–like densities were found in membranes of all species except S. cerevisiae, which is known not to have this particular complex. We conclude that these rectangular densities are in fact complex I or belong to supercomplexes that contain it.
Fig. 3.
Complex I (A) Tomographic slice of proteoliposomes with reconstituted complex I from Y. lipolytica (green arrowheads). The red box indicates a complex I density that was segmented (complex I, green; membrane, blue). Slices through the boxed complex I volume are shown above the rendered image. (B) Tomographic slice and rendered volume of two complex I densities (green) labeled with a quantum dot (red arrow). Tomographic slice of isolated cristae membranes from potato (C) and bovine heart (D) showing different structures for complex I (green arrowheads) and ATP synthase (yellow arrowheads). The particle next to the complex I density in D may be a complex III dimer in a supercomplex. The rendered volume shows a row of ATP synthase dimers (yellow) in the membrane (light blue) next to a density containing complex I (green) of the same shape and size as bovine supercomplex I1III2IV1 (22). Scale bars: A and C, 50 nm; B and D, 20 nm.
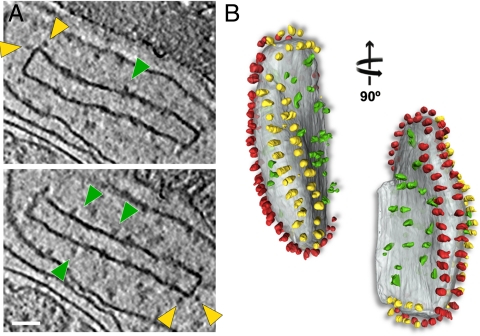
Because of the higher background, we were not able to identify complex I unambiguously in whole mitochondria. However, we were able to visualize complex I in tomograms of isolated cristae membranes from P. anserina, which had maintained their characteristic box-like shape (Fig. 4). Rows of ATP synthase dimers were found along the approximately 90° edges of the lamellar cristae, indicating that the supramolecular arrangement of membrane complexes had been preserved, whereas the flat sides of the box-shaped vesicles contained numerous densities without apparent long-range order. Approximately 30 complex-I-like densities and 108 ATP synthase molecules were present in one such box-like crista vesicle, giving a ratio of complex I to ATP synthase of 1∶3.6, very similar to what has been found biochemically (9). In the cristae, complex I thus occupies the membrane regions on either side of the dimer rows, which are more or less flat. The other two mitochondrial proton pumps, complex III and IV, which are too small to be detected by present cryo-ET techniques, most likely also occur in these membrane regions, and may form supercomplexes with complex I. Indeed, some of the complex I densities in potato and bovine heart resemble the shape and size of the supercomplex (22) (Fig. 3 C and D and Movie S3). We can exclude that, in the mitochondria we investigated, these complexes form respiratory strings (30–32), which would be easily visible by cryo-ET.
Fig. 4.
Isolated crista vesicle from P. anserina. (A) Two tomographic slices of the same box-shaped cristae vesicle showing ATP synthase dimers at approximately 90° bends in the membrane (yellow arrowheads) and complex I densities (green arrowheads) in flat membrane regions of the vesicle. (B) Segmented surface representation. Two rows of ATP synthase dimers (yellow and red) in the membrane (gray) run along the 90° membrane ridges. Irregularly distributed particles of complex I or other respiratory chain complexes (green) are confined to flat membrane regions (see Movie S2). Scale bar, 50 nm.
Functional Implications.
ATP production in mitochondria is powered by the pmf, manifest in the electrochemical transmembrane gradient. The resulting current of protons through the Fo rotor of the ATP synthase drives ATP production by rotary catalysis in the F1 head on the matrix side of the membrane. The pmf is generated by complexes I, III, and IV, the proton pumps of the respiratory chain, which translocate protons across the membrane from the matrix side into the cristae space. The pmf has two components, one resulting from the pH difference across the membrane (ΔpH), and the other from the membrane potential (Δψ). In mitochondria and chloroplasts, the pmf is roughly 180 mV. In chloroplasts, this is mostly accounted for by the ΔpH, whereas in mitochondria the ΔpH contribution is comparatively small. Given that the matrix pH is 7.9 (33), the cytosolic pH is 7.35 (34), and the outer membrane is freely permeable to ions, the nominal ΔpH across the mitochondrial inner membrane is 0.55 units, equivalent to 32 mV. However, in vitro studies indicate that the mitochondrial, chloroplast and bacterial ATP synthases all need a ΔpH close to 2 units, equivalent to approximately 120 mV, and a p-side pH close to 6, to produce enough ATP to sustain life (35–38). We propose that the apparent paradox of a ΔpH that is seemingly insufficient and a p-side pH that is too high for efficient ATP production is resolved by the special geometry of the mitochondrial cristae.
