Abstract
The principles underlying the assembly and structure of complex microbial communities are an issue of long-standing concern to the field of microbial ecology. We previously analyzed the community membership of bacterial communities associated with the green macroalga Ulva australis, and proposed a competitive lottery model for colonization of the algal surface in an attempt to explain the surprising lack of similarity in species composition across different algal samples. Here we extend the previous study by investigating the link between community structure and function in these communities, using metagenomic sequence analysis. Despite the high phylogenetic variability in microbial species composition on different U. australis (only 15% similarity between samples), similarity in functional composition was high (70%), and a core of functional genes present across all algal-associated communities was identified that were consistent with the ecology of surface- and host-associated bacteria. These functions were distributed widely across a variety of taxa or phylogenetic groups. This observation of similarity in habitat (niche) use with respect to functional genes, but not species, together with the relative ease with which bacteria share genetic material, suggests that the key level at which to address the assembly and structure of bacterial communities may not be “species” (by means of rRNA taxonomy), but rather the more functional level of genes.
Keywords: lateral gene transfer, biofilm, ecological model
Metagenomic analysis of environmental microbial communities has revealed an enormous and previously unknown microbial diversity, and expanded our knowledge of their function in a variety of environments (1–5). Much still remains unknown, however, such as the principles underlying the assembly and structure of complex microbial communities, an issue of long-standing concern to the field of microbial ecology. To this aim, several recent studies have supported the “neutral hypothesis” (6–8), a largely stochastic model for community assembly, which assumes that species are ecologically equivalent and that community structure is determined by random processes (9, 10). However, there is also evidence that niche or deterministic processes play a role in community structure (11, 12); thus, both niche and neutral processes are likely to affect the assembly of complex microbial communities.
Support for these models is based on species abundance distributions, and critical functional aspects, such as the assumption of ecological equivalence, have for the most part not been tested. In this study, we examine the encoded functions of an algal-associated bacterial community and link patterns of function to patterns of community assembly. Following the results of an earlier study (13), we investigate these communities in the context of the lottery hypothesis, a model for community “assembly” derived from studies of eukaryotic communities, such as coral reef fish (14). This hypothesis incorporates both neutral and functional aspects and argues that ecological niches are colonized randomly from a pool of species with similar ecological function that can coexist in that niche (14, 15). Available space within that niche is colonized by whichever suitable species happens to arrive there first, meaning that colonization of space is random from within a functionally equivalent group of species. In the context of a bacterial community, this model implies that there are guilds of bacterial species, whose members are functionally equivalent with respect to their ability to colonize a particular niche (e.g., the surface of the seaweed Ulva australis), but that the composition of species in any particular community (e.g., a single U. australis individual) is determined stochastically by recruitment from within those guilds. Importantly, members of a guild can be phylogenetically related or unrelated. If this model is correct, different species from within these guilds should share functional traits, and a core suite of functional genes should be consistently present in all communities of a particular habitat, independent of the taxonomic or phylogenetic composition of its species.
In an earlier study (13) we characterized the bacterial phylogenetic diversity of seawater and U. australis, a member of a common green algal family often found in tidal rock pools or shelves around the world. We found that algal-associated communities were highly distinct from the surrounding seawater communities, but were also highly variable among individual algal samples, with only six operational taxonomic units (of a total of 528) at a 97% sequence identity cut-off occurring on all samples (13). This finding means that each U. australis sample hosts a unique assemblage of species (as defined by 97% 16S rRNA similarity). Given that the recruitment of new community members onto U. australis is most likely to come from the seawater, these results are somewhat contradictory with respect to dominant models of community assembly: the differences between seawater and algal communities imply selective mechanisms of assembly on the algal surface (niche partitioning), and the high variability between algal hosts is consistent with random colonization (e.g., neutral processes).
Here, we analyze the metagenomes of these communities to show that the algal-associated bacterial communities are functionally distinct from seawater communities, but contain a core of functional genes, which are represented across all algal samples. These functions are consistent with the ecology of surface- and host-associated bacteria, and importantly are distributed across a variety of taxa in individual communities, indicating functional redundancy across taxa. This mix of functional and random processes is consistent with the predictions of the competitive lottery model for the assembly of complex microbial communities. Moreover, this functional (niche) partitioning with respect to genes, but not phylogeny, in these assemblages highlights the potential difficulty in using bacterial species to test hypotheses derived from eukaryotic ecology, which focus on species as the critical unit. Given the relative ease with which bacteria share genetic material, the key level at which to address the assembly of these bacterial communities may not be species, but rather the more functional level of genes or gene clusters.
Results and Discussion
Algal-Associated Bacterial Communities Encode a Distinct Functional Profile.
We generated over 681 Mbp of metagenomic shotgun sequencing data from six algal (UA1–UA6) and eight seawater samples (SW3–SW10) (see Materials and Methods and Table S1). Environmental parameters were similar for each sample (see Materials and Methods and Table S2) and we found no evidence for increased viral or eukaryotic DNA between algal samples, indicating that these groups were a minor part of the microbial community at the time of sampling. Chao 1 estimates showed very similar levels of bacterial species diversity (ranging from 225 to 451) for the six algal communities (13). U. australis specimens were in the same developmental stage (i.e., fully matured) and the simple, two-cell layer alga is depauperate in bioactives found in many other algae, making it an ideal choice to minimize effects of host variability. Overall, these observations led us to the conclusion that bacterial communities on the U. australis samples collected existed under similar broad ecological conditions.
