Abstract
Human helicase-like transcription factor (HLTF) exhibits ubiquitin ligase activity for proliferating cell nuclear antigen (PCNA) polyubiquitylation as well as double-stranded DNA translocase activity for remodeling stalled replication fork by fork reversal, which can support damage bypass by template switching. However, a stalled replication fork is surrounded by various DNA-binding proteins which can inhibit the access of damage bypass players, and it is unknown how these proteins become displaced. Here we reveal that HLTF has an ATP hydrolysis-dependent protein remodeling activity, by which it can remove proteins bound to the replication fork. Moreover, we demonstrate that HLTF can displace a broad spectrum of proteins such as replication protein A (RPA), PCNA, and replication factor C (RFC), thereby providing the first example for a protein clearing activity at the stalled replication fork. Our findings clarify how remodeling of a stalled replication fork can occur if it is engaged in interactions with masses of proteins.
Unrepaired DNA lesions are dangerous obstacles for the replication machinery because most of them cannot be accommodated into the active site of the replicative polymerases, thereby blocking the progression of the replication fork. Prolonged stalling might lead to DNA strand breaks, chromosomal rearrangements, or cell death (1–3). To minimize this danger cells have evolved various DNA damage bypass mechanisms that are initiated by exchanging protein components of the normal replication machinery for protein players which either carry out a direct damage bypass or manipulate the stalled fork to generate transitional DNA structures that facilitate damage bypass indirectly. In the first situation, specialized translesion synthesis polymerases that can accommodate even bulky lesions at their active sites take over the 3′ primer end from the accurate replicative polymerase and incorporate either a correct or an incorrect nucleotide opposite the lesion (4, 5). Alternatively, DNA remodeling might lead to the annealing of the stalled nascent strand to the newly synthesized strand of the undamaged sister duplex, resulting in template switch (6–9). Although the exact mechanism and factors of template switch have remained largely unknown, two proposed mechanisms have gained significant attention. One is named—based on the shape of the intermediate DNA structure—chicken foot model, which proposes pairing of the two newly synthesized strands of the sister chromatids by reversal of the stalled fork (9, 10). The other model also suggests pairing of the newly synthesized strands, but assumes that it occurs via a D-loop recombination intermediate (6, 11–13). It is possible that these mechanisms are not mutually exclusive and the choice can be regulated at the level of displacement and exchange of the protein components of the stalled replication machinery for various new players.
It is generally accepted that in eukaryotic cells damage bypass is governed by Rad6 and Rad18, a ubiquitin conjugating and a ubiquitin ligase enzyme, respectively, that form a complex known to monoubiquitylate proliferating cell nuclear antigen (PCNA) at the K164 residue upon DNA damage (14–17). Yeast genetic studies on replication of UV-damaged DNA revealed that PCNA monoubiquitylation is a prerequisite for the operation of at least three downstream branches of the Rad6-Rad18 pathway (4, 18). Two branches include error-free or error-prone translesion synthesis polymerases, whereas the third branch depends on Mms2, Ubc13, and Rad5, for which yeast genetic data indicate template switching mechanism (8). The choice between translesion synthesis and template switching is thought to be dependent on the polyubiquitylation of PCNA, where Rad5, through its really interesting new gene (RING)-domain ubiquitin ligase activity, stimulates the Mms2-Ubc13-dependent synthesis of a lysine-63 linked polyubiquitin chain onto monoubiquitylated PCNA (16, 19–21).
Yeast genetic data also revealed that, in addition to its RING domain, the Rad5 ATPase domain is equally important for replication of damaged DNA (22, 23). Moreover, biochemical data have given strong support for a role of Rad5, and particularly of its ATPase domain, in a fork reversal-dependent mode of template switch, because purified Rad5 was shown to be able to regress stalled replication fork-like structures in an ATP hydrolysis-dependent manner (24). Identification, in human cells, of Rad5 orthologues, helicase-like transcription factor (HLTF) and Snf2 histone linker PHD RING helicase (SHPRH) and their characterization as ubiquitin ligases for PCNA K63 polyubiquitylation provided unique evidence for the conservation of the Rad5-dependent pathway in higher order eukaryotes (25–28).
HLTF, structurally the closest homologue of yeast Rad5, is a molecular motor protein, which—like many other members of the SWItch/Sucrose nonfermentable (Swi2/Snf2) family—does not exhibit a canonical DNA helicase activity, but has an ATP hydrolysis-driven dsDNA translocase activity. This activity provides the ability for HLTF to reverse replication fork-like structures in vitro. Furthermore, examining the movement of replication forks by DNA fiber method revealed that the ATPase activity of HLTF plays a critical in vivo role in the replication of damaged DNA (29). These findings taken together with yeast genetic data on RAD5 suggest a role for HLTF in template switch-dependent error-free damage bypass and are in keeping with its proposed tumor suppressor function (30).
In addition to HLTF, a fork reversal activity has been indicated for a number of other repair proteins such as the Bloom helicase (BLM), HepA-related protein (HARP), and Fanconi anemia complementation group M (FANCM) (31–34), which suggests that multiple pathways might exist for template switch-dependent error-free DNA damage bypass. However, all previous fork reversal assays were carried out on naked replication fork-like structures, whereas in vivo a stalled replication fork contains several ssDNA- and dsDNA-bound proteins such as the polymerases, RPA, replication factor C (RFC), and PCNA, which can interfere with DNA remodeling. It is evident that somehow these proteins have to be displaced from the fork for productive template switch and to give access to new damage bypass protein players. It is possible that they become degraded, but a more reasonable hypothesis is that these proteins are transiently displaced from the damage site by some protein remodeling mechanism.