In a previous study (12), we calculated that the high degree of membrane curvature at the cristae ridges could accommodate a higher charge density for a given membrane potential, resulting in a local ΔpH increase of up to 0.5 units. Furthermore, an earlier modelling study by Cherepanov et al (39) found that protons released on the membrane surface encounter a potential barrier of 0.1 to 0.15 mV. The potential barrier makes it more difficult for the pumped protons to escape to the bulk solvent, resulting in a higher proton concentration, and consequently a lower pH, at distances up to 1 nm from the membrane surface. The effective pH in this boundary layer has been estimated at approximately 6, independent of the pH of the bulk solvent (39). The boundary layer, in combination with the higher charge density at the cristae ridges, would meet both the above requirements for efficient ATP synthesis, of a pH below 6 on the p-side of the membrane, and a ΔpH around 2. Protons, which move along membrane surfaces about 10 times faster than into bulk solvent (40), would thus diffuse preferentially along the membrane from their source at the respiratory chain proton pumps on either side of the cristae ridges toward the proton sinks at the ATP synthase dimer rows (Fig. 5). The rapid consumption of protons by the ATP synthases at the cristae ridges would establish a proton gradient along the membrane, by which protons would tend to flow from source to sink, instead of diffusing equally in all directions. The directional proton flow along the membrane surface would provide both a functional role for the mitochondrial cristae, and explain how the conserved, ubiquitous rows of ATP synthase dimers along the cristae ridges help to optimize ATP synthesis in mitochondria.
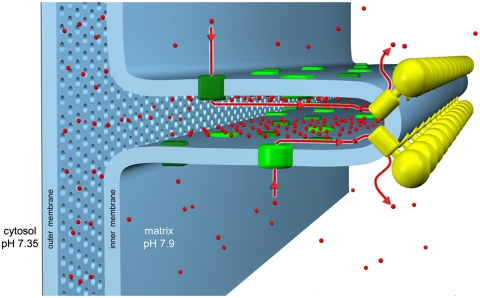
Fig. 5.
Molecular organization of cristae membranes. ATP synthase and complex I occupy different regions of the mitochondrial cristae. The ATP synthase forms dimer rows (yellow) at the cristae tips, whereas the proton pumps of the electron transfer chain (green), in particular complex I, reside predominantly in the adjacent membrane regions. Protons (red) pumped into the cristae space by the electron transport complexes flow back into the matrix through the ATP synthase rotor, driving ATP production. We propose that this conserved arrangement generates a local proton gradient in the cristae space, which would explain how the dimer rows help to optimize mitochondrial ATP synthesis, and provide a functional role for the mitochondrial cristae.
Materials and Methods
Culture Conditions.
Yarrowia lipolytica strain E129 (41), and Sacchromyces cervasiea strain BY4741 (EUROSCARF, Germany) were grown under standard conditions in YPG and YPEG medium respectively. Podospora anserina strain “s” (42) was grown as described (43). Young P. anserina mitochondria were isolated from liquid cultures inoculated with 6-d-old mycelium.
Purification of Mitochondria.
P. anserina and S. cerevisiae mitochondria were isolated as described in refs. 25 or 44, respectively. Y. lipolytica mitochondria were obtained by either method. Bovine heart mitochondria were isolated as described in refs. 45 and 46. Potato mitochondria were obtained as described in ref. 47 followed by a Percoll step gradient (26/46%). Whole mitochondria were fragmented by successive freeze-thaw cycles at -20 °C.
Complex I Reconstitution and Labeling.
Complex I was purified from Yarrowia lipolytica strain PIPO and reconstituted into liposomes as described (48). Labeling was performed on submitochondrial vesicles obtained from Y. lipolytica by osmotic shock. Membrane extracts were incubated for 1 h at 4 °C with a primary antibody (Y37F3) against the matrix arm of complex I from Y. lipolytica (49), followed by a secondary anti-mouse IgG antibody conjugated with a quantum-dot emitting at 605 nm (Invitrogen). Unbound antibodies were removed by an Optiprep step density gradient centrifugation (20/24%, 80,000 × g, 30 min).