Sequencing data were used to create functional community profiles for each sample based on Clusters of Orthologous Groups (COG) (16) and SEED (17) annotations (details in Table S1). This process revealed that the algal-associated communities were functionally distinct from those in the surrounding seawater. Multidimensional scaling (MDS) plots show that the U. australis samples clustered together, and separately from seawater samples (Fig. 1A), and permutational multivariate analysis of variance (PERMANOVA) indicates that this difference is significant (P = 0.001). This distinctiveness was quantitative and multivariate, rather than qualitative, and there were few functions that were consistently present in one environment and consistently absent in the other. As such, the differences between the two environments lie predominantly in the relative abundance of particular functions as defined by COG and SEED annotations. Variation was also observed among seawater or U. australis samples, but this was primarily seen for those samples with the least (order-of-magnitude less) sequence data. When these samples were removed, the remaining algal samples still clustered together quite tightly (Fig. 1B). Bray-Curtis similarity shows that on average there was 15% similarity in species composition across samples (13), compared with 70% similarity in functional composition (COGs), indicating that despite large differences in species composition between hosts [as detected in our previous study (13)], many encoded functions are shared.
Fig. 1.
MDS plots based on Bray-Curtis similarity of functional gene profiles based on COG and SEED annotations of U. australis and planktonic seawater metagenomes. (A) MDS plots for all samples, including UA4, UA6, SW4, SW6, SW8, and SW10, which contained an order-of-magnitude less sequencing than the remaining samples. (B) MDS plots with low coverage samples removed. Samples from each of the respective environments cluster together based on their functional profile.
Functional Core in U. australis-Associated Bacterial Communities.
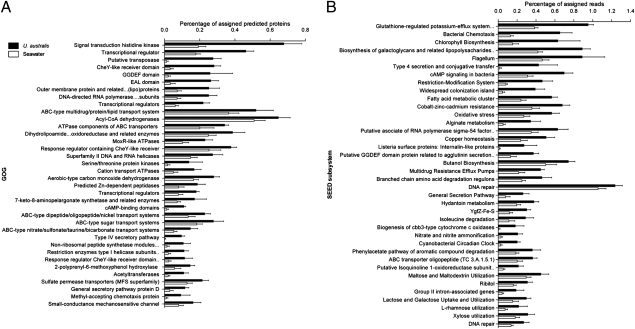
We observed a set of COG and SEED functions that contributed strongly to the difference between the two community types (Fig. S1 and SI Materials and Methods), and that were consistently abundant across U. australis samples. These were defined as the core functions of the U. australis associated bacterial communities (Fig. 2). These functions fit well with the current understanding of the ecology of an algal- or surface-associated bacterial community and could mostly be grouped into broader categories (Table S3) and are summarized here.
Fig. 2.
COG (A) and SEED subsystems (B), which comprise the characteristic functions of the U. australis community, by comparison with planktonic seawater, with SD across the six algal and eight seawater samples analyzed. COGs and SEED subsystems are presented in the order of their contribution to difference (highest to lowest, Top to Bottom) as assessed by SIMPER analysis.
Detection and movement toward the host surface.
Proteins associated with chemotaxis and flagellum-mediated motility were consistently abundant in the U. australis community, and are likely to be important for the detection and movement toward the algal host surface during colonization. Chemotaxis is essential for the development and maintenance of symbiotic, surface associations, as for example in symbiotic Rhizobium sp., which are chemotactically attracted to the flavenoids that induce the nodulation genes necessary for nitrogen fixation in the host plant (18). Flagella-mediated motility is also important for biofilm formation in a range of bacteria (19–22).
Attachment and biofilm formation.
An array of proteins functionally assigned to attachment and biofilm formation were overrepresented across U. australis samples. Functions include homologs of the OmpA protein (COG2197), which is required for adhesion to both mammalian and fish epithelial cells in a range of Proteobacteria (23, 24), Listeria internalin-like proteins, which enhance attachment and biofilm formation (25, 26), and the widespread colonization island, which is essential for biofilm formation, colonization, and pathogenesis in a range of bacteria (27). Proteins related to the production and excretion of galactoglycan, or exopolysaccharide II, were more abundant in the U. australis community, and apart from forming part of the biofilm matrix, is also essential for the establishment and maintenance of symbiosis in several Rhizobium strains (28–32). GGDEF and EAL domain proteins, which are involved in the production and degradation, respectively, of bis-(3′-5′)-cyclic dimeric GMP (cyclic-di-GMP), were also detected at a higher abundance (33, 34). Cyclic-di-GMP is an important secondary messenger, regulating the transition from a motile planktonic to a surface-associated biofilm lifestyle in a range of bacteria (34, 35), for example by up-regulating the production of adhesins and biofilm matrix components (36–39) or downregulating motility genes (34).
Genes encoding for Cbb3-type cytochrome c oxidases, which have a high affinity for oxygen and are associated with microaerobic metabolism in oxygen-limited environments (40), were overrepresented, as well as the function of nitrite and nitrate ammonification, considered the highest energy-yielding respiration systems after oxygen has been depleted (41). The higher abundance of proteins assigned these functions may be related to survival in biofilms, which are known to be spatially heterogeneous, containing pockets of low or no oxygen in some areas (42).
Response to the algal host environment.
Some of the overrepresented functions can be related directly to U. australis’ surface environment. For example, proteins associated with the metabolism of water-soluble polysaccharides produced by Ulva sp., such as rhamnose, xylose, glucose, mannose, and galactose (43), would enable bacteria to use these sugars as a source of carbon and energy and, hence, gain a competitive advantage in colonizing the host surface. Proteins associated with heat and osmotic stress (e.g., COG0668: small conductance mechanosensitive channel) (44–46) may be related to the desiccation of U. australis fronds, which occur in their intertidal habitats at low tide. Macroalgae also defend themselves against bacteria and herbivores by the release of reactive oxygen species, an oxidative burst (47), and the overrepresentation of proteins controlling oxidative stress might represent a protective mechanism for the surface community. Finally, Ulva sp. is known to take up and store heavy metals (48), and the overrepresentation of proteins associated with the export of heavy metals could thus be related to the presence of heavy metals in the algal host.
