Abstract
Ubiquitin mediated protein degradation is crucial for regulation of cell signaling and protein quality control. Poly(ADP-ribose) (PAR) is a cell-signaling molecule that mediates changes in protein function through binding at PAR binding sites. Here we characterize the PAR binding protein, Iduna, and show that it is a PAR-dependent ubiquitin E3 ligase. Iduna’s E3 ligase activity requires PAR binding because point mutations at Y156A and R157A eliminate Iduna’s PAR binding and Iduna’s E3 ligase activity. Iduna’s E3 ligase activity also requires an intact really interesting new gene (RING) domain because Iduna possessing point mutations at either H54A or C60A is devoid of ubiquitination activity. Tandem affinity purification reveals that Iduna binds to a number of proteins that are either PARsylated or bind PAR including PAR polymerase-1, 2 (PARP1, 2), nucleolin, DNA ligase III, KU70, KU86, XRCC1, and histones. PAR binding to Iduna activates its E3 ligase function, and PAR binding is required for Iduna ubiquitination of PARP1, XRCC1, DNA ligase III, and KU70. Iduna’s PAR-dependent ubiquitination of PARP1 targets it for proteasomal degradation. Via PAR binding and ubiquitin E3 ligase activity, Iduna protects against cell death induced by the DNA damaging agent N-methyl-N-nitro-N-nitrosoguanidine (MNNG) and rescues cells from G1 arrest and promotes cell survival after γ-irradiation. Moreover, Iduna facilitates DNA repair by reducing apurinic/apyrimidinic (AP) sites after MNNG exposure and facilitates DNA repair following γ-irradiation as assessed by the comet assay. These results define Iduna as a PAR-dependent E3 ligase that regulates cell survival and DNA repair.
Keywords: PAR binding motif, RING finger, RNF146
Protein ubiquitination is a major regulatory process that controls a variety of cellular functions (1). Covalent modifications of proteins by ubiquitin can either mediate protein interactions or target the proteins for degradation depending on the nature of the ubiquitin modification. Conjugation of ubiquitin to a substrate uses a complex of proteins composed of an E1 ubiquitin activating enzyme, an E2 ubiquitin conjugating enzyme, and an E3 ubiquitin ligase. E3 ligases are involved in substrate recognition and transfer of the ubiquitin molecule to the lysine residue on the substrate. Ubiquitin conjugation is activated and regulated by a few cellular signals (1–3). Phosphorylation is a well studied intracellular signaling motif that marks proteins for the ubiquitination machinery (4). SUMOylation of proteins also appears to be a signal for ubiquitin modification and proteasomal modification (5). Other mechanisms of substrate recognition are not as well characterized.
Poly(ADP-ribose) (PAR) modification (PARsylation) of proteins is an important cellular signaling mechanism (6–8). Proteins are PARsylated by PAR polymerases (PARPs). PARsylation regulates the function of a variety of nuclear proteins. Proteins can be covalently modified by PARP with PAR of different size and complexity, but proteins can also bind PAR noncovalently at specific PAR binding sites to regulate cellular signaling (8, 9). For instance, PAR can act as a cytosolic signaling molecule during parthanatos (PARP1-dependent cell death) (10–13).
RNF146 is a really interesting new gene (RING) finger protein that contains a WWE domain. We previously identified this protein as an N-methyl-D-aspartate (NMDA) glutamate-receptor inducible gene in a genetic screen as clone PLING932 (14). RNF146 was renamed Iduna and was recently shown to possess a PAR binding motif (PBM) and protects against parthanatos via binding to PAR (15). Here we show that Iduna is a PAR-dependent E3 ligase that binds and ubiquitinates both PARsylated and PAR binding proteins via its PBM, marking these proteins for ubiquitin proteasomal degradation. Moreover, Iduna appears to play a prominent role in DNA repair through its PAR-dependent E3 ligase activity.
Results
Iduna Is an E3 Ubiquitin Ligase.
To determine whether Iduna is an E3 ligase, cells were transfected with GFP-Iduna and compared to cells transfected with GFP alone (Fig. S1A in SI Appendix). Following immunoprecipitation of GFP-Iduna or endogenous-Iduna an in vitro ubiquitination assay was performed with recombinant E1, different recombinant E2s (UBCH2, 3, 5a, 5b, 5c, 6, 7, 8, and 10), and ubiquitin. Immunoblotting with antibodies to ubiquitin and GFP reveals that Iduna is ubiquitinated in the presence of UBCH 5a, 5b, 5c, and 6 whereas UBCH 2, 3, 7, 8, and 10 do not support Iduna mediated ubiquitination (Fig. S1A in SI Appendix). The observed ubiquitination is due to Iduna because there is no ubiquitination observed in the absence of Iduna (Fig. S1B in SI Appendix). To confirm that Iduna is autoubiquitinated an in vitro ubiquitination assay was performed with recombinant Iduna, E1, E2 (UBCH 2, 3, 5a, 5b, 5c, 6, 7, 8, and 10), and ubiquitin. In the presence of UBCH 5a, 5b, 5c, Iduna is polyubiquitinated, and in the presence of UBCH 6 Iduna seems to be multimonoubiquitinated because the polyclonal antiubiquitin antibody does not recognize the high molecular weight of autoubiquitinated Iduna catalyzed by UBCH6 (Fig. 1A), although we cannot exclude the possibility that UBCH 6 is capable of supporting Iduna polyubiquitination (Fig. S1A in SI Appendix).