Cryo-ET and Subtomogram Averaging.
Tomography was carried out as described (12, 43). All mitochondria samples were resuspended in trehalose buffer (250 mM trehalose, 10 mM Tris-HCl pH 7.4) immediately before plunge-freezing. Subtomograms were averaged in IMOD (50) after preorientation using manually picked contours which described the positions of the F1-F1 parts relative to the membrane. The final volume was 2-fold averaged.
Supplementary Material
Acknowledgments.
We thank Angelika Horst and Thorsten Blum for assistance in collecting tomograms; Alexandra Werner for preparing mitochondria from P. anserina; Julian Langer, Stephen Marino, Ilka Wittig and Götz Hofhaus for assistance in preparation of bovine heart mitochondria; Deryck Mills for maintaining the electron microscope facility; and Ernst Bamberg for helpful discussions. This work was supported by the Max Planck Society (W.K., K.D., M.S., B.D., and A.R.), the Deutsche Forschungsgemeinschaft (A.W., H.O., and V.Z.), and the Deutsche Forschungsgemeinschaft–funded Cluster of Excellence Frankfurt “Macromolecular Complexes” (W.K., H.O., and J.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103621108/-/DCSupplemental.
References
- 1.Collinson IR, et al. Fo membrane domain of ATP synthase from bovine heart mitochondria: Purification, subunit composition, and reconstitution with F1-ATPase. Biochemistry. 1994;33:7971–7978. doi: 10.1021/bi00191a026. [DOI] [PubMed] [Google Scholar]
- 2.Morgner N, et al. Subunit mass fingerprinting of mitochondrial complex I. Biochim Biophys Acta. 2008;1777:1384–1391. doi: 10.1016/j.bbabio.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Sambongi Y, et al. Mechanical rotation of the c subunit oligomer in ATP synthase (F0F1): Direct observation. Science. 1999;286:1722–1724. doi: 10.1126/science.286.5445.1722. [DOI] [PubMed] [Google Scholar]
- 4.Rubinstein JL, Walker JE, Henderson R. Structure of the mitochondrial ATP synthase by electron cryomicroscopy. EMBO J. 2003;22:6182–6192. doi: 10.1093/emboj/cdg608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandt U. Energy converting NADH: Quinone oxidoreductase (complex I) Annu Rev Biochem. 2006;75:69–92. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- 6.Stock D, Leslie AG, Walker JE. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- 7.Hunte C, Zickermann V, Brandt U. Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science. 2010;329:448–451. doi: 10.1126/science.1191046. [DOI] [PubMed] [Google Scholar]
- 8.Efremov RG, Baradaran R, Sazanov LA. The architecture of respiratory complex I. Nature. 2010;465:441–445. doi: 10.1038/nature09066. [DOI] [PubMed] [Google Scholar]
- 9.Schägger H, Pfeiffer K. The ratio of oxidative phosphorylation complexes I–V in bovine heart mitochondria and the composition of respiratory chain supercomplexes. J Biol Chem. 2001;276:37861–37867. doi: 10.1074/jbc.M106474200. [DOI] [PubMed] [Google Scholar]
- 10.Kagawa Y, Racker E. Partial resolution of the enzymes catalyzing oxidative phosphorylation. IX. Reconstruction of oligomycin-sensitive adenosine triphosphatase. J Biol Chem. 1966;241:2467–2474. [PubMed] [Google Scholar]
- 11.Nicastro D, Frangakis AS, Typke D, Baumeister W. Cryo-electron tomography of neurospora mitochondria. J Struct Biol. 2000;129:48–56. doi: 10.1006/jsbi.1999.4204. [DOI] [PubMed] [Google Scholar]
- 12.Strauss M, Hofhaus G, Schröder RR, Kühlbrandt W. Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 2008;27:1154–1160. doi: 10.1038/emboj.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wittig I, Velours J, Stuart R, Schägger H. Characterization of domain interfaces in monomeric and dimeric ATP synthase. Mol Cell Proteomics. 2008;7:995–1004. doi: 10.1074/mcp.M700465-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Couoh-Cardel SJ, Uribe-Carvajal S, Wilkens S, Garcia-Trejo JJ. Structure of dimeric F1F0-ATP synthase. J Biol Chem. 2010;285:36447–36455. doi: 10.1074/jbc.M110.144907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minauro-Sanmiguel F, Wilkens S, García JJ. Structure of dimeric mitochondrial ATP synthase: Novel F0 bridging features and the structural basis of mitochondrial cristae biogenesis. Proc Natl Acad Sci USA. 2005;102:12356–12358. doi: 10.1073/pnas.0503893102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas D, et al. Supramolecular organization of the yeast F1Fo-ATP synthase. Biol Cell. 2008;100:591–601. doi: 10.1042/BC20080022. [DOI] [PubMed] [Google Scholar]