A number of Swi2/Snf2 proteins have been described as motor subunits of particular chromatin remodeling complexes, which use the energy of ATP hydrolysis to induce local DNA distortion resulting in the alteration of the association of nucleosomes with DNA (35, 36). By analogy, we hypothesized that, upon translocation on the replication fork, HLTF might induce local DNA bending or twisting, which might effect the DNA-binding property of proteins in the stalled replication machinery.
Here we examine whether proteins bound to replication fork-like DNA structures inhibit two distinct fork reversal enzymes, namely HLTF, an Swi2/Snf2 family protein, and BLM, a RecQ family helicase. We provide evidence that HLTF can specifically remodel replication forks bound by either dsDNA- or ssDNA-binding proteins (SSB), a property associated with a unique protein remodeling activity of HLTF. These observations shed light on how masses of proteins surrounding the stalled replication fork can become displaced from the DNA, providing access to new damage bypass players.
Results
HLTF can Regress a Modeled Replication Fork Bound by dsDNA-Binding Protein.
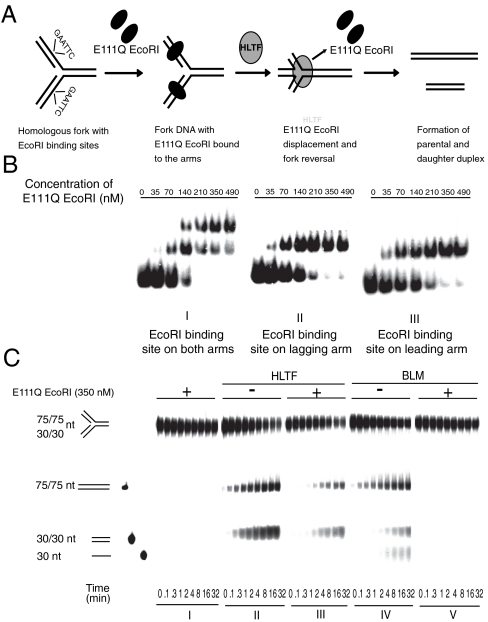
To examine whether HLTF DNA remodeling activity is inhibited by a protein bound to stalled replication fork-like DNA structures, oligonucleotide-based homologous forks were generated where dsDNA-binding proteins can be bound to its arms. To eliminate the possibility of protein-protein interaction between HLTF and DNA-bound protein, an Escherichia coli E111Q EcoRI endonuclease mutant protein was chosen that is selectively defective in DNA cleavage but retains its sequence-specific dsDNA-binding activity (37). An EcoRI recognition sequence was introduced to one or both arms of the homologous fork (Fig. 1A), and the sequence-specific binding of E111Q EcoRI to the homologous fork was confirmed by gel mobility shift experiments (Fig. 1B). The remodeling of these protein-bound DNA structures can be followed by the appearance of 75/75- (parental strands) and 30/30-nt (daughter strands) long dsDNA fragments that would arise upon fork reversal (Fig. 1A) as described earlier (24, 29). For control, we used the BLM, which has also been reported to have fork reversal activity (33) and found that it was completely inhibited by binding of E111Q EcoRI proteins to both arms (Fig. 1C, compare IV to V). In contrast, we found that HLTF retained its fork reversal activity on the same protein-bound fork substrate and only weak inhibition occurred (Fig. 1C, compare II to III, and Fig. S1A for quantification). In addition, if the fork DNA contained only a single EcoRI binding site in one of its arms, HLTF processed the leading or lagging strand protein-bound substrates with similar kinetics (Fig. S2). Moreover, HLTF could also achieve fork reversal when EcoRI was bound on the dsDNA ahead of the fork (Fig. S3). These results suggest that HLTF can facilitate fork remodeling even when the base or fork arms are bound by proteins, which represents the actual scenario when the replication fork is stalled. The lack of a similar activity in BLM helicase reveals the specificity of HLTF.
Fig. 1.
Fork reversal activity of HLTF on modeled replication fork bound by dsDNA-binding protein. (A) Schematic representation of a possible mechanism through which HLTF can coordinately remodel a model replication fork bound by E111Q EcoRI. (B) Gel retardation assay showing sequence-specific binding and formation of stable DNA-protein complex by E111Q EcoRI on oligo-based fork-like structures. Increasing amount of E111Q EcoRI was incubated with homologous fork containing an EcoRI binding site. E111Q EcoRI binding to both the arms of the fork is shown in I, whereas II and III show binding to lagging or leading arm only. (C) Comparison of HLTF and BLM fork reversal activities on homologous fork bound by E111Q EcoRI protein on both the arms. In I, control without BLM or HLTF; II, activity of HLTF on naked fork; III, activity of HLTF on E111Q EcoRI-bound fork; IV, activity of BLM on naked fork; V, activity of BLM on E111Q EcoRI-bound fork. Each lane within the panel represents time points at which samples are collected and are noted at the bottom of the gel. Quantitation is shown in Fig. S1.
Protein Displacement from Modeled Fork Requires the DNA Translocase Activity of HLTF.