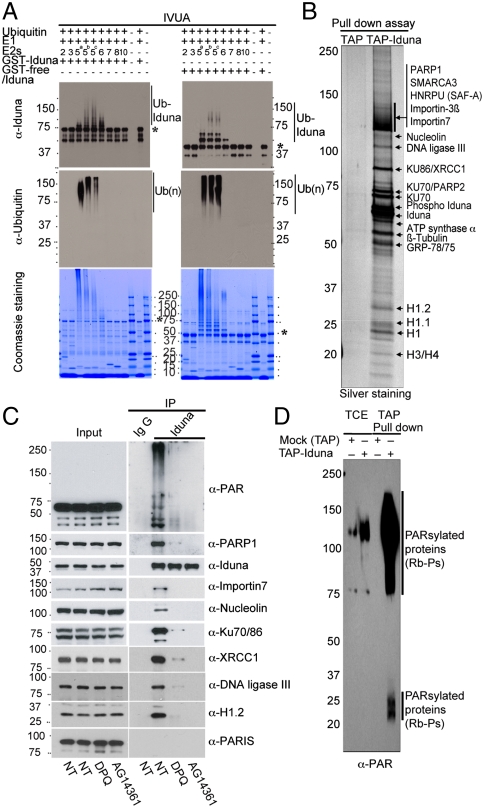
Fig. 1.
Iduna is an ubiquitin E3 ligase that binds PARsylated proteins. (A) Screening of UbcH E2 enzymes for Iduna via an in vitro ubiquitination assay (IVUA) with GST-Iduna (Left) or GST free Iduna (Right). Samples were resolved in 8–16% SDS/PAGE and either stained with coomassie or immunoblotted by anti-Iduna or anti-ubiquitin antibody. (*) indicates unmodified GST-Iduna or GST-free Iduna. (B) Identification of potential Iduna substrates. TAP purification of SK-N-SH cells stably transfected with pNTAP or pNATP-Iduna were resolved in 8–16% SDS/PAGE and silver stained. Mass spectrometric analysis identified 16 proteins as indicated. (C) Iduna interacts with its potential substrates in a PAR-dependent manner. MCF7 cells were preincubated with DMSO or PARP1 inhibitors as indicated and then harvested and lysed. Endogenous Iduna was immunoprecipitated by anti-Iduna antibody from each cell lysate and subjected into immunoblot with appropriate antibodies. IgG was used as a negative control. (D) Iduna strongly binds to PARsylated proteins. TAP or TAP-Iduna pull-down samples were analyzed by immunoblot with anti-PAR antibody. Abbreviations: Ub (n), polyubiquitin chains; Ub-Iduna, poly ubiquitinated Iduna. All experiments were repeated two to three times.
To identify potential Iduna substrates, tandem affinity purification (TAP) was performed with TAP-tagged Iduna (TAP-Iduna) composed of a streptavidin binding peptide (SBP) and calmodulin binding peptide (CBP) fused in frame to the N terminus of Iduna in stably transfected SK-N-SH cells (Fig. 1B). Following the TAP procedure bands were excised and sequenced by mass spectrometry (Table S1 in SI Appendix). Because the TAP results reveal that most of Iduna’s binding proteins are general factors involved in the DNA damage response and Iduna might be a breast cancer risk locus at 6q22.23 (16–18), we elected to perform the remaining studies in the breast cancer MCF-7 cell line. Proteins identified include: PARP1, SMARCA3, HNRPU (SAF-A), Importin-ß3, Importin-7, Nucleoin, DNA ligase III, KU70, KU86, XRCC1, PARP2, Phospho-Iduna, Iduna, ATP-synthase-α, ß-tubulin, GRP-78 and GRP-75, and histones 1.2, 1.1, 1, 3, and 4 (Fig. 1B). Confirmation of the interaction between Iduna and these proteins was performed by immunoprecipitation followed by immunoblot analysis for which there are commercially available antibodies including: PARP1, Importin-7, Nucleoin, DNA ligase III, KU70/86, XRCC1, histone 1.2, Iduna. PARIS (parkin interacting substrate) serves as a negative control (Fig. 1C). To determine whether the interaction between Iduna and its binding partners is dependent on PAR, the PARP inhibitors DPQ or AG14361 were added to the cell culture media before harvest. Both DPQ and AG14361 treatment markedly reduce the interaction between Iduna and its binding partners (Fig. 1C). To confirm that Iduna binds PAR modified proteins the TAP pull-down was probed with antibodies against PAR. TAP pull-down of Iduna markedly enriches for PAR binding proteins as previously reported (Fig. 1D) (15).
Iduna Is a PAR-Dependent E3-Ligase.