Regulation in response to environmental stimuli.
Some of the most overrepresented functions found in the microbial community of U. australis involve regulatory mechanisms in response to environmental stimuli. The high proportion of environmental signal transducers and transcriptional regulators could be indicative of the need to respond to changes in the host environment (e.g., osmotic; see above), to control the complex steps of colonization or biofilm formation and to mediate the interactions with other community members. For example, there are many homologs of COG0642 (histidine kinase), members of which are known to be involved in osmoregulation (49), multidrug export (50), sporulation (51), nitrate reduction (52), cell differentiation (53), and plant virulence (54). Also overrepresented is COG0583 (transcriptional regulator), which contains proteins that regulate transcription in response to plant exudates (55), virulence, motility, and quorum sensing (56). There are many examples of plants and their metabolites affecting gene regulation of associated bacteria (57–61), and it is likely that such regulators play an important part in mediating interactions between U. australis and the surface community.
Lateral gene transfer.
Type IV secretion system (T4SS), transposases, and intron-associated genes were overrepresented in U. australis samples. These functions are associated with lateral gene transfer, a source of dynamic genomic change that allows for rapid ecological adaptation (62), and which would provide a broad mechanism for facilitating the functional similarity of phylogenetically distinct bacteria on the surface of U. australis (see below). Biofilms are ideal environments for lateral gene transfer (63), and an abundance of transposases has been noted in other biofilm communities (64). A recent survey of the transposase families in different taxa suggested that transposases are most often transferred to other organisms within the same habitat and can be shared by distantly related taxa (65), and COG2801 (putative transposase) is a potential source of shared functional traits among different taxa in the U. australis bacterial community. Although T4SSs are also often associated with virulent host/bacterium interactions (66), this system can also mediate symbiotic interactions (67). For example, Mesorhizobium loti R7A encodes a “symbiosis island,” containing a T4SS homologous to the Vir pilus from Agrobacterium tumefaciens. It is believed that this T4SS transfers effector proteins into host plant cells (68). Although speculative, it is plausible that T4SS could be used by the bacterial community of U. australis to mediate symbiotic interactions, such as the transfer of compounds inducing correct morphology of the alga (69).
Defense.
Genes related to defense include multidrug transport, restriction modification systems, nonribosomal peptide synthase modules, and ABC transporters, which have reported links with virulence (70, 71), and are also abundant in the algal surface community. Algal-associated bacteria may protect the host by inhibiting the attachment of other bacteria and biofouling organisms through the production of secondary metabolites (71). Toxic and antibiotic compounds may be transported out of the cell via homologs of ABC transporters known to export multiple drugs (72). The presence of restriction modification systems indicates a need to minimize transduction or transformation, or to only allow for genetic exchange between bacteria that have similar restriction modification systems. All these functions can clearly contribute to the maintenance of the structural and genetic integrity and stability of the surface community, which is of particular importance in a system that is constantly exposed to a large number of potentially invading bacteria (e.g., planktonic secondary colonizers).
These functions are all consistent with the ecological role of the U. australis community and together provide support for the notion of a specific and stable core metagenome, which is functionally adapted to life on the alga’s surface.
Colonization of U. australis: Does Taxonomy Reflect Function?
In eukaryotic models of colonization via a competitive lottery, functional groupings (or “guilds”) often reflect taxonomic groupings (15, 73–76). Alternatively, there may be taxonomic redundancy, in which any given function is distributed broadly across a variety of taxa as opposed to being associated with any particular taxonomic group. To address this question of taxonomic redundancy, amino acid sequences of proteins assigned to six functions, which contributed most to the difference between algal and seawater samples, were analyzed using phylogenetic tree comparisons in Unifrac (77) and a taxonomic last common ancestor algorithm (MEGAN) (78) (see Materials and Methods). A large degree of phylogenetic dissimilarity of the core functions was exemplified by pairwise comparisons of samples, displaying between 65% and 97% dissimilarity (Table S4), indicating that the protein phylogenies for each core function were distinct in each sample. Unifrac P values indicated that for five core functions, the phylogenetic distribution of proteins between samples was not significantly different to that expected by chance (P values were > 0.05). The only exception was the pairwise dissimilarities of as little as 52% at a P value of 0.03 for COG2801 (putative transposase). Transposases are part of mobile genetic elements and therefore are likely transferred between members of the different communities. This finding would make those communities more similar and the differences appear less random with respect to the function of COG2801.
Taxonomic assignment with MEGAN further showed that the core functions were generally widely distributed across major bacterial groups present on U. australis, namely the α- and γ-proteobacteria, Bacteroidetes, and Planctomycetes, suggesting that a broad range of bacteria possess the ability to carry out these functions (Figs. S2–S7). At the level of species or genus, proteins were distributed across a variety of taxa, which differed from sample to sample, and often a function was assigned to a species that was present in one sample only (although it should be noted that proteins are not necessarily from the specific species assigned, but have high similarity to the homologous protein from that species). This finding means that protein functions are derived from distinct lineages that can only be crudely assigned to very high-level taxa (e.g., phyla) or, when a low-level assignment (e.g., species/genus) is possible, there is little overlap between samples. Together the result of the phylogenetic and taxonomic analysis of proteins support the assertion that the core functions are not restricted to a particular taxonomic group. This result also means that different taxa provide the core functions to the community and that members from different taxa could form a functional guild.