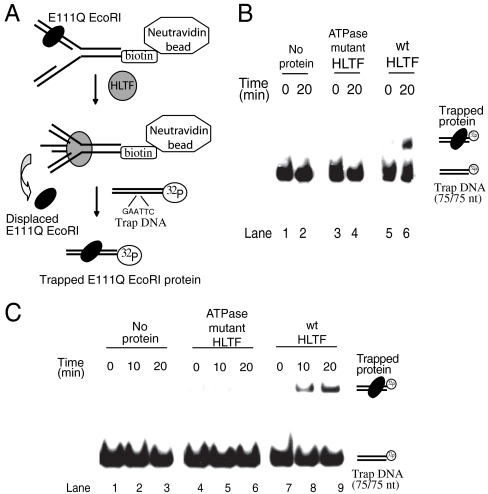
The ability of HLTF to regress a model replication fork in spite of its being covered by proteins suggests that HLTF is able to actively remodel these proteins. To confirm this remodeling further we set up an experimental system in which the actual displacement of E111Q EcoRI protein molecules from the fork can be monitored. As shown in Fig. 2A, biotin-tagged model forks containing an E111Q EcoRI protein bound to its binding site on one of the arms were immobilized on NeutrAvidin beads. Next, the fork regression assay was carried out on the beads by HLTF, where E111Q EcoRI proteins can be released into the supernatant. Finally, the supernatant was examined for the presence of E111Q EcoRI by trapping with a labeled duplex DNA containing a single EcoRI binding site. Thus the actual displacement can be monitored by the appearance of E111Q EcoRI-bound trap DNA in gel mobility shift experiments (Fig. 2B). We found that upon HLTF-dependent DNA remodeling E111Q EcoRI protein molecules were released into the supernatant (Fig. 2B, lane 6). Removal of bound E111Q EcoRI protein from the fork DNA was observed only with wild-type HLTF but was absent in mutant DE557, 558AA HLTF lacking ATPase/dsDNA translocase activity (Fig. 2B, compare lanes 4 and 6). Thus, this result not only provides evidence for HLTF’s ability to remove dsDNA-binding proteins from the replication fork-like DNA structures, but also confirms that this removal is an active process depending on HLTF’s ATPase activity. However, it was not clear whether the displacement of dsDNA-binding protein is due to HLTF fork regression activity or is solely dependent on its dsDNA translocase activity. To answer this question, instead of a modeled fork, a 75/30-mer partial duplex DNA resembling only an arm of the previously used replication fork with an EcoRI binding site was bound to NeutrAvidin beads. This experiment revealed that HLTF can indeed displace a protein from a duplex DNA, and this displacement was only observed with the wild-type HLTF protein but not when the DE557, 558AA HLTF ATPase mutant was used (Fig. 2C, compare lanes 5–6 with 8–9). These results provide evidence that HLTF, along with its ubiquitin ligase and fork regression activity, also has a protein displacing activity.
Fig. 2.
Evidence for dsDNA-binding protein disposal from DNA by HLTF. (A) Experimental setup to prove the actual displacement of dsDNA-binding protein during fork reversal. A homologous fork with a single EcoRI-binding site is bound to NeutrAvidin beads through its biotin tag, and the E111Q EcoRI displaced from the fork is trapped by a 75-mer labeled duplex containing a single EcoRI site. The trap DNA is subjected to gel retardation assay to confirm the binding of E111Q EcoRI. (B) Gel retardation assay showing trapped E111Q EcoRI displaced from a modeled fork. Lanes 1–2 no protein control, 3–4 HLTF ATPase mutant, 5–6 HLTF wild-type protein. Samples were collected at 0 and 20 min and incubated with duplex trap DNA prior to gel retardation assay. (C) Similar assay like in B, except that instead of a modeled fork a 75/30-mer partial duplex DNA was used. Lanes 1–3 no protein control, 4–6 HLTF ATPase mutant, 7–9 HLTF wild-type protein. Samples were collected at 0, 10, and 20 min for each protein sample and incubated with duplex trap DNA prior to gel retardation assay.
HLTF can Remodel Gapped Replication Forks Bound by ssDNA-binding proteins.