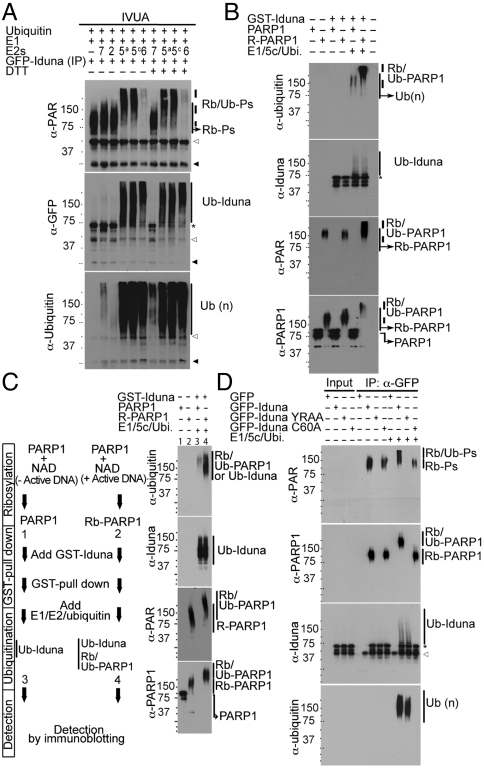
Because Iduna interacts with PAR modified proteins the activity of Iduna ubiquitination of PAR modified proteins was evaluated (Fig. 2). MCF7 cells were transfected with GFP-Iduna, followed by immunoprecipitation with an antibody to GFP. An in vitro ubiquitination assay was performed on the precipitates in the presence of E1, E2 (UBCH 7, 2, 5a, 5c, and 6) and ubiquitin. Immunoblotting with antibodies to PAR reveals that PAR modified proteins are ubiquitinated with the E2s (UBCH 5a, 5c, and 6) whereas there is no ubiquitination with the E2 UBCH 2 or 7 (Fig. 2A). GFP-Iduna is only ubiquitinated in the presence of the E2s UBCH 5a, 5c, and 6. Addition of DTT controls for nonspecific ubiquitination (19) and has no effect on Iduna PAR-dependent ubiquitination. Because PARP1 is a major interacting protein with Iduna (Fig. 1B) (15) and it is the prototypic and prominently modified PAR protein, 2D gel analysis was conducted on the in vitro ubiquitination assay of the GFP-Iduna immunoprecipitate in the presence of E1, UBCH 5a, and ubiquitin to determine whether Iduna ubiquitinates PARP1. Silver staining reveals that GFP-Iduna is shifted to several high molecular weight spots consistent with polyubiqutination (Fig. S2A in SI Appendix). Immunblot analysis with antibodies to PARP1 reveals that PARP1 is similarly shifted to high molecular weight spots consistent with polyubiqutination (Fig. S2B in SI Appendix). To ascertain whether Iduna only ubiquitinates PARsylated PARP1, an in vitro ubiquitination assay in the presence of E1, UBCH 5c, and ubiquitin was utilized to monitor the ubiquitination of non-PARsylated PARP1 versus PARsylated PARP1 (Fig. 2B). PARP1 was PARsylated with biotin-labeled NAD in an in vitro PARsylation reaction. Only PARsylated PARP1 is ubiquitinated by GST-Iduna as revealed by immunoblot analysis with antibodies to ubiquitin (Fig. 2B). A GST pull-down experiment was performed to determine whether Iduna selectively binds and ubiquitinates PARsylated PARP1. Only PARsylated PARP1 binds and is ubiquitinated by Iduna (Fig. 2C).
Fig. 2.
Iduna mediates PARsylation-dependent ubiquitination of its substrates. (A) In vitro ubquitination assay of immunoprecipiated GFP-Iduna and different UbcHE2 enzymes in presence or absence of DTT as indicated. Samples were analyzed by immunoblot with anti-PAR, anti-GFP, and anti-ubiquitin antibodies. White or black arrow heads indicate the immunoglobulin heavy or light chains, respectively. (B) In vitro ubiquitination assay of recombinant PARP1 or PARsylated PARP1 (R-PARP1) by GST-Iduna subjected to immunoblot analysis with indicated antibodies. (C) Iduna binds and/or ubiquitinates PARP1 in a PARsylation-dependent manner. PARP1 or R-PARP1 were incubated with GST-Iduna, followed by GST pull-down and subjected to the in vitro ubiquitination assay (Left) and analyzed by immunoblot (Right). (D) Iduna is a PAR-dependent ubiquitin E3 ligase. In vitro ubiquitination assay of immnuoprecipitated GFP, GFP-Iduna, GFP-Iduna YRAA, and GFP-Iduna C60A analyzed by immunoblot with anti-GFP, anti-ubiquitin, anti-PAR, anti-PARP1, and anti-ubiquitin antibodies. Abbreviations: Rb-P, PARsylated proteins; Rb/Ub-P, PARsylated and polyubiquitinated proteins; Ub (n), polyubiquitin chains; Rb/Ub-PARP1, PARsylated and polyubiquitinated PARP1; Ub-Iduna, poly ubiquitinated Iduna; Rb-PARP1, PARsylated PARP1. All experiments were repeated three times.
Iduna’s E3 Ligase Activity Requires Its RING Domain and PBM.
Two mutations were constructed to disrupt the zinc binding in the RING finger domain of Iduna (H54A and C60A). The ubiquitination activity of Iduna was monitored. MCF7 cells were transfected with GFP-Iduna, GFP-Iduna H54A, or GFP-Iduna C60A, and immunoprecipitation was performed followed by in vitro ubiquitination in the presence of E1, UBCH 5c, and ubiquitin. Iduna possessing point mutations at either H54A or C60A is devoid of ubiquitination activity (Fig. S3A in SI Appendix). An in vitro ubiquitination assay with recombinant GST-Iduna, GST-Iduna H54A, or GST-Iduna C60A also reveals that GFP-Iduna H54A or GFP-Iduna C60A have markedly diminished ubiquitination activity (Fig. S3B in SI Appendix).