Structure of Bacterial Communities: Assembly of Functional Genes or Assembly of Species?
Although our samples are taken over a limited time scale, and thus do not fully accommodate potential successional or historic changes in these communities, the evidence presented here and in our previous study (13) is most consistent with a competitive lottery model for community assembly on the surface of U. australis. Originally proposed for coral reef fish (15, 73), and subsequently applied to plant (74, 75) and parasite communities (76), the lottery model combines functional (niche- or guild-based) and random components as drivers of community structure. Specifically, species with similar trophic or other ecological properties are able to occupy the same niche within an ecosystem, and the particular species that occupies a particular space is then determined by stochastic recruitment. This means that within a group of species with similar ecologies, the “lottery” for space is won by whoever gets there first (15, 79).
In our system, as long as a bacterium has the necessary functional characters (defined here as particular gene functions) to colonize or grow on U. australis, the specific assemblage of species present on any given algal surface is stochastic, determined by which members of the guild happen to be available to colonize from the water column when space becomes available. Although the lottery model was originally proposed for relatively phylogenetically narrow groups of organisms (e.g., families of coral reef fish), we argue here that it also explains community assembly when the species pool spans multiple bacterial phyla.
More broadly and independently of any specific theory of community assembly, our results imply that genes or gene clusters are as or more important than species for understanding community assembly in bacterial systems. Dawkins (80) has famously argued in the context of evolutionary theory that individuals, and by inference collections of individuals such as species, are essentially containers for collections of genes. This approach contrasts with ecological models for community assembly, in which species (or higher taxonomic levels) are the fundamental metric. Such models are largely drawn from studies of eukaryotes, and so this species-focused approach is not surprising, given the general assumption of substantial genetic coherence within eukaryotic species.
However, the utility of the species concept for bacteria has been challenged in a number of ways (81–83), ranging from the level of genetic similarity necessary to define a species, to the extent (or lack thereof) of the genomic coherence of “species,” because of the occurrence of substantial genetic exchange among taxonomically distinct bacteria. Indeed, in our system this frequent genetic exchange was supported by the abundance of functions for horizontal gene transfer (e.g., type IV secretion and transposase). We also found that analysis of functional gene systems revealed a considerable biological pattern that was not evident by focusing only on patterns of species diversity (13). Similar observations have been made for the human microbiome project (84, 85). This finding has at least two implications for studies of microbial community ecology. First, tests of community assembly theory [e.g., neutral theory (8)] using species as the key parameter may be misleading, because, if function does not map onto taxonomy, then accumulation or assembly of species will always appear random. Second, almost all of the theory currently used to understand patterns of microbial diversity is derived from eukaryotic ecology. However, our results raise the general possibility that genes and their functions may be more useful in testing models of community ecology for bacterial communities. This may be one important way in which the ecology of bacteria differs from that of eukaryotes.
Conclusion
This study provides insight into the link between community structure and function for a complex, algal-associated bacterial community. This community contains a consistent functional profile, with features related to a host-associated lifestyle, and functional similarities exist within phylogenetically distinct members from different host individuals. Although the community members on U. australis contain functional similarities, we do not yet know whether they form guilds that are specific to U. australis, or whether they are more generally adapted to other living or inanimate surfaces. Nevertheless, our evidence is consistent with community assembly via a competitive lottery mechanism. This model encompasses both selective and neutral processes, and could apply to other complex host-associated microbial communities, such as the human microbiome, where a consistent core of functional genes is detected across hosts (85), but species-level community composition is highly variable (86). The lack of correspondence between function (as determined by functional gene systems) and phylogeny in this system suggests that genes, rather than species, may be the appropriate parameter for understanding patterns of diversity in many microbial communities.
Materials and Methods
Sampling.
Seawater was collected from Botany Bay (SW3 and SW4: 33°59′S, 151°14′E) on the 3rd of January 2005 and on the 19th of January 2005 from Bare Island (SW5 and SW6: 33°59′S, 151°13′E) and 200 m away in Botany Bay (SW7 and SW8). Seawater was collected again from Bare Island (SW9 and SW10) at the 18th of October 2006 to coincide with the sampling of U. australis at this site. Two-hundred liters of seawater were collected for each sample (exception is 100 L each for SW9 and SW10) from a depth of 2 m, and immediately serially filtered through 20-, 3-, 0.8-, and 0.1-μm filters. U. australis thalli were collected (wet weight: 20 g per sample) from two different rock pools at Bare Island in October 2006 (UA1 and UA2: 33°59′S, 151°13′E) and again on the 7th of February 2007 (UA3 and UA4). Thalli were also collected from two different rock pools ∼9 km away at Shark Point, Clovelly, on the 7th of February 2007 (UA5 and UA6: 33°91′S, 151°26′E). Sampling was performed between 10:00 AM and 12:00 PM after outgoing tides to ensure rock pools were well flushed with the surrounding seawater, and only algae of the same approximate size were collected to ensure that they were in the same developmental stage of their life cycle. Water parameters were also measured (Table S2) and showed similar values across the years. The sampling design and environmental measurements suggest that each sample of U. australis was subjected to similar environmental conditions.
DNA Sequencing, Assembly, and Annotation.
DNA was extracted from the six algal (UA1–UA6) and eight seawater samples (SW3–SW10), which correspond to samples from ref. 13. Bacterial DNA was extracted from the surface of the algal fronds as described previously (87), which leaves the algal host intact and extracts total DNA from the entire surface community. For the filtered seawater samples, DNA was extracted from the 0.1-μm filter, as previously described (88).