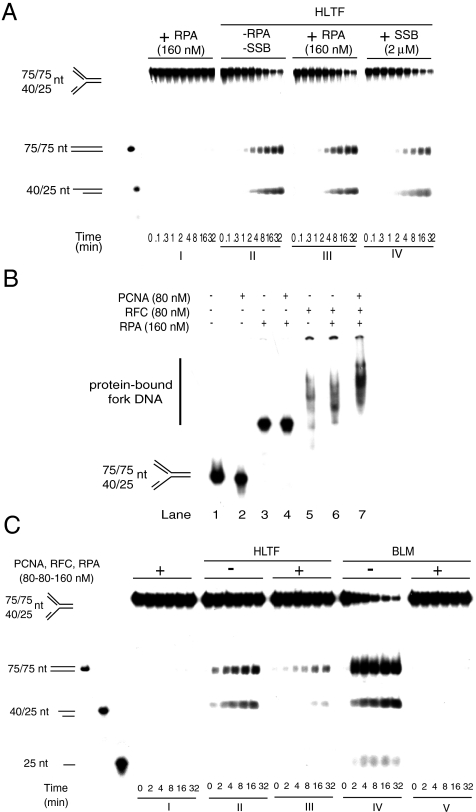
It has been previously reported that the blockage of the replication machinery can lead to uncoupling of leading and lagging strand synthesis, resulting in the generation of ssDNA, which in turn will be occupied by RPA, an SSB (38, 39). One would expect this RPA-ssDNA complex to be dissociated prior to DNA remodeling for a successful damage bypass. To model this situation we generated a 15-nt long ssDNA gap region in the leading strand of fork DNA, where RPA or E. coli SSB was successfully bound, as shown by gel shift experiments (Fig. S4 A and B and Fig. S5). Previous data indicated that some DNA helicases such as Srs2 and BLM have the ability to remove certain proteins bound to ssDNA (40, 41). In concert, we found that RPA or SSB did not inhibit BLM fork reversal activity which can be mechanistically explained by the fact that BLM translocation on ssDNA can displace RPA or SSB (Fig. S4C). However, with a similar mechanism one cannot expect ssDNA-bound protein removal by a dsDNA translocase. Previously, we showed that HLTF cannot translocate on ssDNA but is able to displace the third strand of a partial triple-helix in an ATP-dependent manner, thereby providing evidence that HLTF is a dsDNA translocase (29). Nevertheless, by fork reversal assay we managed to reveal that HLTF can successfully regress ssDNA-binding protein-covered forks (Fig. 3A). RPA and E. coli SSB-covered fork DNAs were processed with similar efficiency, ruling out a role for a potential interaction between RPA and HLTF in this activity (Fig. 3A, compare III and IV). RPA or SSB displacement can be explained by the dsDNA translocase activity of HLTF, assuming that during fork reversal HLTF translocates on the parental duplex DNA, when it concertedly unwinds the arms of the fork and zips the parental strands and the nascent strands and, coordinately with this process, removes the proteins encountered. Altogether, these fork reversal assays on protein-bound substrates reveal the fact that during fork reversal HLTF is also able to remodel SSBs from DNA.
Fig. 3.
Fork reversal activity of HLTF on gapped fork bound by replicative proteins. (A) Fork reversal activity of HLTF on RPA- or SSB-bound substrate. In I, control without HLTF; II, gapped fork without any ssDNA-binding protein; III, RPA bound to gapped fork; IV, SSB bound to gapped fork. Gel shift experiment for confirming RPA and SSB binding to fork DNA is shown in Fig. S4 A and B, respectively. (B) Gel retardation assays for confirming the binding of RPA, PCNA, and RFC to a homologous fork containing a 15-nt gap on its leading arm. Various combinations of RPA (160 nM), PCNA (80 nM), and RFC (80 nM) were incubated with a 15-nt gapped fork. (C) Comparison of HLTF and BLM fork reversal activities on PCNA-, RFC-, and RPA-bound replication-like structures. In I, control without BLM or HLTF; II, activity of HLTF on naked fork; III, activity of HLTF on PCNA-, RFC-, and RPA-bound fork; IV, activity of BLM on naked fork; V, activity of BLM on PCNA-, RFC-, and RPA-bound fork.
HLTF can Dislodge PCNA, RFC, and RPA Complex from DNA Replication Fork.
The above results confirm that HLTF can remodel ssDNA- and dsDNA-binding proteins on stalled replication fork-like DNA structures. However, the proteins we tested were far from the nature and complexity of the proteins which can be found at the stalled replication fork in vivo. To provide further evidence that HLTF can indeed remodel various proteins expected to be present at a stalled replication fork, we examined if HLTF can overcome the inhibitory effect of the complex of the replicative polymerase clamp PCNA, the clamp loader RFC, and RPA bound to a model replication fork substrate. PCNA, RFC, and RPA were bound to a substrate containing a 15-nt long ssDNA region in the leading strand of fork DNA (Fig. 3B). We note that considering the short ssDNA region present in this model fork DNA RPA binding presumably occurred only by its DNA-binding subunit, which, however, was stable as confirmed by gel shift assay (Fig. 3B, lane 3). In control experiments these DNA-binding proteins completely inhibited fork reversal by BLM helicase (Fig. 3C, compare IV and V). Importantly, however, HLTF was able to remodel the fork DNA substrate bound by these protein factors of the replication machinery (Fig. 3C, compare II and III). This activity was similar to what we saw with E. coli E111Q, where HLTF actively removed the protein from DNA and regressed oligo-based fork-like structures. As expected, the yeast Rad5, an orthologue of HLTF, was also able to reverse protein-bound fork DNA indicating the conservation of this particular protein remodeling activity (Fig. S6). To show that DNA size is not limiting, we conducted a similar experiment with a plasmid-based fork structure and found that PCNA and RFC do not prevent HLTF-dependent fork reversal (Fig. S7). Together, these findings suggest that HLTF-dependent fork reversal is not prevented if these replication accessory proteins are bound to stalled replication fork DNA. Hence, we conclude from our results that along with its DNA remodeling activity, HLTF also has a general protein remodeling activity and the two together provide an ability for HLTF to process a protein-covered stalled replication fork.
