Abstract
Enamel is a highly organized hierarchical nanocomposite, which consists of parallel arrays of elongated apatitic crystallites forming an intricate three-dimensional microstructure. Amelogenin, the major extracellular matrix protein of dental enamel, regulates the formation of these crystalline arrays via cooperative interactions with forming mineral phase. Using cryoelectron microscopy, we demonstrate that amelogenin undergoes stepwise hierarchical self-assembly. Furthermore, our results indicate that interactions between amelogenin hydrophilic C-terminal telopeptides are essential for oligomer formation and for subsequent steps of hierarchical self-assembly. We further show that amelogenin assemblies stabilize mineral prenucleation clusters and guide their arrangement into linear chains that organize as parallel arrays. The prenucleation clusters subsequently fuse together to form needle-shaped mineral particles, leading to the formation of bundles of crystallites, the hallmark structural organization of the forming enamel at the nanoscale. These findings provide unique insight into the regulation of biological mineralization by specialized macromolecules and an inspiration for bottom-up strategies for the materials design.
Biominerals are nanostructured composite materials, formed via dynamic interactions between supramolecular assemblies of biomacromolecules and forming mineral phases. Dental enamel is a prime example of a biomineralization system, which serves as a model for understanding basic mechanisms that regulate biomineral formation (1–3). Enamel is the most mineralized tissue in the body, possessing exceptional mechanical properties that combine high hardness with remarkable resilience (4), which are determined largely by its unique structural organization. The building blocks of enamel, the enamel rods, are densely packed arrays of elongated carbonated apatite crystals organized into an intricate interwoven structure (5) (Fig. S1). Although mature enamel contains almost no organic material, matrix proteins comprise roughly a third of the forming enamel volume. The major protein of forming enamel, amelogenin, plays essential roles in regulation of the mineralization and structural organization of this tissue (6, 7). Amelogenin transiently stabilizes amorphous calcium phosphate (ACP) and regulates the formation of parallel arrays of mineral crystallites (8, 9). It is comprised of three domains: an N-terminal tyrosine-rich amelogenin peptide (TRAP), a charged C-terminal hydrophilic telopeptide (C telopeptide), and a central domain rich in X-Y-Pro repeats (2). Amelogenin in solution is globally unfolded with some regions containing extended β-sheets and polyproline type II helices (10, 11). The TRAP and C-telopeptide domains are involved in molecular recognition between amelogenin molecules, which is essential for proper enamel formation (7). Amelogenin self-assembles at pH values above 6.5 forming nanospheres, spherical (12, 13) or oblate (14) structures 10–20 nm in diameter, which are believed to play a key role in the mineralization and structural organization of enamel. When secreted, amelogenin undergoes a series of proteolytic cleavages starting at the C telopeptide (15). Although amelogenin lacking the C telopeptide forms nanospheres similar to those of a full-length protein (16), they loose the ability to assemble into chains (12, 17) and to organize mineral particles (1, 8), suggesting a unique role for the full-length molecule in regulation of mineralization. Recent experiments with transgenic animals also indicate that the full-length molecule is absolutely essential for enamel formation (18).
Results and Discussion
To gain in-depth insight into the molecular mechanisms of amelogenin function in biomineralization, we conducted a cryoelectron microscopy (cryo-EM) study of amelogenin self-assembly and calcium phosphate mineralization using recombinant full-length murine amelogenin rM179 and its truncated version, rM166, that lacks the 13-residue C telopeptide.