- 17.Dudkina NV, Sunderhaus S, Braun H-P, Boekema EJ. Characterization of dimeric ATP synthase and cristae membrane ultrastructure from Saccharomyces and Polytomella mitochondria. FEBS Lett. 2006;580:3427–3432. doi: 10.1016/j.febslet.2006.04.097. [DOI] [PubMed] [Google Scholar]
- 18.Allen R, Schroeder C, Fok A. An investigation of mitochondrial inner membranes by rapid-freeze deep-etch techniques. J Cell Biol. 1989;108:2233–2240. doi: 10.1083/jcb.108.6.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paumard P, et al. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002;21:221–230. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soubannier V, et al. In the absence of the first membrane-spanning segment of subunit 4(b), the yeast ATP synthase is functional but does not dimerize or oligomerize. J Biol Chem. 2002;277:10739–10745. doi: 10.1074/jbc.M111882200. [DOI] [PubMed] [Google Scholar]
- 21.Schägger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schäfer E, Dencher N, Vonck J, Parcej D. Three-dimensional structure of the respiratory chain supercomplex I1III2IV1 from bovine heart mitochondria. Biochemistry. 2007;46:12579–12585. doi: 10.1021/bi700983h. [DOI] [PubMed] [Google Scholar]
- 23.Frey TG, Perkins GA, Ellisman MH. Electron tomography of membrane-bound cellular organelles. Annu Rev Biophys Biomol Struct. 2006;35:199–224. doi: 10.1146/annurev.biophys.35.040405.102039. [DOI] [PubMed] [Google Scholar]
- 24.Perkins GA, Ellisman MH, Fox DA. Three-dimensional analysis of mouse rod and cone mitochondrial cristae architecture: Bioenergetic and functional implications. Mol Vis. 2003;9:60–73. [PubMed] [Google Scholar]
- 25.Gredilla R, Grief J, Osiewacz HD. Mitochondrial free radical generation and lifespan control in the fungal aging model Podospora anserina. Exp Gerontol. 2006;41:439–447. doi: 10.1016/j.exger.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Cabezon E, Butler PJ, Runswick MJ, Walker JE. Modulation of the oligomerization state of the bovine F1-ATPase inhibitor protein, IF1, by pH. J Biol Chem. 2000;275:25460–25464. doi: 10.1074/jbc.M003859200. [DOI] [PubMed] [Google Scholar]
- 27.Dudkina NV, Oostergetel GT, Lewejohann D, Braun H-P, Boekema EJ. Row-like organization of ATP synthase in intact mitochondria determined by cryo-electron tomography. Biochim Biophys Acta . 2010;1797:272–277. doi: 10.1016/j.bbabio.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Allen R. Membrane tubulation and proton pumps. Protoplasma. 1995;189:1–8. [Google Scholar]
- 29.Lapaille M, et al. Atypical subunit composition of the chlorophycean mitochondrial F1FO-ATP synthase and role of Asa7 protein in stability and oligomycin resistance of the enzyme. Mol Biol Evol. 2010;27:1630–1644. doi: 10.1093/molbev/msq049. [DOI] [PubMed] [Google Scholar]
- 30.Bultema JB, Braun H-P, Boekema EJ, Kouril R. Megacomplex organization of the oxidative phosphorylation system by structural analysis of respiratory supercomplexes from potato. Biochim Biophys Acta. 2009;1787:60–67. doi: 10.1016/j.bbabio.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 31.Nubel E, Wittig I, Kerscher S, Brandt U, Schägger H. Two-dimensional native electrophoretic analysis of respiratory supercomplexes from Yarrowia lipolytica. Proteomics. 2009;9:2408–2418. doi: 10.1002/pmic.200800632. [DOI] [PubMed] [Google Scholar]