Large-scale shotgun sequencing was performed and sequences were quality-filtered, assembled, and annotated (details in SI Materials and Methods). Each ORF was searched against the COG database (16) and a matrix of raw counts per COG and per sample was generated. Data were also submitted to the MG-RAST server (89) in the form of unassembled individual reads. Matches to the SEED database (17) with an e-value of less than 10−20 and a minimum alignment length of 100 amino acids were used in a matrix of counts per SEED subsystem per sample.
Statistical Analysis of Metagenomic Datasets and Core Functions.
Matrices of raw counts of COG and SEED functional annotations per U. australis or seawater sample were standardized to account for the unequal sequence coverage between samples. Bray-Curtis similarity matrices were calculated and used to generate MDS plots. PERMANOVA (90) were carried out to compare samples from each environment. Similarity percentage analyses (SIMPER) (91) were carried out to determine the contribution of each COG or SEED subsystem to similarities within, and difference between, environments. The PRIMER-6 package was used for all multivariate statistical analysis (92).
The top 50 COG and SEED subsystems which contributed most to the differences between the two environments (as assessed by SIMPER), were selected for further analysis. After removal of those, which were not consistently more abundant in the algal dataset, the remaining 36 COG and 38 SEED subsystems were defined as comprising the core functions of the U. australis bacterial community. These functions were grouped into broader categories with respect to a surface-associated lifestyle with a living algal host (see SI Materials and Methods for details).
Assessment of Phylogenetic Similarity and Taxonomic Orgin of Core Functions.
Protein sequences from each U. australis sample for the six COGs, which contributed most to the difference from seawater libraries, were aligned in ClustalW 1.83 (93) and maximum likelihood trees were built in RAxML 7.0.4 (94) using 100 bootstraps. Tree files were analyzed with the Unifrac Webserver (http://bmf2.colorado.edu/unifrac/) (77) to calculate the proportion of branch length, which is unique or shared in each environment. The unweighted algorithm was used as each protein sequence was unique. Sequences were also compared against the National Center for Biotechnology Information nonredundant protein database using BLASTP (95) and analyzed with MEGAN 3.7 (78), which uses a last common ancestor algorithm to assign a likely taxonomic origin to each sequence. The predicted origins of proteins were compared to determine if particular functions were associated with particular taxa. Only sequences from samples UA1, UA2, UA3, and UA5 were used in MEGAN analysis; samples UA4 and UA6 contained fewer than 10 sequences for each function because of the lower depth of sequence coverage obtained.
Supplementary Material
Acknowledgments
We thank Matt DeMaere for help with the annotation and Alastair Poore for help with the statistical analysis. This work was funded by the Australian Research Council; the Gordon and Betty Moore Foundation; the US Department of Energy, Office of Science, Office of Biological and Environmental Research (DE-FG02-02ER63453); The Center for Marine Bio-Innovation; and the J. Craig Venter Institute. This is contribution #0053 from the Sydney Institute of Marine Science.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Shotgun sequencing data are available through the Community Cyberinfrastructure for AdvancedMicrobial Ecology Research and Analysis (accession: CAM_PROJ_BotanyBay).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101591108/-/DCSupplemental.
References
- 1.Debroas D, et al. Metagenomic approach studying the taxonomic and functional diversity of the bacterial community in a mesotrophic lake (Lac du Bourget—France) Environ Microbiol. 2009;11:2412–2424. doi: 10.1111/j.1462-2920.2009.01969.x. [DOI] [PubMed] [Google Scholar]
- 2.Hewson I, et al. Metagenomic potential of microbial assemblages in the surface waters of the central Pacific Ocean tracks variability in oceanic habitat. Limnol Oceanogr. 2009;54:1981–1994. [Google Scholar]
- 3.Ram RJ, et al. Community proteomics of a natural microbial biofilm. Science. 2005;308:1915–1920. [PubMed] [Google Scholar]
- 4.van Elsas JD, et al. The metagenomics of disease-suppressive soils— Experiences from the METACONTROL project. Trends Biotechnol. 2008;26:591–601. doi: 10.1016/j.tibtech.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Yooseph S, et al. The Sorcerer II Global Ocean Sampling expedition: Expanding the universe of protein families. PLoS Biol. 2007;5(3):e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ofiteru ID, et al. Combined niche and neutral effects in a microbial wastewater treatment community. Proc Natl Acad Sci USA. 2010;107:15345–15350. doi: 10.1073/pnas.1000604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sloan WT, et al. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ Microbiol. 2006;8:732–740. doi: 10.1111/j.1462-2920.2005.00956.x. [DOI] [PubMed] [Google Scholar]
- 8.Woodcock S, et al. Neutral assembly of bacterial communities. FEMS Microbiol Ecol. 2007;62(2):171–180. doi: 10.1111/j.1574-6941.2007.00379.x. [DOI] [PubMed] [Google Scholar]
- 9.Hubbell SP. The Unified Neutral Theory of Biodiversity and Biogeography. Princeton: Princeton Univeristy Press; 2001. [Google Scholar]
