Abstract
Lipid droplets are ubiquitous cellular structures in eukaryotes and are required for lipid metabolism. Little is currently known about plant lipid droplets other than oil bodies. Here, we define dual roles for chloroplast lipid droplets (plastoglobules) in energy and prenylquinone metabolism. The prenylquinones—plastoquinone, plastochromanol-8, phylloquinone (vitamin K1), and tocopherol (vitamin E)—are partly stored in plastoglobules. This work shows that NAD(P)H dehydrogenase C1 (NDC1) (At5g08740), a type II NAD(P)H quinone oxidoreductase, associates with plastoglobules. NDC1 reduces a plastoquinone analog in vitro and affects the overall redox state of the total plastoquinone pool in vivo by reducing the plastoquinone reservoir of plastoglobules. Finally, NDC1 is required for normal plastochromanol-8 accumulation and is essential for vitamin K1 production.
Keywords: lipidomics, alternative electron flow, high light
Energy production in plants largely relies on oxygenic photosynthesis, implicating linear electron flow from water to NAD(P)H catalyzed by photosystems (PS) I and II. For optimal energy production, however, plants and algae have additional pathways, such as chlororespiration and cyclic electron transport around PSI, allowing the electrons to be recycled into the intersystem electron transport chain (1–4) (Fig. S1). In angiosperms, those alternative electron flows are mediated in part by the proton gradient regulation 5 (PGR5)/PGR5-like 1 (PGRL1) pathway and in part by a multisubunit NAD(P)H:plastoquinone oxidoreductase (NDH) complex with similarity to bacterial and mitochondrial Complex I (1, 5). Both the PGR5/PGRL1 pathway and the NDH complex have the ability to reduce plastoquinone (PQ) by using ferredoxin as the electron donor (6). A similar activity exists in the green alga Chlamydomonas reinhardtii, but this organism does not encode the subunits of the NDH complex (7–9). Instead, a monomeric type II NAD(P)H:quinone oxidoreductase, NDA2, takes over the function of the NDH complex in cyclic electron flow and chlororespiration (10, 11). In Arabidopsis thaliana, seven such type II NAD(P)H dehydrogenase homologs are encoded by three subfamilies (12): two NDAs, four NDBs, and one NDC [termed NAD(P)H dehydrogenase C1 (NDC1)]. Their role, if any, in chloroplast electron pathways is not known. Interestingly, only NDC1 appears to be of cyanobacterial origin (12). A study using GFP-fusion proteins initially localized all seven homologs in mitochondria (12). Later, NDC1 was imported into isolated mitochondria and also chloroplasts, suggesting a dual localization (13, 14). Furthermore, NDC1 was identified in proteomics studies of chloroplast lipid droplets (plastoglobules) (15, 16). The plastoglobules are a site of both prenylquinone metabolism and storage. Recently, a large reservoir of PQ, probably not immediately involved in linear electron transport, was discovered in plastoglobules (17–19). Tocopherol (vitamin E), phylloquinone (vitamin K1), and plastochromanol-8 (PC-8), a chromanol derivative of PQ, are also present in plastoglobules (15, 18–20) (Fig. S2).
Here, we demonstrate that NDC1 functions in a unique electron-flow pathway to the plastoglobule PQ reservoir and thereby determines the overall PQ redox state. Unexpectedly, NDC1 also plays a key role in plastochromanol accumulation and promotes the production of phylloquinone beyond its nonmethylated 2-phytyl-1,4-naphthoquinone precursor.
Results
Chloroplast Lipid Droplet Localization of NDC1.
Earlier studies suggested either a mitochondrial or dual mitochondrial/chloroplast localization for NDC1 (12–14). Two independent proteomics studies identified NDC1 as a chloroplast lipid droplet (plastoglobule) protein (15, 16). Sucrose-gradient flotation of total Arabidopsis chloroplast membranes followed by Western blotting demonstrated cofractionation of NDC1 with the plastoglobule markers PGL35 and PGL40 in the low-density fractions as well as the clear separation from the envelope marker Toc75 and the thylakoid marker LHCB2 (Fig. 1A). Transient expression of NDC1-YFP in Nicotiana benthamiana protoplasts resulted in punctate fluorescence mostly inside the chloroplasts (Fig. 1B). The punctate fluorescence, in most cases, colocalized with that of the neutral lipid dye Nile Red (Fig. 1D). These results are consistent with the lipid droplet/plastoglobule localization of NDC1 indicated by the earlier proteomic studies and the fractionation experiment (Fig. 1A). Nevertheless, both isolated chloroplasts (Fig. 1F) and mitochondria (Fig. 1G) imported synthetic, full-length [35S]-labeled pre-NDC1 in vitro, resulting in protease-protected, mature forms in both cases.
Fig. 1.
NDC1 lipid droplet localization. (A) Western blotting of chloroplast membrane fractions separated by sucrose-gradient flotation. Toc75 and LHCB2 are envelope and thylakoid markers, respectively. PGL35 and PGL40 are plastoglobule markers. CP, total chloroplast; St, stroma. (B) NDC1-YFP under the control of the cauliflower mosaic virus 35S promoter was transiently expressed in tobacco protoplasts. Transformed protoplasts were analyzed by confocal laser microscopy. (C) The neutral lipid dye Nile Red reveals lipid droplets in chloroplasts (plastoglobules). (D) The first merged image shows the superposition of NDC1-YFP and Nile Red in white. (E) The second merged image shows the chlorophyll autofluorescence to visualize chloroplasts. (F) 35S-labeled pre-NDC1 (lane 1) was incubated with isolated chloroplasts in vitro in a time-course experiment (0, 2, 5, and 10 min, lanes 2–5). The imported, lower molecular mass NDC1 was resistant to exogenously added thermolysin protease (lane 6). (G) 35S-labeled pre-NDC1 was incubated with isolated mitochondria in vitro (lane 7). The experiment was analyzed at the 0- and 30-min time points (lanes 8 and 10). The imported, lower molecular mass NDC1 was resistant to exogenously added proteinase K (lanes 9 and 11).
