To uphold genome stability, cells are armed with sophisticated mechanisms to promote timely and accurate duplication of the genome. For instance, stalling of the replication machinery upon encountering blockage, such as a bulky lesion, template DNA with a high tendency for forming secondary structures, or DNA-bound protein(s), ignites a DNA damage response (DDR). The DDR signaling cascade ultimately results in recruitment of many proteins that cooperate to remove or bypass the block, and facilitate replication restart (1). However, in PNAS, Achar et al. (2) report that one protein, HLTF, singlehandedly can accomplish several tasks to deal with replication blocks.
HLTF is one of two mammalian proteins closely related to yeast Rad5, which plays a key role in the error-free branch of the postreplication repair (PRR) pathway of DNA damage tolerance. PRR has two branches, both controlled by the highly conserved Rad6–Rad18 ubiquitin conjugating enzyme complex (3, 4). Rad6–Rad18 monoubiquitinates Lys164 of PCNA, a ring-shaped DNA clamp that tethers DNA polymerase to template DNA. Monoubiquitinated PCNA promotes the translesion DNA synthesis (TLS) branch of PRR in which a specialized polymerase incorporates nucleotides opposite damaged bases, thus allowing lesion bypass and the continuation of DNA replication. TLS is often mutagenic because the same low stringency that allows TLS polymerases to use damaged templates renders them inherently error-prone.
Alternatively, Rad5, together with the Ubc13–Mms2 ubiquitin conjugating complex, governs the error-free branch of damage avoidance. The mechanism of this is not yet well defined, but it is thought to use homologous recombination and DNA template switching by using the undamaged sister chromatid to facilitate replication restart. In one model, this PRR branch involves reversal of the replication fork, wherein the newly replicated strands dissociate from their leading and lagging strand templates and anneal to form a cruciform intermediate, also famously known as the “chicken foot” structure (Fig. 1). Rad5 appears to play a dual role in error-free PRR: (i) Rad5 harbors a ubiquitin ligase activity that, together with Ubc13–Mms2, adds Lys 63-linked polyubiquitin chains to monoubiquitinated PCNA; and (ii) Rad5 is an ATP-dependent DNA translocase that can remodel stalled replication forks to form “chicken feet” in vitro (5). Both of these Rad5 activities are present in HLTF (6–8).
Fig. 1.
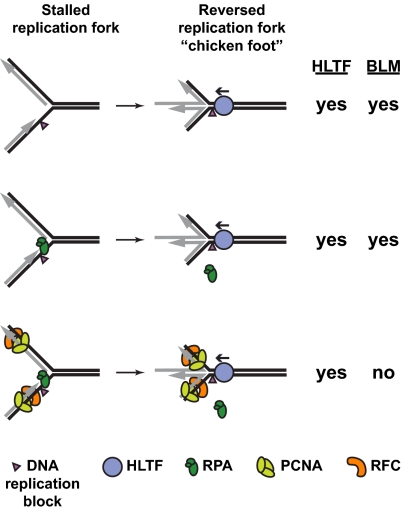
Fork reversal and protein clearing activities of HLTF and BLM. With help from enzymes such as HLTF and BLM, replication blocked on the leading strand can result in a “template switch” whereby the newly synthesized strands (gray) dissociate from their templates (black) and anneal with each other to produce the “chicken foot” structure shown. This facilitates bypass of the block because leading strand DNA synthesis can continue using the undamaged sister chromatid as a template. Whereas HLTF can promote reversal of naked, RPA- and PCNA/RFC-bound model replication forks in vitro, BLM is able only to reverse the naked and RPA-bound forks (2). Blockage of lagging strand replication is dealt with by other mechanisms (1).
The study by Achar et al. (2) is predicated on the premise that stalled replication forks are not naked DNA, but are associated with proteins such as PCNA, the replicative polymerase, and the single-stranded binding protein RPA, that would need to be cleared for fork remodeling and replication restart. By analogy with the chromatin remodeling SWI/SNF family of DNA motor proteins that can move DNA-bound histones (9), Achar et al. (2) asked whether HLTF can displace DNA-bound proteins. They first attached Escherichia coli E111Q EcoRI protein (a catalytically inactive EcoRI restriction enzyme that still binds its cognate DNA target) to model replication forks and then examined its effect on HLTF-stimulated fork reversal. Regardless of whether E111Q EcoRI was bound ahead of or behind the fork, HLTF was readily able to reverse the fork with the same efficiency as naked fork DNA. The authors additionally showed that E111Q EcoRI was ejected from DNA by HLTF, in an ATP-dependent manner; a mutant form of HLTF unable to hydrolyze ATP could not accomplish the feat. Taken together, the results indicate that HLTF can catalyze removal of proteins from DNA via its DNA translocase activity.