- 10.Hubbell SP. Neutral theory and the evolution of ecological equivalence. Ecology. 2006;87:1387–1398. doi: 10.1890/0012-9658(2006)87[1387:ntateo]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 11.Dumbrell AJ, Nelson M, Helgason T, Dytham C, Fitter AH. Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J. 2010;4:337–345. doi: 10.1038/ismej.2009.122. [DOI] [PubMed] [Google Scholar]
- 12.Yang H, Schmitt-Wagner D, Stingl U, Brune A. Niche heterogeneity determines bacterial community structure in the termite gut (Reticulitermes santonensis) Environ Microbiol. 2005;7:916–932. doi: 10.1111/j.1462-2920.2005.00760.x. [DOI] [PubMed] [Google Scholar]
- 13.Burke C, Thomas T, Lewis M, Steinberg P, Kjelleberg S. Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J. 2011;5:590–600. doi: 10.1038/ismej.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sale PF. Reef fish lottery. Nat Hist. 1976;85:60–65. [Google Scholar]
- 15.Munday PL. Competitive coexistence of coral-dwelling fishes: The lottery hypothesis revisited. Ecology. 2004;85:623–628. [Google Scholar]
- 16.Tatusov RL, et al. The COG database: An updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Overbeek R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munoz Aguilar JM, et al. Chemotaxis of Rhizobium leguminosarum biovar phaseoli towards flavonoid inducers of the symbiotic nodulation genes. J Gen Microbiol. 1988;134:2741–2746. [Google Scholar]
- 19.Hossain MM, Tsuyumu S. Flagella-mediated motility is required for biofilm formation by Erwinia carotovora subsp. carotovora. J Gen Plant Pathol. 2006;72:34–39. [Google Scholar]
- 20.Houry A, Briandet R, Aymerich S, Gohar M. Involvement of motility and flagella in Bacillus cereus biofilm formation. Microbiology. 2010;156:1009–1018. doi: 10.1099/mic.0.034827-0. [DOI] [PubMed] [Google Scholar]
- 21.Lemon KP, Higgins DE, Kolter R. Flagellar motility is critical for Listeria monocytogenes biofilm formation. J Bacteriol. 2007;189:4418–4424. doi: 10.1128/JB.01967-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 23.Namba A, et al. OmpA is an adhesion factor of Aeromonas veronii, an optimistic pathogen that habituates in carp intestinal tract. J Appl Microbiol. 2008;105:1441–1451. doi: 10.1111/j.1365-2672.2008.03883.x. [DOI] [PubMed] [Google Scholar]
- 24.Serino L, et al. Identification of a new OmpA-like protein in Neisseria gonorrhoeae involved in the binding to human epithelial cells and in vivo colonization. Mol Microbiol. 2007;64:1391–1403. doi: 10.1111/j.1365-2958.2007.05745.x. [DOI] [PubMed] [Google Scholar]
- 25.Chen BY, Kim TJ, Silva JL, Jung YS. Positive correlation between the expression of inlA and inlB genes of Listeria monocytogenes and its attachment strength on glass surface. Food Biophys. 2009;4:304–311. [Google Scholar]
- 26.Franciosa G, Maugliani A, Scalfaro C, Floridi F, Aureli P. Expression of internalin A and biofilm formation among Listeria monocytogenes clinical isolates. Int J Immunopathol Pharmacol. 2009;22(1):183–193. doi: 10.1177/039463200902200121. [DOI] [PubMed] [Google Scholar]
- 27.Tomich M, Planet PJ, Figurski DH. The tad locus: Postcards from the widespread colonization island. Nat Rev Microbiol. 2007;5:363–375. doi: 10.1038/nrmicro1636. [DOI] [PubMed] [Google Scholar]
- 28.Borthakur D, et al. A mutation that blocks exopolysaccharide synthesis prevents nodulation of peas by Rhizobium leguminosarum but not of beans by R. phaseoli and is corrected by cloned DNA from Rhizobium or the phytopathogen Xanthomonas. Mol Gen Genet. 1986;203:320–323. [Google Scholar]
- 29.González JE, Reuhs BL, Walker GC. Low molecular weight EPS II of Rhizobium meliloti allows nodule invasion in Medicago sativa. Proc Natl Acad Sci USA. 1996;93:8636–8641. doi: 10.1073/pnas.93.16.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hotter GS, Scott DB. Exopolysaccharide mutants of Rhizobium loti are fully effective on a determinate nodulating host but are ineffective on an indeterminate nodulating host. J Bacteriol. 1991;173:851–859. doi: 10.1128/jb.173.2.851-859.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pellock BJ, Cheng HP, Walker GC. Alfalfa root nodule invasion efficiency is dependent on Sinorhizobium meliloti polysaccharides. J Bacteriol. 2000;182:4310–4318. doi: 10.1128/jb.182.15.4310-4318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rolfe BG, et al. Defective infection and nodulation of clovers by exopolysaccharide mutants of Rhizobium leguminosarum bv trifolii. Aust J Plant Physiol. 1996;23:285–303. [Google Scholar]
- 33.Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: Insights into biochemistry of the GGDEF protein domain. J Bacteriol. 2005;187:1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simm R, Morr M, Kader A, Nimtz M, Römling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 35.Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 36.Kulasekara HD, et al. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol Microbiol. 2005;55:368–380. doi: 10.1111/j.1365-2958.2004.04402.x. [DOI] [PubMed] [Google Scholar]
- 37.Lee VT, et al. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol. 2007;65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Römling U. Molecular biology of cellulose production in bacteria. Res Microbiol. 2002;153:205–212. doi: 10.1016/s0923-2508(02)01316-5. [DOI] [PubMed] [Google Scholar]