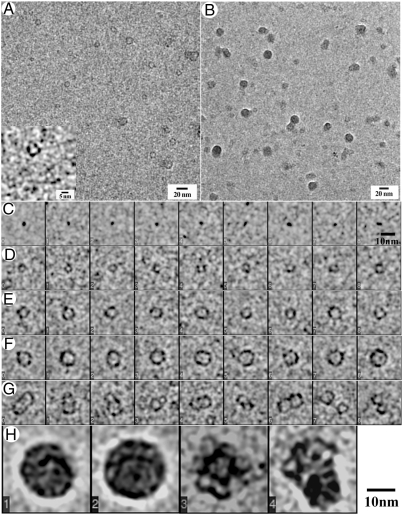
We observed that the self-assembly of rM179 is a hierarchical multistep process. After 1 min of incubation in PBS at pH 8.0, the majority of the protein was present as monomers and oligomers (Fig. 1A). We ascribed the slightly elongated particles less than 3 nm in diameter as monomers (Fig. 1B), whereas rings with diameters ranging from 3 to 10 nm were identified as oligomers (Figs. 1 D–F). Variations in both the size and shape of these oligomers suggest that these assemblies are structurally flexible. Another interesting particle class observed contains pairs and sometimes groups of three or more individual oligomers (Fig. 1G). After 5 min, larger oligomers (6–10 nm in diameter) comprise the major fraction of particles, while smaller aggregates are still numerous (Fig. S2). We also observed a few larger round aggregates, 15–20 nm in diameter, which became the predominant species after 10 min of incubation (Figs. 1 B and H, and Fig. S3 A–D), which were identified as amelogenin nanospheres (12–14). A few short chains of the nanospheres were also observed at this stage (Fig. S3 E–I). Currently, we do not have sufficient data to conclusively determine the morphology of the amelogenin nanospheres (i.e., oblate or spherical), for which additional studies combining cryo-EM and electron tomography will be needed. Importantly, our analysis of the nanospheres revealed that they are not homogeneous but have an internal structure consisting of small doughnut-shaped elements, 5–10 nm in diameter (Fig. 1H and Fig. S3 A–D), structurally similar to amelogenin oligomers found at the earlier time points. These observations provide direct evidence that the nanospheres of full-length amelogenin form via an assembly of intermediate oligomeric subunits, as has been previously proposed (3, 19, 20).
Fig. 1.
Cryo-EM micrographs of rM179 after 1 (A) and 10 min (B) in PBS at pH 8.0. Different classes of rM179 assemblies found in the sample after 1 min of incubation: (C) monomers, (D–F) oligomers of different sizes, (G) pairs of oligomers. After 10 min, (H) nanospheres
In contrast, no intermediate oligomers were observed during the assembly of rM166, the truncated version of murine amelogenin lacking the C telopeptide. In these samples, the major populations of particles observed after 1 min were monomers and random aggregates containing 3–6 individual protein molecules (Fig. S4A). Over a 10-min span, these random aggregates grew in size and started to form nanospheres. At 10 min, 10–20-nm diameter nanospheres were seen (Fig. S4 B and C) that were similar in size to those reported in the literature (12). Unlike rM179 nanospheres, those of rM166 lacked any noticeable internal structure (Fig. S5 B and C). These results indicate that the assembly mechanism of full-length amelogenin is significantly different from that of truncated amelogenin.
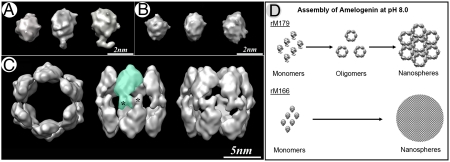
To further explore the mechanism of the amelogenin self-assembly process, we conducted three-dimensional single-particle reconstructions from cryo-EM images of amelogenin rM179 monomers, rM166 monomers, and the most abundant oligomer class of rM179 (53%) containing dodecamers (Fig. 2 and Figs. S5 and S6). A single-particle reconstruction of the rM179 monomer, reveals an overall oval shape with a foot-like extension on one end (Fig. 2A and Movie S1). Significantly, although rM166 monomer is similar in size and shape to the full-size molecule, it lacks the foot-like extension (Fig. 2B and Movie S2). These results strongly suggest that this extension in the full-length amelogenin molecule is its C-terminal hydrophilic telopeptide.
Fig. 2.
Single-particle reconstruction models of rM179 monomers (A), rM166 monomers (B), and rM179 dodecamers (C). Asterisks indicate the extension (C telopeptide) in monomers and dodecamers. Green shading roughly outlines the contour of one monomer in the dodecamer. (D) Models of rM179 self-assembly and rM166 aggregation into nanospheres.
Amelogenin exists in a globally disordered state under acidic conditions (10) and undergoes significant conformational changes upon pH-triggered assembly, manifested by a sizeable increase in β-sheet content (11) (Fig. S7). In both rM179 and rM166 samples, we observed monomers with relatively stable structures at earlier time points, suggesting that pH-triggered conformational changes from disordered to folded states in these proteins might occur prior to assembly. These conformational changes might further trigger specific protein–protein interactions and the self-assembly of amelogenin into oligomers. At the same time, other factors triggering the assembly process cannot be ruled out, such as a decrease in the electrostatic repulsion and enhancement of hydrophobic interactions between amelogenin molecules at pH values around the protein isoelectric point.