Discussion
Previously we have shown that HLTF and its yeast homologue, Rad5, have a DNA translocase activity enabling them to carry out replication fork reversal by concertedly unwinding the leading and lagging strand arms of the fork and then annealing together the nascent strands and the parental strands (24, 29). This finding provided unique biochemical evidence for the capability of HLTF and Rad5 to facilitate error-free damage bypass by switching the template from the damaged leading strand to the newly synthesized undamaged nascent strand of the lagging arm. In addition to Rad5 and HLTF, the number of enzymes with proven fork reversal activity is continuously growing (31, 33, 34, 42, 43), which raises the possibility that many parallel pathways might exist for fork reversal and template switching. However, it has not been examined what happens to the huge protein complex present at the stalled replication fork, collectively referred to as replication machinery. It is evident that this complex or at least its particular elements have to be displaced from the fork for rescue mechanisms such as template switching to occur smoothly. For this obvious problem, however, no solution has been proposed, mainly because previous fork reversal assays were carried out using naked oligonucleotide- or plasmid-based fork DNA substrates. Possible hypotheses involve either removing particular proteins from the stalled fork by some unknown remodeling factor, or in situ degradation of these proteins.
Here we provide evidence for the first scenario and show that HLTF has a unique protein remodeling activity. We recognized this activity by investigating various dsDNA- or ssDNA-binding protein covered modeled replication fork structures and by asking if bound proteins have any inhibitory effects on fork reversal by HLTF and BLM that belong to the Swi2/Snf2 and RecQ family, respectively (29, 33). To our surprise, neither a complex of human replication proteins consisting of RPA, RFC, and PCNA nor E. coli EcoRI protein, used as a site-specific dsDNA-binding model protein, posed a big challenge for HLTF fork reversal activity, whereas they completely inhibited BLM helicase-dependent fork reversal. These experiments suggested that HLTF has a general protein remodeling activity, which can relocate DNA-bound proteins or might completely break off DNA-protein interaction. Supporting the second assumption, using NeutrAvidine bead-bound biotin-DNA substrates, we managed to show that HLTF is indeed able to disrupt DNA-protein interactions, as revealed by the appearance of displaced proteins relocated from the solid bead-DNA-protein fraction into the supernatant. Thus we suggest that on a protein-bound stalled replication fork, HLTF cannot only facilitate a mere DNA-protein structural readjustment such as forcing the backtrack of the PCNA ring, but it is also able to remove inhibitory proteins from DNA and that both of these mechanisms might play a role in productive protein exchange and damage bypass.
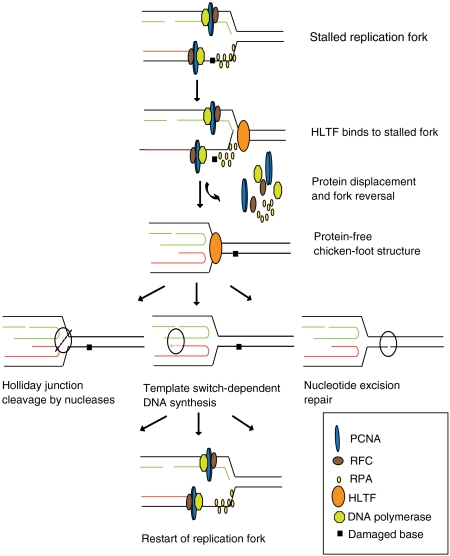
As shown on a schematic model in Fig. 4, we propose that HLTF has a protein cleansing function at stalled replication forks, which is a prerequisite for successful fork remodeling leading finally to replication through the lesion. We hypothesize that remodeling proteins at the stalled replication fork and fork reversal can provide an opportunity either for a DNA polymerase to extend the originally blocked 3′ DNA end using the newly synthesized sister strand as a template, or for excision repair to remove the lesion, or for recombination mediated replication to restart after cleavage of the reversed fork. It would be interesting to examine if HLTF protein displacement/DNA remodeling activity can operate on other structures, e.g., on D-loop intermediates of synthesis-dependent strand annealing mechanism, which has also been proposed to function in filling in ssDNA gaps that are left behind during replication (18).
Fig. 4.
Model for the role of HLTF in remodeling protein-covered stalled replication forks. (A) Stalled replication fork at an unrepaired DNA lesion has to undergo severe remodeling such that it can clear up most of its protein content, allowing other DNA repair machinery to get access to the DNA. We suggest that one such mechanism through which a fork can clear up its protein content can be facilitated by HLTF. HLTF can have dual functions because it can not only clear up the proteins, but also can give rise to a four-way junction intermediate called chicken foot. This four-way junction can then be used by other subsequent repair pathways like (i) Holliday junction resolvases, which can resolve a four-way junction through their nuclease activity, (ii) template switch-dependent DNA synthesis, where a DNA polymerase extends the 3′ OH end of the leading nascent strand by copying from the nascent lagging strand, (iii) nucleotide excision repair pathway, where a short ssDNA segment is removed creating a single-strand gap in the DNA, which is subsequently filled in by DNA polymerase using the undamaged strand as template. Once the lesion is repaired or bypassed, the stalled replication fork readopts its original structure and the progression of DNA replication is reestablished.
For a number of DNA helicases such as yeast Srs2, human RECQ5, and BLM, displacement activity for the ssDNA-binding Rad51 protein has been reported (40, 41, 44–47). The specificity of these enzymes is ensured by their physical interaction with Rad51, and their mechanism can be explained by their ssDNA translocase activity by which they might break into the Rad51-ssDNA interface upon ATP hydrolysis-dependent ssDNA translocation, resulting in breaking off the Rad51-ssDNA interaction. Being a dsDNA translocase, HLTF can be distinguished from these canonical DNA helicases, and we propose that the mechanism of protein remodeling by HLTF is more related to the action of Swi2/Snf2 chromatin remodeling enzymes.