Photosynthetic Parameters of the ndc1 Mutant.
We isolated two homozygous transfer-DNA insertion lines for the NDC1 gene: Salk_024063 and GABI_614F03, named ndc1-1 and ndc1-3, respectively. Immunoblotting with specific antibodies indicated the absence of the NDC1 protein and confirmed the homozygous knockout nature of the two mutants (Fig. S3A). Neither of the lines had a visible phenotype (Fig. S3B).
We measured several chlorophyll fluorescence parameters in WT and ndc1-1 leaves (hereafter called ndc1) and also, for comparison purposes, in leaves of the ndho mutant (hereafter called ndh1) lacking the NDH complex (21): the maximal quantum yield of PSII photochemistry (Fv/Fm) was determined in the dark, whereas the actual PSII quantum yield (ΔF/Fm′) and the nonphotochemical quenching of chlorophyll fluorescence were measured at different photon flux densities (PFDs) (Fig. S4 A and B). None of those parameters differed between WT and the ndc1 mutant. We also analyzed alternative electron flows, which donate electrons from the stroma to the PQ pool, by using measurements of chlorophyll fluorescence (Fig. S4C), P700 redox state (Fig. S4D), and chlorophyll thermoluminescence (Fig. 2). The postillumination rise in chlorophyll fluorescence, which is indicative of PQ and primary quinone acceptor (QA) reduction specifically mediated by the NDH complex (21), remained unchanged in ndc1 compared with WT (Fig. S4C). We also measured the postillumination reduction of oxidized P700, the PSI reaction center pigment (22). Although a slight increase in the half life (t½) of P700 reduction was observed in the ndh1 mutant, indicating a slowdown of the electron donation to P700, the ndc1 mutation had no effect on this parameter (Fig. S4D). Fig. 2A shows the afterglow thermoluminescence band, induced by far-red preillumination, that corresponds to a heat-induced back electron transfer to the PQ pool in the dark (23). In Arabidopsis, this pathway has been shown to reflect mainly the NDH activity (24, 25), as confirmed here by the strong reduction of this band in ndh1. Again, no difference was found between WT and ndc1. From those results, we can conclude that NDC1 and NDH do not fulfill a similar function, and therefore NDC1 is probably not directly involved in cyclic or chlororespiratory electron flows (Fig. S1) under standard growth conditions.
Fig. 2.
Thermoluminescence measurements in WT, ndc1, and ndh1 mutant leaves of Arabidopsis. (A) Far-red light-induced thermoluminescence signal in WT Arabidopsis leaves and in mutant leaves (ndc1, ndh1) grown under standard conditions. The band peaking at temperature >35 °C is the so-called afterglow band. The shoulder observed at lower temperature (∼20 °C) is a B band corresponding to the S2/3 QB− charge recombination. The data are representative traces of at least six separate experiments. (B) Immunodetection of NDC1 in total extracts of WT and two ndc1 mutant lines (ndc1-1 and ndc1-3) under normal and high-light growth conditions. The upper band, weakly visible in the WT under low-light conditions and more strongly under high-light conditions, corresponds to the NDC1 protein. The band is absent from the ndc1-1 and ndc1-3 mutants. The lower band is a nonspecific, cross-reacting signal. (C) Effects of DPI (15 and 30 μM) on the afterglow thermoluminescence band of WT and ndh1 mutant leaves acclimated for 7 d to high light. Data are representative traces of three separate experiments; (D) Afterglow thermoluminescence band measured in leaves of the ndh1 mutant and the ndc1 ndh1 mutant acclimated for 7 d to high light. Data are representative traces of at least six separate experiments.
We also analyzed plants previously exposed to high-light conditions that induce accumulation of plastoglobules (26) and increase expression of the NDC1 gene in the WT (Fig. 2B) and ndh1 mutant (Fig. S5) (27). In WT, high light shifted the afterglow band toward lower temperatures compared with low light (Fig. 2 A and C). This band shift is indicative of an activation of the cyclic electron pathway: electrons are rapidly transferred to the secondary quinone acceptor (QB) at room temperature, leading to charge recombination between QB− and the S states and to the merging of the afterglow band with the B band (28, 29). This activation was not observed in ndh1 mutant leaves. Rather surprisingly, high light induced the appearance of a unique band in this mutant, peaking at ∼42 °C (Fig. 2 C and D). This band was insensitive to antimycin A (Fig. S6) but showed a high sensitivity to diphenyleneiodonium (DPI), a known inhibitor of type II NAD(P)H:quinone oxidoreductases (7) (Fig. 2C). Moreover, its amplitude was strongly reduced in the ndc1 ndh1 double mutant (Fig. 2D), indicating the participation of NDC1 in this nonphotochemical electron flow.
In Vitro Activity of NDC1.