Previously we showed that Iduna contains a consensus PBM in its WWE domain and that alanine substitution of the hydrophobic and basic amino acids 156Y and 157R to alanine to create an Iduna YRAA mutant disrupts PAR binding to Iduna (15). MCF7 cells were transfected with GFP-Iduna, GFP-Iduna C60A, and GFP-Iduna YRAA. After 48 hr Iduna was immunoprecipitated with antibodies to GFP followed by immunoblot analysis with antibodies to PAR. Iduna and Iduna C60A bind PAR whereas Iduna YRAA is incapable of binding PAR (Fig. S3C in SI Appendix). To confirm that Iduna binding to PARP1 is dependent on PARsylation of PARP1, binding of automodified PARP1 with 32P-NAD was monitored in a GST pull-down experiment with histone H3 as a positive control (Fig. S4 A and B in SI Appendix). GST-Iduna and GST-Iduna-C60A pulls down PARsylated PARP1 whereas GST-Iduna YRAA fails to pull down PARsylated PARP1. Treatment of the extract prior to GST pull-down with PAR glycohydrolase (PARG), which degrades PAR, eliminates this interaction (Fig. S4A in SI Appendix). Confirmation that PARG is active is the demonstration that PARG dose dependently removes PAR from PARsylated PARP1 (Fig. S4B in SI Appendix). An electrophoretic mobility shift assay reveals that the GST tagged PAR binding proteins histone H3, Iduna, Iduna C60A bind PAR but Iduna YRAA does not (Fig. S4C in SI Appendix). Iduna and Iduna-C60A bind to a range of PAR polymers of varying length similar to H3, whereas Iduna-YRAA fails to bind to PAR, as determined by phosphorimager detection of radiolabeled PAR polymer bound to Iduna, Iduna-C60A, and H3 after separation by Tris-borate-EDTA PAGE (Fig. S4D in SI Appendix).
To determine if PAR binding is required for Iduna ubiquitination, MCF7 cells were transfected with GFP-Iduna, GFP-Iduna C60A, and GFP-Iduna YRAA. Following immunoprecipitation with antibodies to GFP an in vitro ubiquitination assay was performed. Iduna YRAA fails to bind PARsylated PARP1 whereas Iduna and Iduna C60A bind PARsylated PARP1. Immunoblot analysis of the immunoprecipitates with antibodies to PAR and ubiquitin reveals that only GFP-Iduna is capable of polyubiquitination of PARsylated PARP1, whereas Iduna YRAA autoubiquitinates itself (Fig. 2D). Thus, Iduna has PAR-dependent and -independent E3 ligase activity.
To determine whether free PAR can activate Iduna ubiquitination, an in vitro ubiquitination assay containing Iduna, E1, UBCH 5c, and ubiquitin was performed. Iduna autoubiquitination is increased with increasing concentrations of PAR and the addition of PARG in a dose-dependent manner reduces Iduna autoubiquitination to baseline (Fig. S5A in SI Appendix). In the same in vitro ubiquitination reaction, PARP1 ubiquitination was monitored in the presence of Iduna and Iduna YRAA. PARP1 ubiquitination is dose-dependently increased by Iduna in the presence of PAR and decreased by the addition of PARG. Iduna YRAA fails to ubiquitinate PARP1 in the presence of PAR (Fig. S5B in SI Appendix).
Mass spectrometry analysis was performed to ascertain the conjugation mode, the site of PAR-dependent ubiquitination of PARP1, and autoubiquitination of Iduna. In the absence of PAR, Iduna autoubiquitination occurs on lysines 85, 95, and 176 via K11 and K48 ubiquitin linkages (Table S2 in SI Appendix) whereas in the presence of PAR, lysines 131 and 176 are ubiquitinated via K6, K33, and K48 ubiquitin linkages (Table S2 in SI Appendix). High resolution mass spectrometry also indicated that PARP1 was ubiquitinated on 24 different lysines via K11 and K48 ubiquitin linkages (Table S3 in SI Appendix).
To ascertain if Iduna ubiquitinates other proteins in a PAR-dependent fashion, an in vitro ubiquitination assay was performed (Fig. S6 in SI Appendix). In the presence of E1, UbcH 5c, Iduna and free PAR polymer, Iduna ubiquitinates the nuclear proteins XRCC1, KU70, DNA ligase III, PARP1, but not the cytosolic ATP subunit α (Fig. S6 in SI Appendix). The ubiquitination is PAR-dependent because the addition of PARG to the reaction ablates the ubiquitination.
Iduna Targets PARsylated PARP1 for Ubiquitin Proteasomal Degradation.