In general, proteins in the Swi2/Snf2 family have been considered as chromatin remodeling enzymes for nucleosome displacement (35, 36). Mechanically, most of these enzymes can interact with dsDNA as well as with histones, usually present in their particular posttranslationally modified forms. By translocation on dsDNA they can induce local DNA distortion which contributes to nucleosome remodeling. Interestingly, the Swi2/Snf2-related Rdh54 and the Rad54 DNA branch migrating proteins exhibit not only nucleosome remodeling activity, but can displace Rad51 as well (48–51). Also, from the Swi2/Snf2 family members Mot1 has been reported to have a nonnucleosomal protein remodeling activity. Mot1 is able to displace the TATA box-binding protein (TBP) from DNA, thereby providing a regulatory check for transcription. Mot1 does not detectably bind to DNA on its own, but the cooperative interaction between Mot1 and TBP can stabilize the ternary complex and, subsequently, the ATP hydrolysis-dependent translocation of Mot1 on dsDNA into the TBP-DNA interface can result in TBP dissociation (52). Whereas Mot1 is specific to TBP protein removal, which is ensured, at least partly, by its interaction with TBP, our study identifies HLTF as a more general protein remodeling enzyme. Support for this notion is provided by the ability of HLTF to remodel not only components of the replication machinery such as PCNA, RFC, and RPA with which its interaction cannot be ruled out, but also an E. coli dsDNA-binding protein, namely EcoRI E111Q, as well as E. coli SSB with which its physical interaction is highly unlikely. Because the ATPase mutant HLTF is impaired in protein remodeling, we suggest that it is local twisting and bending of DNA induced by ATP hydrolysis-dependent HLTF translocation on dsDNA that constitutes the main force for breaking up protein-DNA interactions.
The discovered coordinated protein displacing/DNA remodeling activity of HLTF further extends the repertoire of the enzymatic capabilities of the intensively examined Swi2/Snf2 protein family, and raises the question of whether other members also exhibit similar activities. Thus, it would be interesting to test other Swi2/Snf2 proteins such as the Rad54, HARP, and FANCM fork reversal enzymes for general protein remodeling activity on various DNA structures such as stalled replication fork.
Finally, in a high percentage of various cancers, for example over 40% in colon cancers, HLTF expression is silenced or various Swi2/Snf2 domain deletion mutant HLTF proteins are expressed, which suggests that HLTF can be a tumor suppressor (53, 54). Because HLTF was also reported to be a transcription factor, one acceptable explanation for this finding could be that tumor suppression is because of its effect on gene expression. The other possibility is that as a PCNA polyubiquitin ligase, HLTF has a regulatory role for providing error-free damage bypass (25, 27, 30). In addition to these possibilities, now we suggest that the described coordinated protein displacing and DNA remodeling activity of HLTF can also be important for providing genome stability.
Materials and Methods
Proteins and DNA Substrates.
Purification of wild-type HLTF, ATPase mutant DE557,558AA HLTF, BLM, E. coli E111Q EcoRI endonuclease mutant protein, PCNA, RFC, RPA, and the generation of modeled fork substrates are described in SI Materials and Methods.
Protein-Bound DNA Substrates and Gel Shift Assay.
To generate an E111Q EcoRI-bound fork, a homologous fork containing EcoRI binding site(s) (1 nM) was preincubated prior to fork reversal assay with purified E111Q EcoRI (350 nM) in a binding buffer containing 20 mM Tris pH 7.5, 150 mM NaCl, 1 mM DTT, 0.1 mg/mL BSA, 10% glycerol at 37 °C for 15 min. For ssDNA protein-bound substrates, a gap substrate containing a 15-nt gap on the leading arm of the fork (1 nM) was incubated with human RPA (160 nM) or E. coli SSB (2 μM) in binding buffer containing 50 mM Hepes 7.8, 150 mM NaCl, 2 mM DTT, 10 mM MgCl2, 0.1 mg/mL BSA and 10% glycerol at room temperature for 15 min (used for Fig. 3A and Fig. S4). To generate PCNA-, RFC-, and RPA-bound substrates (used for Fig. 3B and Fig. S7), first PCNA (80 nM), next RFC (80 nM), and finally RPA (160 nM) were added to the gap substrates ( 1 nM) at 0 °C in a buffer containing 20 mM Tris·HCl pH 7.5, 150 mM NaCl, 1 mM MgCl2, 1 mM DTT, 1 mM ATP, 0.1 mg/mL BSA, and 10% glycerol followed by incubation at 37 °C for 15 mins. The protein-bound DNA substrates were divided and immediately used for fork reversal assay and for confirming protein binding by gel shift assay. For gel shift assay, samples were loaded onto a 4% native polyacrylamide gel containing acrylamide and N,N bis-acrylamide in 30∶0.8 ratio, 0.5x Tris-borate and 2.5% glycerol before gel electrophoresis at 4 °C in 0.5x Tris-borate buffer containing no EDTA.
Fork Reversal Assay.