NDC1 is a predicted NADH/NADPH:quinone reductase, and the thermoluminescence experiments supported such a role in vivo. For in vitro experimentation, purified recombinant NDC1 protein was incubated with NADPH as the electron donor and decyl-PQ (Fig. 3A). The decyl derivative is more soluble because of the replacement of the highly hydrophobic isoprenoid chain. We tracked the reactions by recording the oxidation of NAD(P)H at 340 nm. For decyl-PQ, we measured Km = 9 μM and Vmax = 46 μmol/mg per min (Fig. 3A). DPI inhibited decyl-PQ reduction in the presence of NAD(P)H with a half-maximal inhibition at ∼2.5 μM (Fig. 3B). Isolated plastoglobules (Fig. 3C) and decyl-ubiquinone (with NADH as the electron donor) (Fig. 3D) also functioned as NDC1 substrates in vitro.
Fig. 3.
NDC1 enzyme activity and prenylquinone quantification in the ndc1 and ndh1 mutants. (A) NDC1 activity was measured by monitoring NADPH oxidation in the presence of increasing concentrations of decyl-PQ (a PQ analog). (B) Increasing concentrations of the inhibitor DPI were added to the assay in the presence of 100 μM decyl-PQ. The kinetic parameters were calculated with SigmaPlot software. (A and B) Each data point represents two (B) and three (A) experimental replicates. (C and D) NDC1 activity was measured in the presence of isolated plastoglobules (C) and in the presence of decyl-ubiquinone and NADH (D). The arrows indicate the time point of addition of purified NDC1. (E–H) Prenylquinones in leaves determined by HPLC. (E) Total PQ. The white bar indicates the fraction of oxidized PQ. (F) Percentage reduction of the PQ pool. (G) PC-8. (H) α-Tocopherol in WT Arabidopsis leaves and ndc1 or ndh1 mutant leaves grown under standard conditions. Data are mean values of at least three separate experiments.
To determine how NDC1 affects prenyllipid levels in leaf extracts, we targeted PQ (Fig. 3 E and F), its derivative PC-8 (Fig. 3G), and α-tocopherol (Fig. 3H) for quantification by using HPLC coupled with UV and fluorescence detection systems. Neither tocopherol nor PQ were diminished in ndc1 relative to WT. Interestingly, the PQ pool was substantially more oxidized in ndc1 (50% oxidation) compared with WT (30% oxidation) (Fig. 3 E and F). Another striking result was the strong decrease of PC-8 in ndc1 leaves (Fig. 3G). In contrast, the ndh1 mutant did not show any significant change in the PQ redox state or in the PC-8 concentration.
Untargeted Lipidomic Analysis of the ndc1 Mutant.
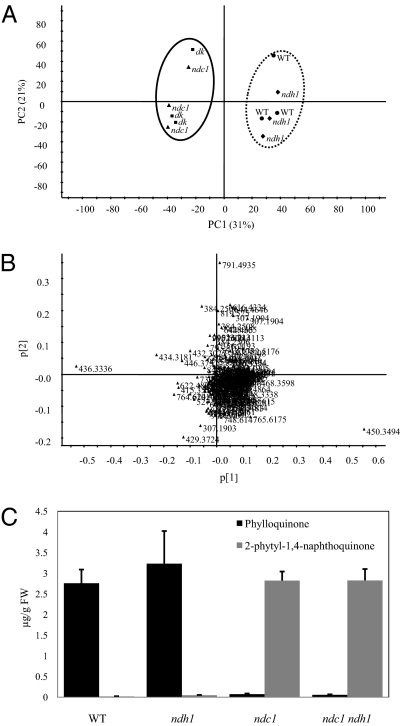
To determine the more general role of NDC1 in chloroplast metabolism, we carried out untargeted lipidomics analyses (Fig. 4). Total lipid extracts of WT, ndc1 (both ndc1-1 and ndc1-3), ndh1 single mutants, and the ndc1 ndh1 double mutant were prepared and injected into an ultra HPLC–quadrupole time-of-flight mass spectrometer (UHPLC-QTOFMS). The data obtained were subjected to multivariate analysis to determine differences in lipid content between samples (Fig. 4A). Using principal component analysis, we observed two distinct groups (Fig. 4A): one containing WT and the ndh1 mutant and the other containing the ndc1 and ndc1 ndh1 mutants. Principal component analysis loadings were then investigated and revealed that two variables were mostly contributive to the discrimination between both groups: m/z 450.3494 at 4.99 min for WT and ndh1 and m/z 436.3336 at 4.64 min for ndc1 and ndc1 ndh1 (Fig. 4B and Table S1). The ion at m/z 450.3494 was associated to the molecular formula C31H46O2 (error = 0.9 ppm). A high-resolution MS/MS experiment gave a main fragment at m/z 185.0604, corresponding to the raw formula C12H9O2 (error = 0.5 ppm) and a smaller fragment at m/z 225.0910 (C15H13O2, error 2.7 = ppm). These molecular and fragment ions were typical of phylloquinone, a finding that was further confirmed by the injection of a standard compound that yielded identical mass spectra and eluted at identical retention times (Fig. S7). The ion at m/z 436.3336 corresponded to the molecular formula C30H44O2 (error = 1.1 ppm) and could be caused by the demethylated form of phylloquinone (2-phytyl-1,4-naphthoquinone). The loss of a methyl group was confirmed by MS/MS, which generated a main fragment at m/z 171.0446 (C11H7O2, error = 0.0 ppm) together with a smaller fragment at m/z 211.0760 (C14H11O2, error = 0.5 ppm). No standard compound was available for this molecule, and the ion at m/z 436.3336 was thus tentatively annotated as 2-phytyl-1,4-naphthoquinone. The latter was further confirmed by using the AtmenG mutant known to accumulate 2-phytyl-1,4-naphthoquinone instead of phylloquinone (20): as in ndc1, the peak at m/z 436.3336 was present, whereas phylloquinone was almost absent (Fig. S7). Phylloquinone levels in the whole plant were statistically similar in WT and ndh1 mutant with 2.76 and 3.23 μg/g of fresh weight (FW), respectively, whereas 2-phytyl-1,4-naphthoquinone was almost completely absent from both lines (0.07 and 0.06 μg/g of FW in WT and ndh1, respectively). Conversely, in ndc1 and ndc1 ndh1 mutants, concentrations of phylloquinone were very low (0.02 and 0.05 μg/g of FW, respectively) and those of 2-phytyl-1,4-naphthoquinone were much higher, with 2.82 and 2.83 μg/g of FW, respectively. Interestingly, the levels of 2-phytyl-1,4-naphthoquinone in ndc1 and ndc1 ndh1 mutants were similar to those of phylloquinone in WT and ndh1 mutant (Fig. 4C).