Remarkably, the rM179 dodecamer structure reveals six dimers aligned as a double-ring barrel (Fig. 2C and Movie S3), with the charged C telopeptides forming a rim in the equatorial plane of the structure. The dimer subunits are connected via the foot-like extensions observed in the full-length monomer, suggesting that the C termini form the dimer interface. This observation is consistent with a previous proposal that C-terminal domains of amelogenin bind to each other via electrostatic interactions between alternating negatively and positively charged amino acids (21). Furthermore, our dimerization model explains why intermediate oligomeric structures are absent in the rM166 sample, which instead forms aggregates and nanospheres lacking internal structure. We propose that there are two different mechanisms involved in the assembly of the full-length amelogenin and aggregation of the protein-lacking C telopeptide (Fig. 2D). We believe that the full-length molecules form dimers via molecular recognition between the C telopeptide, whereas less specific interactions between other parts of the molecule are responsible for the assembly of dimers into oligomers and further assembly of oligomers into nanospheres. In contrast, interactions between rM166 monomers are likely brought about through nonspecific hydrophobic bonds between individual monomers leading to their aggregation. These findings have important mechanistic implications. Specifically, they indicate that the removal of C telopeptide results in a loss of the ability of hierarchical assembly through intermediate oligomers to higher order structures
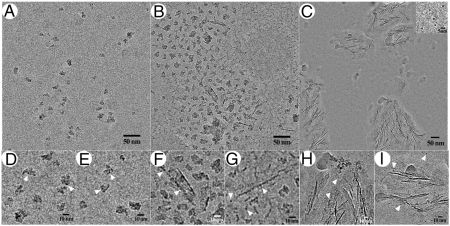
To better understand the impact of structural differences between full-length and truncated amelogenin on mineral formation and organization, we have conducted a series of mineralization experiments with and without rM179 and rM166. In the control samples, 10 min into the mineralization reaction, isolated mineral nanoparticles were observed (Fig. S8A). The average size of these nanoparticles after defocus correction was 9.2 Å (SD = 2.2 Å, N = 55) similar to the prenucleation clusters which were recently demonstrated by Sommerdijk and coworkers in a calcium phosphate mineralization system using cryo-EM (22, 23). The idea of roughly spherical clusters, 9.5 Å in diameter, with the chemical formula of Ca9(PO4)6 was originally proposed by Posner and Betts in the 1970s based upon their studies of calcium phosphate mineralization using the X-ray radial distribution method (24). These clusters, often identified as Posner clusters, are thought to be building blocks of both amorphous calcium phosphate as well as apatitic mineral phases (24, 25), which is of particular importance for biomineralization systems where metastable amorphous phases often serve as precursors to crystalline minerals (26, 27). After 10 min in the presence of rM179, similar prenucleation clusters formed, which often assembled into small groups (Figs. 3 A,D and E, and Fig. S9A). After 30 min, in the control sample, spherical mineral aggregates 50–80 nm in diameter formed, likely by aggregation of the prenucleation clusters (Fig. S8B). However, in the presence of rM179, a very different product was observed at the same reaction time: In addition to small randomly organized groups of the prenucleation clusters, needle-like structures, roughly 1–2-nm across appeared (Fig. 3B and Fig. S9B). A close examination of these structures revealed that they are composed of the aligned prenucleation clusters, some of which were starting to fuse together (Figs. 3 F and G, and Fig. S9B). The ability of rM179 to stabilize the prenucleation clusters and guide their assembly into linear chains that subsequently fuse into the needle-shaped mineral particles is consistent with the view that the enamel crystals form by controlled assembly of prenuclation clusters (8, 28) via a nonclassical crystallization mechanism (29). Finally, after 2 h in the control experiments, large aggregates of plate-like mineral particles were observed (Fig. S8C). At the same time point in the presence of rM179, aligned bundles of thin needle-shaped mineral particles formed, in addition to the stabilized individual prenucleation clusters (Fig. 3C and Fig. S10). Similar mineral bundles formed in the presence of full-length amelogenin have been previously reported in dehydrated samples (1, 8, 28). A close examination of the needle-shaped mineral particles in the bundles (Figs. 3 H and I, and Fig. S10), reveals the mode of growth of these mineral structures. Specifically, our data indicate that, although the central portions of the particles in the bundles appear as electron dense needles, their tips are comprised of individual prenucleation clusters that attach to the growing tips and later fuse to become a part of the needle-like mineral particle (Fig. 3I and Fig. S10 B and C). Interestingly, the growing tips of the needle-shaped mineral particles are surrounded by envelopes of low-density material, which we interpret as the protein matrix, guiding assembly of the prenucleation clusters. The electron diffraction analysis of these mineralization products suggests that at this stage mineral remains amorphous (Fig. S10D) in agreement with our earlier in vitro mineralization studies, which show that the mineral crystallization occurs later (8, 28). In contrast to rM179, rM166 was not able to stabilize prenucleation clusters or guide their organization. Specifically, at 10 and 30 min, spherical mineral aggregates approximately 30 nm in diameter formed, similar to those observed in the absence of protein (Fig. S8 D and E). After 120 min, aggregates of randomly oriented elongated mineral particles were found (Fig. S8C). In the case of rM166, we have not observed any evidence that the mineral growth occurs via assembly of prenucleation clusters and believe that in this case mineralization occurs via classical crystallization pathway, whereas the elongated shape of the mineral particles is dictated by preferential binding of amelogenin to certain mineral surfaces as it was previously proposed (1, 30, 31).