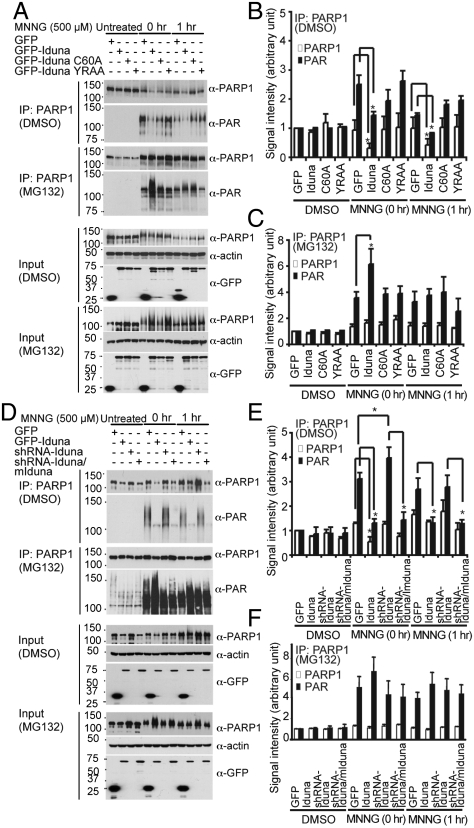
To determine whether Iduna targets PARsylated PARP1 for ubiquitin proteasomal degradation, stably transfected MCF7 cells expressing GFP-Iduna, GFP-Iduna C60A, and GFP-Iduna YRAA and cells stably expressing a shRNA to Iduna were examined (Fig. 3). shRNA for human Iduna effectively knocks down the expression of human Iduna and mouse Iduna serves to rescue Iduna knockdown (Fig. S7 in SI Appendix). PARP1 was activated with the DNA damaging agent N-methyl-N-nitro-N-nitrosoguanidine (MNNG) followed by immunoprecipitation of PARP1. MNNG treatment did not change the overall levels of PARP1. However there is a shift in its molecular weight due to autoPARsylation, and there is an almost threefold increase in PARsylated PARP1 immediately following the MNNG treatment (Fig. 3 A and B). GFP-Iduna leads to a significant reduction in PARP1 and PARsylated PARP1. GFP-Iduna C60A or Iduna YRAA have no effect on PARP1 or PARsylated PARP1 levels (Fig. 3 A and B). One hr post-MNNG treatment total PARP1 levels and PARsylated PARP1 levels are significantly reduced by GFP-Iduna, but not by GFP-Iduna C60A or YRAA. In the presence of the proteasome inhibitor, MG132, Iduna fails to diminish the levels of PARP1 and PARsylated PARP1 (Fig. 3A and C) confirming that Iduna targets PARsylated PARP1 for ubiquitin proteasomal degradation. The effect of knockdown of Iduna with shRNA on the levels of PARP1 and PARsylated PARP1 was evaluated. shRNA to Iduna prevents the reduction in PARP1 and PARsylated PARP1 following MNNG treatment (Fig. 3 D and E). An shRNA-resistant mouse Iduna decreases the levels of PARP1 and PAR modified PARP1 in the presence of the shRNA to human Iduna indicating that the effects observed with shRNA Iduna are specific (Fig. 3 D and E). In the presence of MG132 the levels of PARP1 and PARsylated PARP1 remain elevated following MNNG treatment (Fig. 3 D and F). These results taken together suggest that Iduna ubiquitinates PARP in a PAR-dependent manner leading to its proteasomal degradation.
Fig. 3.
PARsylation-dependent PARP1 degradation by Iduna. (A) Stable MCF7 cell lines expressing GFP, GFP-Iduna, GFP-Iduna C60A, or GFP-Iduna YRAA were exposed to DMSO or MNNG (500 μM) for 15 min with or without MG132. PARP1 was immunoprecipitated at 0 or 1 hr after the MNNG challenge. PARP1 and PARsylated-PARP1 were monitored by immunoblot with anti-PARP1 and anti-PAR antibodies. (B) Quantification of PARP1 and PARsylated-PARP1 in the absence of MG132. (C) Quantification of PARP1 and PARsylated-PARP1 in presence of MG132. Quantifications were normalized with respect to actin levels. (D) Levels of immunoprecipitated PARP1 and PARsylated PARP1 after exposure to DMSO or MNNG (500 μM) for 15 min with or without MG132 GFP in MCF7 cell lines stably expressing GFP-Iduna, shRNA-Iduna, or shRNA-Iduna/GFP-mouse Iduna (mIduna) at 0 or 1 hr after the MNNG challenge. (E) Quantification of PARP1 and PARsylated PARP1 normalized to actin in absence of MG132. (F) Quantification of the PARP1 and PARsylated-PARP1 normalized to actin in presence of MG132. Data represents mean ± s.e.m., n = 3, * P < 0.05 by ANOVA with Tukey-Kramer’s post hoc test. All experiments were repeated two to three times.
Iduna Regulates the DNA Damage Response.
The PAR-dependent association and ubiquitination of known DNA repair factors PARP1, PARP2, XRCC1, KU70, and DNA ligase III suggested a possible role for Iduna in the DNA damage response. To investigate the role of Iduna in the DNA damage response the recruitment of GFP-Iduna to sites of DNA damage induced by laser microirradiation was assessed (Fig. 4). GFP-Iduna begins to translocate to the nucleus and concentrate at the microirradiation site immediately after the laser microirradiation (Fig. 4 A and B). The translocation of GFP-Iduna peaks between 3 to 4 min (Fig. 4 A and B). The recruitment of GFP-Iduna to laser-induced DNA breaks requires PARP activation because the PARP inhibitor AG14361 blocks the translocation of Iduna (Fig. 4 A and B). PAR binding to Iduna is also required for the translocation because the Iduna YRAA mutant, which is defective for PAR binding, is not recruited to laser-induced DNA breaks (Fig. 4 A and B). GFP-Iduna localizes to sites of laser-induced DNA breaks, as marked by γH2AX immunostaining (Fig. 4C).
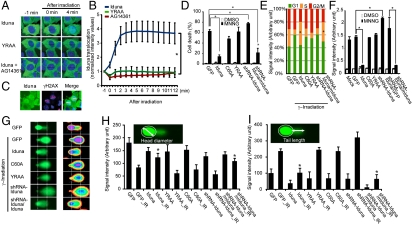
Fig. 4.