Fig. 4.
Untargeted lipidomics of ndc1 mutants. (A) Methanolic extracts from leaves of ndc1, ndh1, and ndc1 ndh1 (dk) mutants lines were analyzed with UHPLC-QTOFMS. (A) Principal component analysis score plots (PC1 × PC2) with their percentages of explained variance, based on normalized data from negative ion UHPLC-QTOFMS analyses. (B) Corresponding loading plot showing the most relevant variables responsible for the separation found in the score plot (436.3336 on far left corresponds to 2-phytyl-1,4-naphthoquinone; 450.3494 on far right corresponds to phylloquinone). (C) Quantification of phylloquinone and 2-phytyl-1,4-naphthoquinone in WT (Col0) and ndc1, ndh1, and ndc1 ndh1 mutant plants obtained from a standard solution of phylloquinone. Data represent the mean of two independent experiments (n = 3 each).
AtMENG Expression in the ndc1 Mutant.
The absence of phylloquinone and its apparent replacement by the demethyl form in the ndc1 mutant lines is surprising. It is known that AtMENG is required for the final methylation step of phylloquinone biosynthesis (20). We used RT-PCR to determine whether AtMENG is still expressed in the ndc1 and ndc1 ndh1 mutants (Fig. S8). We detected no significant differences in the AtMENG expression levels between the WT and the ndc1, ndh1, and ndc1 ndh1 mutants, indicating that AtMENG is normally expressed in the mutant background and strongly suggesting that the active enzyme is also present.
Discussion
This study supports the function of NDC1 in chloroplasts. In chloroplast fractionation experiments, NDC1 was almost exclusively detected in low-density plastoglobule-containing fractions (Fig. 1A). Moreover, NDC1-YFP fluorescence colocalized with Nile Red staining (mostly in chloroplasts), emphasizing its association with lipid droplets (Fig. 1D). Nevertheless, this study also confirms in vitro import into both chloroplasts and mitochondria (Fig. 1 F and G) and does not exclude the dual localization of NDC1. A possible explanation for these apparently disparate findings is that NDC1 accumulates predominantly in chloroplasts and only at lower levels in mitochondria. A potential function in mitochondria was supported by the ability of purified, recombinant NDC1 to use decyl-ubiquinone together with NADH as a substrate (Fig. 3D). Overall, our observations favor the function of NDC1 in chloroplasts: although no effect on linear or cyclic electron transfer in chloroplasts was observed (Fig. S4), a unique afterglow thermoluminescence band (indicative of electron flow to PQ) appeared after highlight treatment of the ndh1 mutant (Fig. 2). Its characteristics (insensitivity to antimycin A, sensitivity to DPI, and disappearance in the ndc1 ndh1 double mutant) indicate that NDC1 functions in the chloroplast in a unique pathway of nonphotochemical PQ reduction parallel to cyclic and chlororespiratory electron flow (Fig. S1). Likely, this pathway corresponds to NDC1-catalyzed electron flow toward the PQ contained in the plastoglobules (17, 18) (Fig. S1). In support of this hypothesis, purified plastoglobules functioned as a quinone-containing substrate and accepted electrons from NADPH and the recombinant NDC1 enzyme in vitro (Fig. 3C). In agreement with this data, the PQ pool was significantly more oxidized in the ndc1 mutant than in the WT (Fig. 3 E and F).
Decyl-PQ functioned as an in vitro substrate of recombinant NDC1 using NADPH as the electron donor (Fig. 3A). The ability of NDC1 to also use purified plastoglobules as a substrate was probably attributable to the PQ contained inside plastoglobules. This reaction may therefore reflect an in vivo scenario in which NDC1 binds to the plastoglobule surface thereby gaining access to and reducing the PQ substrate inside and resulting in a higher overall reduction of the PQ pool in leaves.