- 39.Tischler AD, Camilli A. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol. 2004;53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pitcher RS, Brittain T, Watmough NJ. Cytochrome cbb(3) oxidase and bacterial microaerobic metabolism. Biochem Soc Trans. 2002;30:653–658. doi: 10.1042/bst0300653. [DOI] [PubMed] [Google Scholar]
- 41.Strohm TO, Griffin B, Zumft WG, Schink B. Growth yields in bacterial denitrification and nitrate ammonification. Appl Environ Microbiol. 2007;73:1420–1424. doi: 10.1128/AEM.02508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Beer D, Stoodley P, Roe F, Lewandowski Z. Effects of biofilm structures on oxygen distribution and mass transport. Biotechnol Bioeng. 1994;43:1131–1138. doi: 10.1002/bit.260431118. [DOI] [PubMed] [Google Scholar]
- 43.Lahaye M, Axelos MAV. Gelling properties of water soluble polysaccharides from proliferating marine green seaweeds (Ulva spp) Carbohydr Polym. 1993;22:261–265. [Google Scholar]
- 44.Hurst AC, et al. MscS, the bacterial mechanosensitive channel of small conductance. Int J Biochem Cell Biol. 2008;40:581–585. doi: 10.1016/j.biocel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 45.Miticka H, et al. Transcriptional analysis of the rpoE gene encoding extracytoplasmic stress response sigma factor sigmaE in Salmonella enterica serovar Typhimurium. FEMS Microbiol Lett. 2003;226:307–314. doi: 10.1016/S0378-1097(03)00600-1. [DOI] [PubMed] [Google Scholar]
- 46.Vanaporn M, Vattanaviboon P, Thongboonkerd V, Korbsrisate S. The rpoE operon regulates heat stress response in Burkholderia pseudomallei. FEMS Microbiol Lett. 2008;284:191–196. doi: 10.1111/j.1574-6968.2008.01216.x. [DOI] [PubMed] [Google Scholar]
- 47.Pohnert G. Chemical defense strategies of marine organisms. In: Schulz S, editor. Chemistry of Pheromones and Other Semiochemicals I, Topics in Current Chemistry. Vol 239. Berlin: Springer-Verlag Berlin; 2004. pp. 179–219. [DOI] [PubMed] [Google Scholar]
- 48.Gaudry A, et al. Heavy metals pollution of the Atlantic marine environment by the Moroccan phosphate industry, as observed through their bioaccumulation in Ulva lactuca. Water Air Soil Pollut. 2007;178:267–285. [Google Scholar]
- 49.Yoshida T, Phadtare S, Inouye M. Functional and structural characterization of EnvZ, an osmosensing histidine kinase of E. coli. In: Simon MI, Crane BR, Crane A, editors. Two-Component Signaling Systems, Pt B, Methods in Enzymology. Vol 423. San Diego: Elsevier Academic Press Inc; 2007. pp. 184–202. [DOI] [PubMed] [Google Scholar]
- 50.Nagakubo S, Nishino K, Hirata T, Yamaguchi A. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J Bacteriol. 2002;184:4161–4167. doi: 10.1128/JB.184.15.4161-4167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perego M, Cole SP, Burbulys D, Trach K, Hoch JA. Characterization of the gene for a protein kinase which phosphorylates the sporulation-regulatory proteins Spo0A and Spo0F of Bacillus subtilis. J Bacteriol. 1989;171:6187–6196. doi: 10.1128/jb.171.11.6187-6196.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiang RC, Cavicchioli R, Gunsalus RP. Identification and characterization of narQ, a second nitrate sensor for nitrate-dependent gene regulation in Escherichia coli. Mol Microbiol. 1992;6:1913–1923. doi: 10.1111/j.1365-2958.1992.tb01364.x. [DOI] [PubMed] [Google Scholar]
- 53.Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci USA. 2006;103:10935–10940. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin SG, Roitsch T, Ankenbauer RG, Gordon MP, Nester EW. The VirA protein of Agrobacterium tumefaciens is autophosphorylated and is essential for vir gene regulation. J Bacteriol. 1990;172:525–530. doi: 10.1128/jb.172.2.525-530.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cai T, et al. Host legume-exuded antimetabolites optimize the symbiotic rhizosphere. Mol Microbiol. 2009;73:507–517. doi: 10.1111/j.1365-2958.2009.06790.x. [DOI] [PubMed] [Google Scholar]
- 56.Maddocks SE, Oyston PCF. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology. 2008;154:3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 57.Mark GL, et al. Transcriptome profiling of bacterial responses to root exudates identifies genes involved in microbe-plant interactions. Proc Natl Acad Sci USA. 2005;102:17454–17459. doi: 10.1073/pnas.0506407102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pothier JF, Wisniewski-Dyé F, Weiss-Gayet M, Moënne-Loccoz Y, Prigent-Combaret C. Promoter-trap identification of wheat seed extract-induced genes in the plant-growth-promoting rhizobacterium Azospirillum brasilense Sp245. Microbiology. 2007;153:3608–3622. doi: 10.1099/mic.0.2007/009381-0. [DOI] [PubMed] [Google Scholar]
- 59.Spaepen S, Das F, Luyten E, Michiels J, Vanderleyden J. Indole-3-acetic acid-regulated genes in Rhizobium etli CNPAF512. FEMS Microbiol Lett. 2009;291:195–200. doi: 10.1111/j.1574-6968.2008.01453.x. [DOI] [PubMed] [Google Scholar]
- 60.Spaepen S, Vanderleyden J, Remans R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol Rev. 2007;31:425–448. doi: 10.1111/j.1574-6976.2007.00072.x. [DOI] [PubMed] [Google Scholar]
- 61.Yuan ZC, Haudecoeur E, Faure D, Kerr KF, Nester EW. Comparative transcriptome analysis of Agrobacterium tumefaciens in response to plant signal salicylic acid, indole-3-acetic acid and gamma-amino butyric acid reveals signalling cross-talk and Agrobacterium–plant co-evolution. Cell Microbiol. 2008;10:2339–2354. doi: 10.1111/j.1462-5822.2008.01215.x. [DOI] [PubMed] [Google Scholar]