Fork reversal assays with HLTF (10 nM) and BLM (10 nM) were carried out in buffer H containing 20 mM Tris·HCl, pH 7.5, 150 mM or 100 mM NaCl, 5 mM MgCl2, 5 mM ATP, 0.1 mg/mL BSA, 1 mM DTT and 10% glycerol with 0.5 nM 32P-labeled naked fork DNA or fork DNA preincubated with DNA-binding proteins as described in the gel shift section. We note that all fork reversal assays were carried out at 100 mM as well as 150 mM NaCl concentration because HLTF exhibits higher fork reversal activity at 150 mM NaCl (used for figures with HLTF) whereas BLM at 100 mM NaCl (used for figures with BLM), but at both salt concentrations similar results were reached allowing the same conclusion. Reaction mixtures were incubated at 37 °C for the time indicated in the figures, followed by adding equal volumes of stop buffer containing 20 mM EDTA, 1% sodium dodecyl sulfate, 10% glycerol, and 0.02% bromophenol blue before further incubation for 5 min. DNA samples were loaded onto 10% native polyacrylamide gels, and the products were separated by electrophoresis using 1x Tris-borate buffer containing no EDTA.
Protein Displacement Assay.
A homologous fork or a 75/30-mer partial duplex DNA with an EcoRI binding site was generated, in which one of the oligonucleotides was biotinylated (HomF-biotin). E111Q EcoRI (350 nM) was allowed to bind to HomF-biotin (1 nM), followed by binding of this protein-bound fork (200 μL) to 50 μL of NeutrAvidin beads (PIERCE-29200) before vigorous washing. Next, fork reversal assay was carried out on the bead-bound fork/ E111Q EcoRI substrate using wild-type HLTF (50 nM) or ATPase mutant DE557,558AA HLTF (50 nM). Ten microliters of supernatant fractions were collected at each time point and incubated with labeled trap dsDNA (0.5 nM) containing a single EcoRI binding site. The displacement of E111Q EcoRI protein was followed by the appearance of a shift due to its binding to trap dsDNA in a gel retardation assay.
Supplementary Material
Acknowledgments.
We thank Andras Blastyak for purifying the E111Q EcoRI protein and sharing his expertise on translocase assays. Our research was supported by Howard Hughes Medical Institute Grant 55005612 and Hungarian Science Foundation Grants OTKA 77495 and TAMOP-4.2.2/08/1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 13881.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101951108/-/DCSupplemental.
References
- 1.Boulton SJ. Helicase and translocases required for the maintenance of genome stability. DNA Repair (Amst) 2010;9(3):201. doi: 10.1016/j.dnarep.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 2.Boulton SJ. DNA repair: Decision at the break point. Nature. 2010;465:301–302. doi: 10.1038/465301a. [DOI] [PubMed] [Google Scholar]
- 3.Ciccia A, Elledge SJ. The DNA damage response: Making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 5.Shachar S, et al. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009;28:383–393. doi: 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gangavarapu V, Prakash S, Prakash L. Requirement of RAD52 group genes for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:7758–7764. doi: 10.1128/MCB.01331-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higgins NP, Kato K, Strauss B. A model for replication repair in mammalian cells. J Mol Biol. 1976;101:417–425. doi: 10.1016/0022-2836(76)90156-x. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Lawrence CW. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc Natl Acad Sci USA. 2005;102:15954–15959. doi: 10.1073/pnas.0504586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein HL. Reversal of fortune: Rad5 to the rescue. Mol Cell. 2007;28:181–183. doi: 10.1016/j.molcel.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Atkinson J, McGlynn P. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 2009;37:3475–3492. doi: 10.1093/nar/gkp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein HL. Molecular biology: DNA endgames. Nature. 2008;455:740–741. doi: 10.1038/455740a. [DOI] [PubMed] [Google Scholar]
- 12.Berdichevsky A, Izhar L, Livneh Z. Error-free recombinational repair predominates over mutagenic translesion replication in E. coli. Mol Cell. 2002;10:917–924. doi: 10.1016/s1097-2765(02)00679-2. [DOI] [PubMed] [Google Scholar]
- 13.Adar S, Izhar L, Hendel A, Geacintov N, Livneh Z. Repair of gaps opposite lesions by homologous recombination in mammalian cells. Nucleic Acids Res. 2009;37:5737–5748. doi: 10.1093/nar/gkp632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bailly V, Lamb J, Sung P, Prakash S, Prakash L. Specific complex-formation between yeast Rad6 and Rad18 proteins—a potential mechanism for targeting Rad6 ubiquitin-conjugating activity to DNA-damage sites. Genes Dev. 1994;8:811–820. doi: 10.1101/gad.8.7.811. [DOI] [PubMed] [Google Scholar]
- 15.Bailly V, Lauder S, Prakash S, Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J Biol Chem. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- 16.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 17.Prakash S, Sung P, Prakash L. DNA repair genes and proteins of Saccharomyces cerevisiae. Annu Rev Genet. 1993;27:33–70. doi: 10.1146/annurev.ge.27.120193.000341. [DOI] [PubMed] [Google Scholar]
- 18.Torres-Ramos CA, Prakash S, Prakash L. Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:2419–2426. doi: 10.1128/MCB.22.7.2419-2426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broomfield S, Chow BL, Xiao W. MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc Natl Acad Sci USA. 1998;95:5678–5683. doi: 10.1073/pnas.95.10.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haracska L, Torres-Ramos CA, Johnson RE, Prakash S, Prakash L. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:4267–4274. doi: 10.1128/MCB.24.10.4267-4274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 22.Gangavarapu V, et al. Mms2-Ubc13-dependent and -independent roles of Rad5 ubiquitin ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:7783–7790. doi: 10.1128/MCB.01260-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson RE, Prakash S, Prakash L. Yeast DNA repair protein RAD5 that promotes instability of simple repetitive sequences is a DNA-dependent ATPase. J Biol Chem. 1994;269:28259–28262. [PubMed] [Google Scholar]