This interpretation is in good agreement with the results of Kruk and coworkers (18, 30) who reported a large PQ reservoir that is not immediately implicated in photosynthesis and may be located in plastoglobules. Moreover, they proposed that this PQ reservoir plays a role as a lipid antioxidant in the thylakoid membrane. In such a scenario, NDC1 may function in the regeneration of reduced PQ upon oxidation, which would require the diffusion of PQ between plastoglobules and the thylakoid membrane, the compartment where PQ would likely function as an antioxidant in addition to its role as an electron carrier in photosynthesis. The diffusion of lipid compounds between plastoglobules and the thylakoid membranes has been proposed earlier, and physical connections between the outer lipid leaflet of the thylakoid membrane and the plastoglobule polar lipid monolayer were observed (17, 18, 26, 31).
Using HPLC and UHPLC-QTOFMS, we observed that the PC-8 concentration was strongly diminished in ndc1 mutants (Fig. 3G). PC-8 is derived from PQ by the activity of VTE1, the tocopherol cyclase, present in the plastoglobule (18, 19, 32). For the cyclase to close the chromanol ring in dimethylphytylquinone (the precursor of γ-tocopherol) or in PQ, the quinol groups must preferentially be present in the reduced form (33). Thus, the reduction of PC-8 levels may reflect the redox state of the plastoglobule PQ reservoir, which is more oxidized than in WT. Interestingly, α-tocopherol was present at WT levels in the ndc1 mutant, indicating that NDC1 is not required for its accumulation.
The untargeted lipidomics experiments led to unexpected discoveries (Fig. 4). The most prominent differences between WT and the ndc1 mutants via UHPLC-MS were the almost complete absence of phylloquinone and the appearance of an additional peak corresponding in mass to the 2-phytyl-1,4-naphthoquinone, precursor of phylloquinone, in ndc1 mutants. It is known that the complete absence of phylloquinone in Arabidopsis causes a lethal albino phenotype (34). However, in the AtmenG mutant (like in ndc1), its immediate precursor, 2-phytyl-1,4-naphthoquinone, still accumulates (20). Surprisingly, the AtmenG mutant has no visible phenotype, and it is therefore plausible that the 2-phytyl-1,4-naphthoquinone precursor functionally replaces phylloquinone. It is unclear why phylloquinone does not accumulate in the ndc1 mutant. In RT-PCR experiments, AtMENG was expressed at the same level in both ndc1 mutants and WT, suggesting that the enzyme is still present. However, AtMENG activity may be down-regulated at the enzyme level, for instance, by the PQ redox state of the plastoglobules. Another possibility would be that, in the absence of NDC1, AtMENG is no longer correctly recruited to the site of its activity, which may implicate plastoglobules.
There are some indications that plastoglobules play a role in phylloquinone metabolism. A fraction of the total phylloquinone pool is present in the plastoglobules. In the AtmenG mutant, the 2-phytyl-1,4-naphthoquinone precursor of phylloquinone accumulated to a large extent in plastoglobules (20). Both the multiactivity protein PHYLLO and AtMENG gave punctate fluorescence when transiently expressed as GFP-fusion proteins in Arabidopsis protoplasts, suggesting a plastoglobule localization (20, 35).
In conclusion, NDC1 represents a unique electron input device for PQ. Unlike C. reinhardtii NDA2 (10, 11), it does not seem to be directly implicated in cyclic/chlororespiratory electron flows in thylakoids, but rather it plays a role in electron transfer to the plastoglobule PQ pool—a role that appears to be essential for prenylquinone metabolism.
Materials and Methods
Plant Materials and Growth Conditions.
Arabidopsis plants were grown on soil (Ricoter) or in vitro on 0.8% phyto agar (Duchefa) containing 0.5× Murashige and Skoog medium (Duchefa) under short-day conditions (8 h light, 16 h dark, 150 μmol⋅m−2⋅s−1). For high-light experiments, plants aged 4 wk were acclimated for 7 d by increasing PFD to 750 μmol⋅m−2⋅s−1. Pea (Pisum sativum) seeds were germinated on soil after one night in water and grown in a chamber for 2 wk under a long-day conditions (16 h light, 8 h dark). Nicotiana tabacum cv. petit Havana plants used for protoplast isolation were grown for 4–5 wk on 0.5× Murashige and Skoog medium under long-day conditions.
Chlorophyll Fluorescence and Thermoluminescence.
Chlorophyll fluorescence emission from attached leaves was measured with a PAM-2000 fluorometer (Walz) as previously described (21, 36). Thermoluminescence measurements were performed on leaf discs (1 cm in diameter) with a custom-made apparatus that has been described previously (24). The sample was cooled at 1 °C for 40 s, then it was illuminated for 30 s with far-red light (>715 nm, 3 W⋅m−2). Immediately after interrupting far-red illumination, temperature was increased from 1 °C to 70 °C at a rate of 0.5 °C⋅s−1, and luminescence was measured during heating with a photomultiplier tube.
Prenyllipid Quantification.
Tocopherols, PQ (reduced and oxidized), and PC-8 were extracted, separated, and quantified by HPLC using the method described in refs. 18 and 30. The HPLC column was a Phenomenex C18 reverse-phase column (Kinetex, 2.6 μm, 100 × 4.6 mm).
P700 Redox Changes.
The redox state of P700, the reaction center of PSI, was measured in attached leaves using a dual-wavelength emitter/detector ED-P700DW-E (Walz) connected to a PAM-101 unit, as previously described (8). P700 was oxidized by illuminating the leaf with far-red light (>715 nm, 25 W⋅m−2), and the rate of P700 reduction was measured in the dark after switching off the far-red light.
Chloroplast Fractionation and Western Blot Analysis.