Iduna protects against DNA damage. (A) Recruitment of stably expressed GFP-Iduna to sites of laser (405 nm) microirradiation induced DNA damage in MCF7 cells. GFP-Iduna YRAA does not translocate to the damage site. The PARP inhibitor AG14361 blocks GFP-Iduna recruitment. (B) Comparative quantitative analysis of GFP-Iduna, GFP-Iduna-YRAA, and GFP-Iduna plus PARP inhibitor AG14361 kinetics after DNA damage. (C) GFP-Iduna localizes to sites of DNA damage as indicated by colocalization with γH2AX immunostaining. (D) Stable MCF7 cell lines expressing GFP, GFP-Iduna, GFP-Iduna C60A, GFP-Iduna YRAA , shRNA-Iduna, or shRNA-Iduna/GFP-mouse Iduna (mIduna) were treated with DMSO or MNNG (500 μM) for 15 min. After 24 hr, the cells were stained with Hoechst 33342 and propidium iodide (PI), and dead cells were counted by automated computer-assisted program. (E) Stable MCF7 cell lines were γ-irradiated at 2 Gy as indicated. Cells were collected 16 hr after irradiation and then DNA content was measured by flow cytometry. The percentage of each cell cycle phase was measured by FlowJo software using the Dean-Jett-Fox model. (F) Stable MCF7 lines as indicated were treated with either DMSO or MNNG. After 1 h, genomic DNA was isolated and then AP sites on genomic DNA were labeled with biotin by Aldehyde Reactive Probe (ARP) reagent. Biotin-labeled AP sites were quantified using an avidin—biotin assay. (G) Stable MCF7 lines were γ-irradiated at 2 Gy as indicated. After 15 min, cells were collected and then subjected to the comet assay. (H) Quantification of head diameter after comet assay. (I) Quantification of tail length after comet assay. Data represents mean ± s.e.m., n = 3, * P < 0.05 by ANOVA with Tukey-Kramer’s post hoc test. All experiments were repeated three to four times.
The sensitivity of MCF7 cells to DNA damage induced by MNNG or γ-irradiation in the setting of Iduna overexpression and shRNA Iduna knockdown was assessed (Fig. 4 D and E). Iduna overexpression dramatically rescues MCF7 cells from MNNG-induced cell death (Fig. 4D). The rescue requires Iduna’s E3 ubiquitin ligase activity because the Iduna C60A mutant that lacks E3 ligase activity is not protective (Fig. 4D). Moreover, PAR binding of Iduna is also required because Iduna YRAA that lacks PAR binding is also not protective (Fig. 4D). shRNA knockdown of Iduna enhances MNNG toxicity, which is reversed by overexpressing mouse Iduna that is resistant to shRNA Iduna knockdown (Fig. 4D). Following γ-irradiation of MCF7 cells, Iduna overexpression rescues cells from G1 arrest in the cell cycle and promotes cell survival (Fig. 4E). The rescue requires Iduna’s E3 ubiquitin ligase activity because the Iduna C60A mutant that lacks E3 ligase activity is not protective (Fig. 4E). Moreover, PAR binding of Iduna is also required as Iduna YRAA that lacks PAR binding is also not protective (Fig. 4E). shRNA knockdown of Iduna has comparable effects to the GFP control following γ-irradiation, which is reversed by overexpression of mouse Iduna that is resistant to shRNA Iduna knockdown (Fig. 4E).
To ascertain if Iduna may be involved in DNA repair, the level of apurinic/apyrimidinic (AP) sites, which are one of the major types of DNA lesions formed during the course of base excision and repair, was assessed (Fig. 4F). Following DNA damage induced by MNNG there is an eightfold increase in the number of AP sites that is completely prevented by Iduna overexpression (Fig. 4F). The prevention of the increase in AP sites by Iduna following MNNG requires PAR binding of Iduna because Iduna YRAA, which lacks PAR binding, still leads to an eightfold increase in AP sites (Fig. 4F). Moreover, Iduna’s E3 ubiquitin ligase activity is required for the reduction in AP sites because Iduna C60A, which is devoid of E3 ubiquitin ligase activity, fails to reduce the number of AP sites induced by MNNG treatment (Fig. 4F). shRNA knockdown of Iduna increases the number of AP sites by almost 14-fold after DNA damage induced by MNNG (Fig. 4F). Overexpression of mouse Iduna that is resistant to the shRNA knockdown of Iduna prevents the increase in the number of AP sites (Fig. 4F).
To confirm that Iduna facilitates DNA repair, the alkaline comet assay was performed. The comet assay detects DNA fragmentation by monitoring DNA integrity by SYBR green staining during electrophoresis of cells (20). Cells with intact DNA have compact circular staining, whereas cells with DNA damage have bright tails that resemble comets. MCF7 cells were treated with γ-irradiation (2 Gy) in the setting of Iduna overexpression and shRNA Iduna knockdown (Fig. 4 G–I). Iduna overexpression dramatically prevents the reduction in head diameter and increase in tail length in MCF7 cells treated with γ-irradiation compared to GFP control MCF7 cells (Fig. 4 G–I). These effects require Iduna’s E3 ubiquitin ligase activity because the Iduna C60A mutant that lacks E3 ligase activity does not prevent the reduction in head diameter and increase in tail length (Fig. 4 G–I). Moreover, PAR binding of Iduna is also required because Iduna YRAA that lacks PAR binding also does not prevent the reduction in head diameter and increase in tail length (Fig. 4G–I). shRNA knockdown of Iduna enhances the reduction in head diameter and increase in tail length in MCF7 cells treated with γ-irradiation compared to GFP control MCF7 cells, which is reversed by overexpression of mouse Iduna that is resistant to shRNA Iduna knockdown (Fig. 4 G–I). The data are summarized in Fig. S8 in SI Appendix.