Fig. 3.
Cryo-EM micrographs of calcium phosphate mineralization in the presence of full-length amelogenin rM179. (A, D, and E) 10 min in the reaction; (B, F, and G) 30 min in the reaction; and (C, H, and I) 120 min in the reaction. (C, Inset) Isolated mineral prenucleation clusters. Arrowheads in D–I point to individual prenucleation clusters.
Our observation that full-length amelogenin possesses a unique ability to stabilize and organize prenucleation clusters is in agreement with earlier in vitro mineralization experiments (1, 8, 28) and studies of transgenic animals, which demonstrate that the absence of the C telopeptide leads to alterations in amelogenin assembly and structural defects of enamel (7). Our observations correlate well with the initial mineralization events in vivo. Our earlier studies of amelogenesis demonstrate that the first mineral deposited is amorphous and it is comprised of elongated mineral particles organized into parallel arrays similar to those observed in our mineralization experiments with full-length amelogenin (26). The fact that the shape and organization of the mineral particles in vivo is determined prior to their crystallization indicates that the enamel formation occurs via nonclassical crystallization pathway and that organic matrix plays the major role in the regulation of mineral phase shape and structural organization. Furthermore, nanometer-sized mineral particles have been previously observed in developing enamel (32). Additional studies are needed, however, to identify the prenucleation clusters in vivo and clarify their role in the mineralization process.
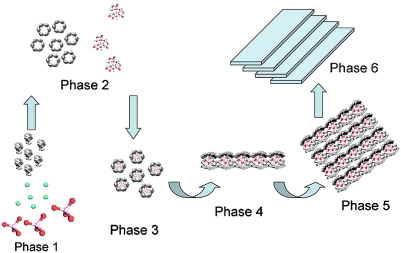
Although the exact relationship between full-length amelogenin assemblies and forming mineral particles remains unresolved, our present findings suggest that the prenucleation clusters may be stabilized inside the double-barrel structured oligomers of full-length amelogenin. According to the structure of amelogenin dodecamer proposed here, the charged C telopeptides form an equatorial band with a high density of hydrophilic charged amino acids in this area that can interface with the prenucleation clusters via electrostatic interactions. The oligomers holding the prenucleation clusters can then assemble into linear chains bringing them into close proximity and trigger aggregation-based mineral growth (33) (Fig. 4). The question remains, however, how is it that amelogenin alone assembles into nanospheres, whereas in the mineralization experiments it guides a formation of parallel arrays of mineral particles. Most likely the mineral interactions change the mode of amelogenin assembly, leading to different structures than those formed without mineral. Our earlier experiments (1) that showed that full-length amelogenin was able to induce mineral organization only when protein assembly and mineralization occurred simultaneously, whereas preformed amelogenin assemblies did not have any effect on mineral organization, support this notion. We propose, therefore, that mineral interactions change the mode of amelogenin assembly to guide formation of parallel arrays of mineral particles, whereas amelogenin alone assembles into nanospheres.
Fig. 4.