Chloroplast fractionation was carried out as described in ref. 14. Western blot analysis was carried out as described in ref. 14. Anti-NDC1 serum was used at 1/1,000 dilution on 5% milk/TBS.
In Vivo Targeting.
Transient transformation of protoplasts was done by using the polyethylene glycol method (37, 38) with reduced cellulase (1%) and macerozyme (0.25%; Serva). Then, 20 μg of pEarlyGate101-NDC1-YFP was used to transform 500,000 tobacco protoplasts. Fluorescence was monitored with a Leica TCS SP5 confocal microscope at 48–80 h after transformation. Nile Red was used at 10 μg/mL (38).
In Vitro Import into Isolated Mitochondria and Chloroplasts.
The radioactive full-length NDC1 protein was produced in a coupled transcription–translation system (TNT T7 Quick Coupled Transcription/Translation System; Promega) using pET21-NDC1-H6 vector according to the manufacturer's instructions. Import of [35S]-labeled fusion protein into purified pea mitochondria was performed according to ref. 39. Import into purified Arabidopsis chloroplasts was performed according to ref. 40. All experiments were resolved by SDS/PAGE and analyzed by PhosphorImaging.
Production of Anti-AtNDC1 Antibody.
Full-length recombinant NDC1-H6 protein was purified under denaturing conditions by nickel nitrilotriacetic acid affinity chromatography (Qiagen) according to the manufacturer's recommendations. Polyclonal antibodies were produced in rabbit (Eurogentec) and were affinity-purified against NDC1-H6 coupled to Affi-Gel 10 (Bio-Rad) according to the supplier's recommendations.
NDC1 Enzyme Assay.
Activity was measured in 1 mL of 50 mM Hepes buffer [+KOH (pH 7.2)] containing 200 μM NADH and decyl-prenylquinone concentrations from 0.5 and 750 μM. The reaction was started by the addition of 18 μg of nickel nitrilotriacetic acid affinity-purified mature NDC1-H6, and NADPH absorbance decay was measured at 340 nm.
Extraction and LC/MS Analysis.
Plants (6-wk-old, n = 6) were ground to a fine powder under liquid nitrogen. Samples (100 mg) were extracted in 500 μL of MeOH for 2 min in a bead mill (MM 200; Retsch) and centrifuged (4 min at 14,000 × g), and the supernatant was recovered. The untargeted analysis of lipids was carried out with an Acquity UPLC system from Waters coupled to a Synapt G2 MS QTOF from Waters equipped with an atmospheric pressure chemical ionization source. The separation was performed at 60 °C on an Acquity BEH C18 column (50 × 2.1 mm, 1.7 μm) at a flow rate of 500 μL/min under the following conditions (A, water; B, MeOH): 80–100% B for 6 min, holding at 100% B for 3 min, followed by reequilibration at 80% for 2 min. The QTOF was operated at a resolution of 20,000 FWHM in MS positive and negative ion modes over an m/z range of 225–1,200 in centroid mode. Scan time was 0.5 s. The corona current was set to 12 μA, and the cone voltage was set to 40 V. The source temperature was maintained at 120 °C, the atmospheric pressure chemical ionization probe was at 370 °C, and desolvation and cone gas flows were set to 800 L/hr and 20 L/hr, respectively. Accurate mass measurements (< 2 ppm) were obtained by infusing a solution of leucin-enkephalin at 400 ng/mL at a flow rate of 10 μL/min through the Lock Spray probe (internal calibration). MS/MS product ion spectra were obtained for phylloquinone and its demethylated form by using precursor ions at m/z 450.3 and 436.3, respectively. The collision energy was set to 25 eV, and argon was used as collision gas at a flow rate of 2.1 mL/min.
Absolute quantification of phylloquinone and 2-phytyl-1,4-naphthoquinone was performed with calibration curves obtained from standard solutions of phylloquinone at 100 ng/mL, 250 ng/mL, 1,000 ng/mL, and 2,500 ng/mL
Data Preprocessing and Multivariate Analysis.
Peak picking and data processing including multivariate analysis were performed with MarkerLynx XS software (Waters), enabling the generation of a list of variables characterized by their m/z, retention time, and intensity. Peak areas of the extracted variables were normalized by dividing them by the sum of the intensities of all detected variables in each sample. Normalized variables were then Pareto-scaled and analyzed by principal component analysis.
Real-Time RT-PCR.
Total RNA was extracted from Col0 control plants and mutant lines with the RNeasy Mini Kit (Qiagen). Then, 1 μg of total DNase-treated RNA was reverse-transcribed with the M-MLV reverse transcriptase (Promega). Real-time PCR was performed with SYBR Green (Thermo Scientific) on an iCycler with ACT2 as a reference. Specific primers to respective genes were as follows: ACT2 forward, 5′-TGGAATCCACGAGACAACCTA-3′ and reverse, 5′-TTCTGTGAACG ATTCCTGGAC-3′); AtMENG forward, 5′- ATTCGATGCGGTTACGATGG-3′ and reverse, 5′- ACTCCTTTGCAAGATCATAAAC-3′; and NDC1 forward, 5′-AGCTTGATTGGTGAAATGCC-3′ and reverse, 5′-CTGCGGTTATGCAGGAGTAG-3′.
Supplementary Material
Acknowledgments
We thank Dr. Kruk for the kind gift of purified PQ standards. F.E.K. thanks the Université de Neuchâtel, SystemsX Plant Growth in a Changing Environment, and National Center of Competence in Research Plant Survival and acknowledges support from Swiss National Science Foundation Grant 31003A_127380.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104790108/-/DCSupplemental.