Discussion
Our findings indicate that Iduna is a PAR-dependent ubiquitin E3 ligase that regulates cell survival and the DNA damage response. We recently reported that Iduna protects the brain from glutamate excitotoxicity and stroke by interfering with PAR induced cell death (parthanatos) via Iduna’s PBM (15). Iduna contains a PBM specified by a sequence of approximately 20 amino acids containing N-terminal basic amino acids and a C-terminal region containing alternating hydrophobic and basic amino acids (9, 15, 21). Mutating key residues in Iduna’s PBM (Y156A and R157A) eliminates its protective function in DNA damage induced cell death, glutamate excitotoxicity, and stroke as well as its ubiquitin E3 ligase activity, thus coupling Iduna’s protective function to its ubiquitin E3 ligase activity. Consistent with this notion is our observation that the E3 ligase inactive mutant, Iduna C60A, is not protective against DNA damage induced cell death. Iduna is thought to regulate cell survival via the prevention of the release of apoptosis inducing factor (AIF) after cellular injury (15). Because Iduna does not directly inhibit PARP1 activity (15) its protective effects are likely to be mediated through the PAR-dependent ubiquitination and ubiquitin proteasomal dependent elimination of a cell-death effector that is PARsylated and/or contains a PBM.
A number of proteins contain PBMs, suggesting a broad role for PAR in regulating protein function and expression (9, 21). However, the prominent role of basic amino acids as determinants of PAR binding in the PBM raised questions of specificity related to the general affinity of basic amino acids within the PBM for charged polymers (6). Our results define the PBM as a functional motif that acts a molecular switch to turn on Iduna’s E3 ligase activity. Consistent with this idea is our observation that Iduna’s autoubiquitination and ubiquitination of PAR binding proteins is increased by PAR. It is likely that PAR binding activates Iduna by inducing changes in the structure of Iduna that relieve steric hindrance. AIF was recently shown to contain a similar PBM that is separate from its DNA binding domain, and upon PAR binding AIF is released from the mitochondria to induce cell death (11). Together, these results indicate that the PBM is a functionally important protein motif. Because the PBM is commonly found in human proteins, we hypothesize that the PAR-dependent molecular switch mechanism found for Iduna will be extended to other PAR binding proteins.
Iduna potently regulates the DNA damage response (Fig. S8 in SI Appendix). Iduna’s participation in the DNA damage response is likely to be complex as it also inhibits the translocation of AIF, which is required for DNA fragmentation in certain cell death paradigms such as parthanatos (22). Although Iduna is prominently localized to the cytosol, it partially translocates to the nucleus after cellular injury (15). We show here that it becomes enriched on chromatin in response to UV-induced DNA damage placing it in position to regulate the DNA damage response. PARP1 is also recruited to DNA damage sites and, through PARsylation of itself and other proteins at sites of DNA damage, it is thought to facilitate DNA repair through chromatin relaxation. PAR also acts as a loading platform that recruits multiple repair factors through noncovalent interactions. In addition to PBM-containing proteins that accumulate at DNA damages sites in a PAR-dependent fashion (e.g., MRE11, NBS1, CHD4) other proteins will bind PAR through specialized modules such as the PAR-binding zinc-finger (PBZ) domain (e.g., APFL, CHFR) or the macrodomain (e.g., ALC1) (6, 23). The ability of Iduna to target PARsylated and PAR binding proteins for ubiquitin proteasomal degradation via its WWE domain adds a new level of complexity to the role of PAR and protein stability in the DNA damage response.
While this work was in preparation, another study reported that Iduna functions as a PAR-dependent E3 ligase, supporting our findings. In contrast, they find that Iduna regulates axin and Wnt signaling linking tankyrase-dependent PARsylation to ubiquitination (24). Because tankyrase is thought to add short and low complexity PAR to proteins whereas PARP1 adds long and complex PAR to proteins, the range of Iduna actions is probably only limited by its localization to subcellular compartments (8). Thus, Iduna is likely to regulate a variety of cellular processes where PARsylation plays a role. For instance, Iduna was originally identified in a screen for NMDA receptor-induced cell survival genes (14). In the brain, Iduna is protective, and this protection requires PAR binding (15). We suspect that Iduna’s PAR-dependent ubiquitin E3 ligase activity is required for this protection. Future studies will be focused on identifying Iduna substrates, determining the extent of Iduna’s actions and its potential dynamic range of activation by PAR polymer.
Other proteins with RING finger containing proteins with PAR-binding domains such as CHFR or other WWE containing proteins, such as HUWE1, TRIP12, DTX1 are likely to function as PAR-dependent E3 ligases (25, 26). Thus Iduna likely represents the first in its class of PAR-dependent E3 ligases. Collectively these data define a PAR-dependent ubiquitin E3 ligase and indicate a mechanism by which cells use PAR-dependent interactions to regulate protein levels.
Materials and Methods
Plasmids and Antibodies.
To generate Iduna’s mutant plasmids, site-directed mutagenesis was carried out using the QuickChange site-directed mutagenesis kit (Stratagene) as detailed in SI Materials and Methods in SI Appendix.
Lentiviral Preparations for Over Expression.