- 62.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 63.Wuertz S, et al. In situ quantification of gene transfer in biofilms. In: Doyle RJ, editor. Microbial Growth in Biofilms, Biological Aspects, Methods in Enzymology. Vol 336. San Diego: Academic Press Inc.; 2001. pp. 129–143. [DOI] [PubMed] [Google Scholar]
- 64.Brazelton WJ, Baross JA. Abundant transposases encoded by the metagenome of a hydrothermal chimney biofilm. ISME J. 2009;3:1420–1424. doi: 10.1038/ismej.2009.79. [DOI] [PubMed] [Google Scholar]
- 65.Hooper SD, Mavromatis K, Kyrpides NC. Microbial co-habitation and lateral gene transfer: What transposases can tell us. Genome Biol. 2009;10(4):R45. doi: 10.1186/gb-2009-10-4-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Juhas M, Crook DW, Hood DW. Type IV secretion systems: Tools of bacterial horizontal gene transfer and virulence. Cell Microbiol. 2008;10:2377–2386. doi: 10.1111/j.1462-5822.2008.01187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sullivan JT, et al. Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J Bacteriol. 2002;184:3086–3095. doi: 10.1128/JB.184.11.3086-3095.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hubber A, Vergunst AC, Sullivan JT, Hooykaas PJ, Ronson CW. Symbiotic phenotypes and translocated effector proteins of the Mesorhizobium loti strain R7A VirB/D4 type IV secretion system. Mol Microbiol. 2004;54:561–574. doi: 10.1111/j.1365-2958.2004.04292.x. [DOI] [PubMed] [Google Scholar]
- 69.Nakanishi K, Nishijima M, Nomoto AM, Yamazaki A, Saga N. Requisite morphologic interaction for attachment between Ulva pertusa (Chlorophyta) and symbiotic bacteria. Mar Biotechnol (NY) 1999;1(1):107–111. doi: 10.1007/pl00011744. [DOI] [PubMed] [Google Scholar]
- 70.Liu ZY, Jacobs M, Schaff DA, McCullen CA, Binns AN. ChvD, a chromosomally encoded ATP-binding cassette transporter-homologous protein involved in regulation of virulence gene expression in Agrobacterium tumefaciens. J Bacteriol. 2001;183:3310–3317. doi: 10.1128/JB.183.11.3310-3317.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wenzel SC, Müller R. Formation of novel secondary metabolites by bacterial multimodular assembly lines: Deviations from textbook biosynthetic logic. Curr Opin Chem Biol. 2005;9:447–458. doi: 10.1016/j.cbpa.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 72.van Veen HW, Konings WN. The ABC family of multidrug transporters in microorganisms. Biochim Biophys Acta. 1998;1365(1-2):31–36. doi: 10.1016/s0005-2728(98)00039-5. [DOI] [PubMed] [Google Scholar]
- 73.Sale PF. Recruitment, loss and coexistence in a guild of territorial coral-reef fishes. Oecologia. 1979;42:159–177. doi: 10.1007/BF00344855. [DOI] [PubMed] [Google Scholar]
- 74.Laurie H, Mustart PJ, Cowling RM. A shared niche? The case of the species pair Protea obtusifolia Leucadendron meridianum. Oikos. 1997;79:127–136. [Google Scholar]
- 75.Lavorel S. Ecological diversity and resilience of Mediterranean vegetation to disturbance. Divers Distrib. 1999;5:3–13. [Google Scholar]
- 76.Janovy J, Jr, Clopton RE, Percival TJ. The roles of ecological and evolutionary influences in providing structure to parasite species assemblages. J Parasitol. 1992;78:630–640. [PubMed] [Google Scholar]
- 77.Lozupone C, Hamady M, Knight R. UniFrac—An online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kelley SE. Experimental studies of the evolutionary significance of sexual reproduction. 5. A field-test of the sib-competition lottery hypothesis. Evolution. 1989;43:1054–1065. doi: 10.1111/j.1558-5646.1989.tb02550.x. [DOI] [PubMed] [Google Scholar]
- 80.Dawkins R. The Selfish Gene. Oxford: Oxford University Press; 1976. [Google Scholar]
- 81.Doolittle WF, Zhaxybayeva O. On the origin of prokaryotic species. Genome Res. 2009;19:744–756. doi: 10.1101/gr.086645.108. [DOI] [PubMed] [Google Scholar]
- 82.Gevers D, et al. Opinion: Re-evaluating prokaryotic species. Nat Rev Microbiol. 2005;3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- 83.Konstantinidis KT, Ramette A, Tiedje JM. The bacterial species definition in the genomic era. Philos Trans R Soc Lond B Biol Sci. 2006;361:1929–1940. doi: 10.1098/rstb.2006.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Qin JJ, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587:4153–4158. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hamady M, Knight R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 2009;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Burke C, Kjelleberg S, Thomas T. Selective extraction of bacterial DNA from the surfaces of macroalgae. Appl Environ Microbiol. 2009;75:252–256. doi: 10.1128/AEM.01630-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shaw AK, et al. It’s all relative: Ranking the diversity of aquatic bacterial communities. Environ Microbiol. 2008;10:2200–2210. doi: 10.1111/j.1462-2920.2008.01626.x. [DOI] [PubMed] [Google Scholar]
- 89.Meyer F, et al. The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26(1):32–46. [Google Scholar]
- 91.Clarke KR. Non-parametric multivariate analyses of changes in community structure. Austral Ecol. 1993;18(1):117–143. [Google Scholar]
- 92.Clarke KR, Gorley RN. PRIMER v6: User Manual/Tutorial. 2006. (PRIMER-E, Plymouth) [Google Scholar]
- 93.Chenna R, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 95.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.