References
- 1.Shikanai T. Cyclic electron transport around photosystem I: Genetic approaches. Annu Rev Plant Biol. 2007;58:199–217. doi: 10.1146/annurev.arplant.58.091406.110525. [DOI] [PubMed] [Google Scholar]
- 2.Rumeau D, Peltier G, Cournac L. Chlororespiration and cyclic electron flow around PSI during photosynthesis and plant stress response. Plant Cell Environ. 2007;30:1041–1051. doi: 10.1111/j.1365-3040.2007.01675.x. [DOI] [PubMed] [Google Scholar]
- 3.Johnson GN. Physiology of PSI cyclic electron transport in higher plants. Biochim Biophys Acta. 2011;1807:384–389. doi: 10.1016/j.bbabio.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 4.Cardol P, Forti G, Finazzi G. Regulation of electron transport in microalgae. Biochim Biophys Acta. 2011;1807:912–918. doi: 10.1016/j.bbabio.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Munekage Y, et al. PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell. 2002;110:361–371. doi: 10.1016/s0092-8674(02)00867-x. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto H, Peng L, Fukao Y, Shikanai T. An Src homology 3 domain-like fold protein forms a ferredoxin binding site for the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Cell. 2011;23:1480–1493. doi: 10.1105/tpc.110.080291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mus F, Cournac L, Cardettini V, Caruana A, Peltier G. Inhibitor studies on non-photochemical plastoquinone reduction and H2 photoproduction in Chlamydomonas reinhardtii. Biochim Biophys Acta. 2005;1708:322–332. doi: 10.1016/j.bbabio.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Ravenel J, Peltier G, Havaux M. The cyclic electron pathways around photosystem I in Chlamydomonas reinhardtii as determined in vivo by photoacoustic measurements of energy storage. Planta. 1994;193:251–259. [Google Scholar]
- 9.Peltier G, Cournac L. Chlororespiration. Annu Rev Plant Biol. 2002;53:523–550. doi: 10.1146/annurev.arplant.53.100301.135242. [DOI] [PubMed] [Google Scholar]
- 10.Jans F, et al. A type II NAD(P)H dehydrogenase mediates light-independent plastoquinone reduction in the chloroplast of Chlamydomonas. Proc Natl Acad Sci USA. 2008;105:20546–20551. doi: 10.1073/pnas.0806896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desplats C, et al. Characterization of Nda2, a plastoquinone-reducing type II NAD(P)H dehydrogenase in Chlamydomonas chloroplasts. J Biol Chem. 2009;284:4148–4157. doi: 10.1074/jbc.M804546200. [DOI] [PubMed] [Google Scholar]
- 12.Michalecka AM, et al. Arabidopsis genes encoding mitochondrial type II NAD(P)H dehydrogenases have different evolutionary origin and show distinct responses to light. Plant Physiol. 2003;133:642–652. doi: 10.1104/pp.103.024208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elhafez D, et al. Characterization of mitochondrial alternative NAD(P)H dehydrogenases in Arabidopsis: Intraorganelle location and expression. Plant Cell Physiol. 2006;47:43–54. doi: 10.1093/pcp/pci221. [DOI] [PubMed] [Google Scholar]
- 14.Carrie C, et al. Type II NAD(P)H dehydrogenases are targeted to mitochondria and chloroplasts or peroxisomes in Arabidopsis thaliana. FEBS Lett. 2008;582:3073–3079. doi: 10.1016/j.febslet.2008.07.061. [DOI] [PubMed] [Google Scholar]
- 15.Vidi PA, et al. Tocopherol cyclase (VTE1) localization and vitamin E accumulation in chloroplast plastoglobule lipoprotein particles. J Biol Chem. 2006;281:11225–11234. doi: 10.1074/jbc.M511939200. [DOI] [PubMed] [Google Scholar]
- 16.Ytterberg AJ, Peltier JB, van Wijk KJ. Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol. 2006;140:984–997. doi: 10.1104/pp.105.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bréhélin C, Kessler F. The plastoglobule: A bag full of lipid biochemistry tricks. Photochem Photobiol. 2008;84:1388–1394. doi: 10.1111/j.1751-1097.2008.00459.x. [DOI] [PubMed] [Google Scholar]
- 18.Szymańska R, Kruk J. Plastoquinol is the main prenyllipid synthesized during acclimation to high light conditions in Arabidopsis and is converted to plastochromanol by tocopherol cyclase. Plant Cell Physiol. 2010;51:537–545. doi: 10.1093/pcp/pcq017. [DOI] [PubMed] [Google Scholar]
- 19.Zbierzak AM, et al. Intersection of the tocopherol and plastoquinol metabolic pathways at the plastoglobule. Biochem J. 2010;425:389–399. doi: 10.1042/BJ20090704. [DOI] [PubMed] [Google Scholar]
- 20.Lohmann A, et al. Deficiency in phylloquinone (vitamin K1) methylation affects prenyl quinone distribution, photosystem I abundance, and anthocyanin accumulation in the Arabidopsis AtmenG mutant. J Biol Chem. 2006;281:40461–40472. doi: 10.1074/jbc.M609412200. [DOI] [PubMed] [Google Scholar]