- 32.Wittig I, Carrozzo R, Santorelli F, Schägger H. Supercomplexes and subcomplexes of mitochondrial oxidative phosphorylation. Biochim Biophys Acta. 2006;1757:1066–1072. doi: 10.1016/j.bbabio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 33.Llopis J, McCaffery JM, Miyawaki A, Farquhar MG, Tsien RY. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc Natl Acad Sci USA. 1998;95:6803–6808. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kneen M, Farinas J, Li Y, Verkman AS. Green fluorescent protein as a noninvasive intracellular pH indicator. Biophys J. 1998;74:1591–1599. doi: 10.1016/S0006-3495(98)77870-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiedenmann A, Dimroth P, von Ballmoos C. Functional asymmetry of the F(0) motor in bacterial ATP synthases. Mol Microbiol. 2009;72:479–490. doi: 10.1111/j.1365-2958.2009.06658.x. [DOI] [PubMed] [Google Scholar]
- 36.Junesch U, Graber P. The rate of ATP-synthesis as a function of delta pH and delta psi catalyzed by the active, reduced H(+)-ATPase from chloroplasts. FEBS Lett. 1991;294:275–278. doi: 10.1016/0014-5793(91)81447-g. [DOI] [PubMed] [Google Scholar]
- 37.Forster K, et al. Proton transport coupled ATP synthesis by the purified yeast H(+)-ATP synthase in proteoliposomes. Biochim Biophys Acta. 2010;1797:1828–1837. doi: 10.1016/j.bbabio.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 38.Fischer S, Graber P. Comparison of ΔpH- and Δφ-driven ATP synthesis catalyzed by the H(+)-ATPases from Escherichia coli or chloroplasts reconstituted into liposomes. FEBS Lett. 1999;457:327–332. doi: 10.1016/s0014-5793(99)01060-1. [DOI] [PubMed] [Google Scholar]
- 39.Cherepanov DA, Feniouk BA, Junge W, Mulkidjanian AY. Low dielectric permittivity of water at the membrane interface: Effect on the energy coupling mechanism in biological membranes. Biophys J. 2003;85:1307–1316. doi: 10.1016/S0006-3495(03)74565-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heberle J, Riesle J, Thiedemann G, Oesterhelt D, Dencher NA. Proton migration along the membrane surface and retarded surface to bulk transfer. Nature. 1994;370:379–382. doi: 10.1038/370379a0. [DOI] [PubMed] [Google Scholar]
- 41.Barth G, Gaillardin C. In: Yarrowia lipolytica. Non-conventional Yeasts in Biotechnology: A Handbook. Wolf K, editor. Berlin: Spinger-Verlag; 1996. pp. 313–388. [Google Scholar]
- 42.Rizet G. Impossibility of obtaining uninterrupted and unlimited multiplication of the ascomycete Podospora anserina. C R Hebd Seances Acad Sci. 1953;237:838–840. (in French) [PubMed] [Google Scholar]
- 43.Brust D, et al. Cyclophilin D links programmed cell death and organismal aging in Podospora anserina. Aging Cell. 2010;9:761–775. doi: 10.1111/j.1474-9726.2010.00609.x. [DOI] [PubMed] [Google Scholar]
- 44.Daum G, Bohni PC, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- 45.Hackenbrock CR. Energy-linked ultrastructural transformations in isolated liver mitochondria and mitoplasts. Preservation of configurations by freeze-cleaving compared to chemical fixation. J Cell Biol. 1972;53:450–465. doi: 10.1083/jcb.53.2.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crane FL, Glenn JL, Green DE. Studies on the electron transfer system. IV. The electron transfer particle. Biochim Biophys Acta. 1956;22:475–487. doi: 10.1016/0006-3002(56)90058-0. [DOI] [PubMed] [Google Scholar]
- 47.Pavlov PF, Rudhe C, Bhushan S, Glaser E. In vitro and in vivo protein import into plant mitochondria. Methods Mol Biol. 2007;372:297–314. doi: 10.1007/978-1-59745-365-3_22. [DOI] [PubMed] [Google Scholar]
- 48.Drose S, Galkin A, Brandt U. Proton pumping by complex I (NADH:ubiquinone oxidoreductase) from Yarrowia lipolytica reconstituted into proteoliposomes. Biochim Biophys Acta. 2005;1710:87–95. doi: 10.1016/j.bbabio.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 49.Zickermann V, et al. Functional implications from an unexpected position of the 49-kDa subunit of NADH:ubiquinone oxidoreductase. J Biol Chem. 2003;278:29072–29078. doi: 10.1074/jbc.M302713200. [DOI] [PubMed] [Google Scholar]
- 50.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.