The protein mutated in Bloom syndrome (BLM), a member of the RecQ DNA helicase family, is also known to reverse model replication forks in vitro, although, notably, this is not a general feature of RecQ family helicases (10, 11). However, unlike HLTF, BLM did not mediate fork reversal when the substrate was E111Q EcoRI-bound, suggesting that HLTF is unique in its ability to displace replication fork-bound proteins. The authors propose that translocation of HLTF along DNA induces a local change in DNA topology that leads to the ejection of bound proteins, akin to some chromatin remodeling enzymes that displace histones (9). In support of this idea, Achar et al. (2) demonstrated that fork remodeling per se is not required for protein displacement, as E111Q EcoRI can be readily removed by HLTF from linear dsDNA. Thus, it will be interesting to examine whether the protein-clearing function of HLTF is important in other processes such as transcription in which HLTF deficiency causes a defect (12).
Interestingly, Achar et al. (2) further show that HLTF-mediated protein displacement is not limited to dsDNA binding proteins. Replication blockage can lead to uncoupling of leading and lagging strand DNA synthesis, causing an accumulation of ssDNA on the blocked strand that is promptly occupied by the abundant ssDNA-binding protein RPA (1). To model this scenario, a replication fork-like substrate with an ssDNA gap ahead of the “leading” strand was loaded with RPA (or E. coli single-stranded binding protein) and tested for fork reversal. Again, HLTF-mediated fork reversal was unhindered—a surprise considering that HLTF cannot travel on ssDNA (8). One explanation, albeit curious, for this result is that HLTF travels toward the fork from the parental duplex DNA ahead of the fork. Upon reaching the junction, HLTF simultaneously initiates fork reversal and RPA displacement (Fig. 1).
Finally, to mimic more closely an in vivo, protein-bound stalled replication fork, two additional replication factors, PCNA and RFC, which positions PCNA onto the primer–template junction, were loaded on the fork DNA substrate. Again, HLTF (as well as yeast Rad5), but not BLM, was found to regress the fork (2) (Fig. 1).
Thus, HLTF, in addition to its fork reversal and ubiquitin ligase activities, can displace DNA-bound proteins while translocating on dsDNA. The mechanism by which HLTF accomplishes protein-DNA remodeling will be an interesting direction for future studies. Does HLTF translocation alter local DNA topology like other SWI/SNF family proteins? HLTF can completely eject proteins from DNA, as shown for E111Q EcoRI, to play a “cleansing” function (2). However, as PCNA encircles DNA, HLTF likely mediates backward sliding of the ring-shaped molecule along the DNA. In this fashion, PCNA and its associated polymerases at a blocked fork may be retained near the fork for replication restart when the block has been cleared (Fig. 1).
Notably, unlike the inhibition observed for the E111Q EcoRI- and PCNA/RFC/RPA-bound forks, BLM readily regressed the RPA or single-stranded binding protein-bound fork (2) (Fig. 1). BLM was found to interact directly with RPA (13), so it remains possible that this interaction facilitates BLM-mediated fork reversal of RPA-bound substrates. Alternatively, or in addition, because BLM likely translocates on ssDNA as it unwinds the duplex (14), it may displace proteins bound to the ssDNA strand on which it travels. The mechanism of protein clearing by HLTF is quite different from how RecQ helicases, including RECQ5, that can strip the recombinase RAD51 from ssDNA (15), displace DNA-bound proteins.
That HLTF efficiently displaces E11Q EcoRI from DNA implies that this DNA translocase can remove proteins in its path without specifically interacting with them. This property of HLTF distinguishes it from other SWI/SNF protein
One protein, HLTF, singlehandedly can accomplish several tasks to deal with replication blocks.
remodelers that must interact with the target proteins they displace. For instance, physical interaction between the Rad51 recombinase and the SWI/SNF-like Rad54 or Rdh54 is required for efficient displacement of Rad51 from duplex DNA by either DNA motor protein (16, 17). Similarly, MotI, which is also SWI/SNF-like, requires interaction with TBP to pry TBP from gene promoters; MotI first weakens the interaction of TBP with DNA by a translocation-induced DNA topology change, then inserts a protein “latch” into the DNA-binding groove of TBP to prevent rebinding of TBP to DNA (18).