- 21.Rumeau D, et al. New subunits NDH-M, -N, and -O, encoded by nuclear genes, are essential for plastid Ndh complex functioning in higher plants. Plant Cell. 2005;17:219–232. doi: 10.1105/tpc.104.028282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maxwell PC, Biggins J. Role of cyclic electron transport in photosynthesis as measured by the photoinduced turnover of P700 in vivo. Biochemistry. 1976;15:3975–3981. doi: 10.1021/bi00663a011. [DOI] [PubMed] [Google Scholar]
- 23.Miranda T, Ducruet JM. Characterization of the chlorophyll thermoluminescence afterglow in dark-adapted or far-red-illuminated plant leaves. Plant Physiol Biochem. 1995;33:689–699. [Google Scholar]
- 24.Havaux M, Rumeau D, Ducruet JM. Probing the FQR and NDH activities involved in cyclic electron transport around Photosystem I by the ‘afterglow’ luminescence. Biochim Biophys Acta. 2005;1709:203–213. doi: 10.1016/j.bbabio.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 25.Lintala M, et al. Comparative analysis of leaf-type ferredoxin-NADP oxidoreductase isoforms in Arabidopsis thaliana. Plant J. 2009;57:1103–1115. doi: 10.1111/j.1365-313X.2008.03753.x. [DOI] [PubMed] [Google Scholar]
- 26.Bréhélin C, Kessler F, van Wijk KJ. Plastoglobules: Versatile lipoprotein particles in plastids. Trends Plant Sci. 2007;12:260–266. doi: 10.1016/j.tplants.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida K, Noguchi K. Differential gene expression profiles of the mitochondrial respiratory components in illuminated Arabidopsis leaves. Plant Cell Physiol. 2009;50:1449–1462. doi: 10.1093/pcp/pcp090. [DOI] [PubMed] [Google Scholar]
- 28.Ducruet JM, Roman M, Havaux M, Janda T, Gallais A. Cyclic electron flow around PSI monitored by afterglow luminescence in leaves of maize inbred lines (Zea mays L.): Correlation with chilling tolerance. Planta. 2005;221:567–579. doi: 10.1007/s00425-004-1464-6. [DOI] [PubMed] [Google Scholar]
- 29.Apostol S, Szalai G, Sujbert L, Popova LP, Janda T. Non-invasive monitoring of the light-induced cyclic photosynthetic electron flow during cold hardening in wheat leaves. Z Naturforsch C. 2006;61:734–740. doi: 10.1515/znc-2006-9-1021. [DOI] [PubMed] [Google Scholar]
- 30.Kruk J, Karpinski S. An HPLC-based method of estimation of the total redox state of plastoquinone in chloroplasts, the size of the photochemically active plastoquinone-pool and its redox state in thylakoids of Arabidopsis. Biochim Biophys Acta. 2006;1757:1669–1675. doi: 10.1016/j.bbabio.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 31.Austin JR, 2nd, Frost E, Vidi PA, Kessler F, Staehelin LA. Plastoglobules are lipoprotein subcompartments of the chloroplast that are permanently coupled to thylakoid membranes and contain biosynthetic enzymes. Plant Cell. 2006;18:1693–1703. doi: 10.1105/tpc.105.039859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mène-Saffrané L, Jones AD, DellaPenna D. Plastochromanol-8 and tocopherols are essential lipid-soluble antioxidants during seed desiccation and quiescence in Arabidopsis. Proc Natl Acad Sci USA. 2010;107:17815–17820. doi: 10.1073/pnas.1006971107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grütter C, Alonso E, Chougnet A, Woggon WD. A biomimetic chromanol cyclization leading to α-tocopherol. Angew Chem Int Ed Engl. 2006;45:1126–1130. doi: 10.1002/anie.200503123. [DOI] [PubMed] [Google Scholar]
- 34.Shimada H, et al. Inactivation and deficiency of core proteins of photosystems I and II caused by genetical phylloquinone and plastoquinone deficiency but retained lamellar structure in a T-DNA mutant of Arabidopsis. Plant J. 2005;41:627–637. doi: 10.1111/j.1365-313X.2004.02326.x. [DOI] [PubMed] [Google Scholar]
- 35.Gross J, et al. A plant locus essential for phylloquinone (vitamin K1) biosynthesis originated from a fusion of four eubacterial genes. J Biol Chem. 2006;281:17189–17196. doi: 10.1074/jbc.M601754200. [DOI] [PubMed] [Google Scholar]
- 36.Havaux M, et al. Vitamin B6 deficient plants display increased sensitivity to high light and photo-oxidative stress. BMC Plant Biol. 2009;9:130. doi: 10.1186/1471-2229-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koop HU, et al. Integration of foreign sequences into the tobacco plastome via polyethylene glycol-mediated protoplast transformation. Planta. 1996;199:193–201. doi: 10.1007/BF00196559. [DOI] [PubMed] [Google Scholar]
- 38.Greenspan P, Mayer EP, Fowler SD. Nile Red: A selective fluorescent stain for intracellular lipid droplets. J Cell Biol. 1985;100:965–973. doi: 10.1083/jcb.100.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore AL, Fricaud AC, Walters AJ, Whitehouse DG. Isolation and purification of functionally intact mitochondria from plant cells. Methods Mol Biol. 1993;19:133–139. doi: 10.1385/0-89603-236-1:133. [DOI] [PubMed] [Google Scholar]
- 40.Agne B, et al. The acidic A-domain of Arabidopsis TOC159 occurs as a hyperphosphorylated protein. Plant Physiol. 2010;153:1016–1030. doi: 10.1104/pp.110.158048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.