- 24.Blastyak A, et al. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Motegi A, et al. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc Natl Acad Sci USA. 2008;105:12411–12416. doi: 10.1073/pnas.0805685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Motegi A, et al. Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. J Cell Biol. 2006;175:703–708. doi: 10.1083/jcb.200606145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unk I, et al. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc Natl Acad Sci USA. 2008;105:3768–3773. doi: 10.1073/pnas.0800563105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Unk I, et al. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc Natl Acad Sci USA. 2006;103:18107–18112. doi: 10.1073/pnas.0608595103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blastyak A, Hajdu I, Unk I, Haracska L. Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. Mol Cell Biol. 2010;30:684–693. doi: 10.1128/MCB.00863-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Unk I, Hajdu I, Blastyak A, Haracska L. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair (Amst) 2010;9:257–267. doi: 10.1016/j.dnarep.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 31.Driscoll R, Cimprich KA. HARPing on about the DNA damage response during replication. Genes Dev. 2009;23:2359–2365. doi: 10.1101/gad.1860609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prakash R, et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: Implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ralf C, Hickson ID, Wu L. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J Biol Chem. 2006;281:22839–22846. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- 34.Collis SJ, Boulton SJ. FANCM: Fork pause, rewind, and play. EMBO J. 2010;29:703–705. doi: 10.1038/emboj.2009.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Durr H, Flaus A, Owen-Hughes T, Hopfner KP. Snf2 family ATPases and DExx box helicases: Differences and unifying concepts from high-resolution crystal structures. Nucleic Acids Res. 2006;34:4160–4167. doi: 10.1093/nar/gkl540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright DJ, King K, Modrich P. The negative charge of Glu-111 is required to activate the cleavage center of EcoRI endonuclease. J Biol Chem. 1989;264:11816–11821. [PubMed] [Google Scholar]
- 38.Cordeiro-Stone M, Zaritskaya LS, Price LK, Kaufmann WK. Replication fork bypass of a pyrimidine dimer blocking leading strand DNA synthesis. J Biol Chem. 1997;272:13945–13954. doi: 10.1074/jbc.272.21.13945. [DOI] [PubMed] [Google Scholar]
- 39.Svoboda DL, Vos JM. Differential replication of a single, UV-induced lesion in the leading or lagging strand by a human cell extract: Fork uncoupling or gap formation. Proc Natl Acad Sci USA. 1995;92:11975–11979. doi: 10.1073/pnas.92.26.11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colavito S, Prakash R, Sung P. Promotion and regulation of homologous recombination by DNA helicases. Methods. 51:329–335. doi: 10.1016/j.ymeth.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krejci L, et al. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 42.Kanagaraj R, Saydam N, Garcia PL, Zheng L, Janscak P. Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006;34:5217–5231. doi: 10.1093/nar/gkl677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Machwe A, Xiao L, Groden J, Orren DK. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry. 2006;45:13939–13946. doi: 10.1021/bi0615487. [DOI] [PubMed] [Google Scholar]
- 44.Hu Y, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwendener S, et al. Physical interaction of RECQ5 helicase with RAD51 facilitates its anti-recombinase activity. J Biol Chem. 2010;285:15739–15745. doi: 10.1074/jbc.M110.110478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seong C, Colavito S, Kwon Y, Sung P, Krejci L. Regulation of Rad51 recombinase presynaptic filament assembly via interactions with the Rad52 mediator and the Srs2 anti-recombinase. J Biol Chem. 2009;284:24363–24371. doi: 10.1074/jbc.M109.032953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bugreev DV, Rossi MJ, Mazin AV. Cooperation of RAD51 and RAD54 in regression of a model replication fork. Nucleic Acids Res. 2011;39:2153–2164. doi: 10.1093/nar/gkq1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chi P, et al. Yeast recombination factor Rdh54 functionally interacts with the Rad51 recombinase and catalyzes Rad51 removal from DNA. J Biol Chem. 2006;281:26268–26279. doi: 10.1074/jbc.M602983200. [DOI] [PubMed] [Google Scholar]
- 50.Kwon Y, Chi P, Roh DH, Klein H, Sung P. Synergistic action of the Saccharomyces cerevisiae homologous recombination factors Rad54 and Rad51 in chromatin remodeling. DNA Repair (Amst) 2007;6:1496–1506. doi: 10.1016/j.dnarep.2007.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kwon Y, et al. ATP-dependent chromatin remodeling by the Saccharomyces cerevisiae homologous recombination factor Rdh54. J Biol Chem. 2008;283:10445–10452. doi: 10.1074/jbc.M800082200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sprouse RO, Brenowitz M, Auble DT. Snf2/Swi2-related ATPase Mot1 drives displacement of TATA-binding protein by gripping DNA. EMBO J. 2006;25:1492–1504. doi: 10.1038/sj.emboj.7601050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Debauve G, Capouillez A, Belayew A, Saussez S. The helicase-like transcription factor and its implication in cancer progression. Cell Mol Life Sci. 2007;65:591–604. doi: 10.1007/s00018-007-7392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moinova HR, et al. HLTF gene silencing in human colon cancer. Proc Natl Acad Sci USA. 2002;99:4562–4567. doi: 10.1073/pnas.062459899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.