The report by Achar et al. (2) is significant because it demonstrates a protein-clearing activity for a DNA translocase that is also capable of catalyzing replication fork regression. Interestingly, in addition to HLTF and BLM, several other DNA motor/translocase proteins have recently been shown to have fork regression activity, including FANCM (deficient in Fanconi anemia complementation group M) and another RecQ helicase, WRN (deficient in Werner syndrome) (19). Can these proteins also displace DNA-bound proteins during fork regression? Neither fork regression nor protein remodeling activity has been reported for the closest mammalian homologue of HLTF, SHPRH (4). A direct comparison, and potential combinatorial effect, of the activities of HLTF and SHPRH in this regard may be interesting for future in vitro studies, as HTLF and SHPRH interact, and appear to oppose each other in vivo to promote specialized TLS pathways specific for different types of DNA damage (20). One might imagine that HLTF-mediated protein-clearing is not indiscriminate in vivo, but rather there are DNA-bound proteins refractory to eviction by HLTF and/or mechanisms that regulate HLTF access to chromatin. Importantly, fork regression ideally should proceed far enough to bypass the cause of the replication block, but not further, so a mechanism that limits replication fork reversal must exist, whether by a protein-bound factor or a dedicated inhibitor. Finally, important to note is that fork reversal normally only occurs in replication checkpoint-defective cells (1), and so the protein displacement function of HLTF in nondamaged cells may have another function, such as in transcription (12). Future studies will clarify these important issues.
Acknowledgments
Work on DNA repair and DNA motor proteins in our laboratory is supported by research grants from the United States National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
See companion article on page14073.
References
- 1.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 2.Achar YJ, Balogh D, Haracska L. Coordinated protein and DNA remodeling by human HLTF on stalled replication fork. Proc Natl Acad Sci USA. 2011;108:14073–14078. doi: 10.1073/pnas.1101951108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ulrich HD. The RAD6 pathway: Control of DNA damage bypass and mutagenesis by ubiquitin and SUMO. ChemBioChem. 2005;6:1735–1743. doi: 10.1002/cbic.200500139. [DOI] [PubMed] [Google Scholar]
- 4.Unk I, Hajdú I, Blastyák A, Haracska L. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair (Amst) 2010;9:257–267. doi: 10.1016/j.dnarep.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 5.Blastyák A, et al. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Motegi A, et al. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc Natl Acad Sci USA. 2008;105:12411–12416. doi: 10.1073/pnas.0805685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Unk I, et al. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc Natl Acad Sci USA. 2008;105:3768–3773. doi: 10.1073/pnas.0800563105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blastyák A, Hajdú I, Unk I, Haracska L. Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. Mol Cell Biol. 2010;30:684–693. doi: 10.1128/MCB.00863-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorch Y, Maier-Davis B, Kornberg RD. Mechanism of chromatin remodeling. Proc Natl Acad Sci USA. 2010;107:3458–3462. doi: 10.1073/pnas.1000398107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ralf C, Hickson ID, Wu L. The Bloom's syndrome helicase can promote the regression of a model replication fork. J Biol Chem. 2006;281:22839–22846. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- 11.Popuri V, et al. The Human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J Biol Chem. 2008;283:17766–17776. doi: 10.1074/jbc.M709749200. [DOI] [PubMed] [Google Scholar]
- 12.Debauve G, Capouillez A, Belayew A, Saussez S. The helicase-like transcription factor and its implication in cancer progression. Cell Mol Life Sci. 2008;65:591–604. doi: 10.1007/s00018-007-7392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brosh RM, Jr, et al. Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity. J Biol Chem. 2000;275:23500–23508. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- 14.Bachrati CZ, Hickson ID. RecQ helicases: Suppressors of tumorigenesis and premature aging. Biochem J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Y, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Solinger JA, Kiianitsa K, Heyer WD. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol Cell. 2002;10:1175–1188. doi: 10.1016/s1097-2765(02)00743-8. [DOI] [PubMed] [Google Scholar]
- 17.Chi P, et al. Yeast recombination factor Rdh54 functionally interacts with the Rad51 recombinase and catalyzes Rad51 removal from DNA. J Biol Chem. 2006;281:26268–26279. doi: 10.1074/jbc.M602983200. [DOI] [PubMed] [Google Scholar]
- 18.Wollman P, et al. Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP. Nature. 2011 doi: 10.1038/nature10215. 10.1038/nature10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atkinson J, McGlynn P. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 2009;37:3475–3492. doi: 10.1093/nar/gkp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin J-R, Zeman MK, Chen JY, Yee MC, Cimprich KA. SHPRH and HLTF act in a damage-specific manner to coordinate different forms of postreplication repair and prevent mutagenesis. Mol Cell. 2011;42:237–249. doi: 10.1016/j.molcel.2011.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]