Invitrogen ViraPower lentiviral packaging system was employed for high-titer viral preparations for effective transduction as detailed in SI Materials and Methods in SI Appendix.
Stable Cell Lines.
MCF7 stable cells expressing GFP, GFP-Iduna, GFP-Iduna C60A and GFP-Iduna YRAA, were established by infection using each lentiviral particles. Iduna knockdown MCF7 cells were selected by puromycin after transfection of RNAi TRC clones from Open Biosystem as detailed in SI Materials and Methods in SI Appendix.
Tandem Affinity Purification.
Iduna’s substrates were isolated using the Interplay mammalian TAP system (Stratagene) as detailed in SI Materials and Methods in SI Appendix.
Mass Spectrometric Analysis.
Mass spectrometry analysis was performed by the Taplin Biological Mass Spectrometry Facility (Harvard Medical School).
In Vitro Ubiquitination Assay.
The autoubiquitination activity of GST-Iduna, GST free Iduna, E1 (50 nM), UbcHs (50 nM) and Iduna (IP samples or recombinant protein) were assessed as detailed in SI Materials and Methods in SI Appendix.
Synthesis of [32P] and Biotin-Labeled PARP1 and Purification of PARP-Free PAR Polymer.
Automodified PARP1 and free PAR polymer were purified as previously described (15) and as detailed in SI Materials and Methods in SI Appendix.
Cellular and Biochemical Assays.
PAR pull downs, EMSA, two-dimensional gel electrophoresis–Western blot (2DE-WB), in vivo PARP1 stability assays, cell-death assays, immunoprecipitations, comet assays, cell cycle analysis, and determination of apurinic/apyrimidinic (AP) sites were performed as detailed in SI Materials and Methods in SI Appendix.
Supplementary Material
Acknowledgments.
This work was supported by grants from the National Institutes of Health (NS039148, NS067525, DA000266, and NS051764) and the McKnight Endowment for the Neurosciences. S.S.A. is an American Heart Research postdoctoral fellow. G.G.P. was supported by a Canadian Institutes of Health Research grant and holds a Canada research chair in proteomics. T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108799108/-/DCSupplemental.
References
- 1.Ciechanover A. The ubiquitin-proteasome pathway: On protein death and cell life. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Fiore PP, Polo S, Hofmann K. When ubiquitin meets ubiquitin receptors: A signalling connection. Nat Rev. 2003;4:491–497. doi: 10.1038/nrm1124. [DOI] [PubMed] [Google Scholar]
- 3.Hochstrasser M. New structural clues to substrate specificity in the “ubiquitin system”. Mol Cell. 2002;9:453–454. doi: 10.1016/s1097-2765(02)00486-0. [DOI] [PubMed] [Google Scholar]
- 4.Harper JW. A phosphorylation-driven ubiquitination switch for cell-cycle control. Trends Cell Biol. 2002;12:104–107. doi: 10.1016/s0962-8924(01)02238-3. [DOI] [PubMed] [Google Scholar]
- 5.Prudden J, et al. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–4101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krishnakumar R, Kraus WL. The PARP side of the nucleus: Molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): Novel functions for an old molecule. Nat Rev. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 9.Gagne JP, et al. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrabi SA, et al. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci USA. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, et al. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos) Sci Signal. 2011;4:ra20. doi: 10.1126/scisignal.2000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu SW, et al. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc Natl Acad Sci USA. 2006;103:18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu SW, et al. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 14.Hong SJ, Li H, Becker KG, Dawson VL, Dawson TM. Identification and analysis of plasticity-induced late-response genes. Proc Natl Acad Sci USA. 2004;101:2145–2150. doi: 10.1073/pnas.0305170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andrabi SA, et al. Iduna protects the brain from glutamate excitotoxicity and stroke by interfering with parthanatos (poly (ADP-ribose) dependent cell death) Nat Med. 2011 doi: 10.1038/nm.2387. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gold B, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22. Proc Natl Acad Sci USA. 2008;105:4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirchhoff T, et al. The 6q22. 33 locus and breast cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 2009;18:2468–2475. doi: 10.1158/1055-9965.EPI-09-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menachem TD, Laitman Y, Kaufman B, Friedman E. The RNF146 and ECHDC1 genes as candidates for inherited breast and ovarian cancer in Jewish Ashkenazi women. Fam Cancer. 2009;8:399–402. doi: 10.1007/s10689-009-9255-7. [DOI] [PubMed] [Google Scholar]
- 19.Woelk T, et al. Molecular mechanisms of coupled monoubiquitination. Nat Cell Biol. 2006;8:1246–1254. doi: 10.1038/ncb1484. [DOI] [PubMed] [Google Scholar]
- 20.Olive PL, Durand RE, Banath JP, Johnston PJ. Analysis of DNA damage in individual cells. Methods Cell Biol. 2001;64:235–249. doi: 10.1016/s0091-679x(01)64016-0. [DOI] [PubMed] [Google Scholar]
- 21.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Dawson VL, Dawson TM. Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos. Exp Neurol. 2009;218:193–202. doi: 10.1016/j.expneurol.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chou DM, et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci USA. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, et al. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat Cell Biol. 2011;13:623–629. doi: 10.1038/ncb2222. [DOI] [PubMed] [Google Scholar]
- 25.Ahel I, et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 26.Aravind L. The WWE domain: A common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem Sci. 2001;26:273–275. doi: 10.1016/s0968-0004(01)01787-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.