A scheme depicting the proposed mechanism by which the amelogenin assemblies manipulate the arrangement of prenucleation clusters into organized mesostructures. (Phase 1) Mixing of mineral ions (calcium, green; phosphate, red) and the full-length amelogenin monomers. (Phase 2) At physiological pH values, amelogenin oligomers and mineral prenucleation clusters are formed. (Phase 3) Amelogenin assemblies and prenucleation clusters then assemble into the proposed composite structures. (Phase 4) Protein-mineral nanoparticles assemble into linear chains. This latter process will require some rearrangement of the amelogenin assemblies. (Phase 5) The linear protein-mineral nanoparticle chains organize into parallel arrays. (Phase 6) Prenucleation clusters fuse, leading to the formation of bundles of elongated crystallites of secretory enamel.
Our present results provide very detailed insight into the mechanisms of amelogenin self-assembly and its effects on calcium phosphate mineralization in vitro. Although these results seem to fit well with the in vivo situation, these data must be interpreted very carefully. Unlike recombinant protein, amelogenin in developing enamel is phosphorylated at Ser16. Our earlier studies have shown that phosphorylated amelogenin is a very strong inhibitor of mineralization and can stabilize amorphous calcium carbonate for relatively long periods of time (8, 17). Hence, although recombinant nonposphorylated amelogenin in vitro seems to recapitulate some aspects of in vivo enamel formation, such as transient stabilization of ACP and formation of parallel arrays of crystallites (1, 8, 26), native phosphorylated protein behaves very differently. At this point, we can only surmise that other proteins in forming enamel modify the strong inhibitory potential of the phosphorylated amelogenin. Alternatively, amelogenin might be dephosphorylated and undergo proteolytic degradation in the extracellular space. Additional studies are needed to examine these possibilities.
In conclusion, we directly demonstrate that full-length amelogein undergoes hierarchical stepwise assembly, first forming oligomers which in turn assemble into higher-order structures. We have developed a structural model of amelogenin oligomers based on the single-particle analysis of the vitrified amelogenin samples and have shown that its C telopeptide plays a critical role in the organization of the oligomers. We also demonstrate that the full-length amelogenin can stabilize prenucleation clusters and regulate the formation of highly organized mineral structures by arranging the prenucleation clusters into linear chains organized into parallel arrays. The prenucleation clusters in the chains fuse together leading to the formation of bundles of elongated mineral particles. Furthermore, our observations imply that the ability of the protein to self-assemble is crucial to its function in the organization of the mineral phase. These results provide unique insights into the mechanisms by which biological macromolecules regulate mineral formation at the nanoscale and can aid in the design of bioinspired approaches toward bottom-up materials design.
Materials and Methods
Self-Assembly Experiments.
Mouse recombinant amelogenins, rM179 and rM166, were produced in bacteria Escherichia coli and purified, as described elsewhere (34). Protein stock solutions were prepared by dissolution of the lyophilized recombinant mouse amelogenins in distilled water at the concentration of 10 mg/mL. The solution was adjusted to pH 3.5 using HCl and kept for at least 24 h at 4 °C prior to the experiments to ensure complete dissolution of the protein.
Aliquots of stock solution were added to 4 mM PBS with pH adjusted to 8.0, on ice to establish a final protein concentration of 100 μg/mL. Lacey carbon transmission electron microscope (TEM) grids meshed 400 (Electron Microscopy Sciences) were placed on top of small droplets (20 to 50 μL) of the protein solutions and incubated for different time intervals from 1 to 30 min at room temperature. The grids were then blotted and immediately plunge-frozen in ethane slush cooled by liquid nitrogen using a Vitrobot automated plunge freezer (Gatan). The grids were transferred cold to the microscope. The data were recorded using a Gatan 4k × 4k CCD camera in an FEI Tecnai F20 TEM with field-emission gun operating at 200 kV accelerating voltage. The images were taken under low-dose mode at 20 e/Å2 to minimize radiation damage to the samples. To enhance the image contrast, underfocusing in the range of 1–3 μm was used to record the images. The calibrated magnification was 70,093, resulting in a pixel size of 2.14 Å.
Single-Particle Reconstruction.
Three-dimensional single-particle reconstructions of amelogenin monomers and oligomers were performed using a single-particle image processing software package EMAN (35). For rM179 monomer and dodecamer reconstruction, 725 and 530 particles were selected, respectively; 788 particles were selected for rM166 reconstruction. The initial models were built using 100 randomly selected raw particles. The initial orientation of individual particles was randomly assigned within the corresponding asymmetry unit. A projection-matching algorithm was then used to determine the center and orientation of raw particles in the iterative refinement until convergence. For the rM179 monomer, no symmetry was imposed during the reconstruction. D6 symmetry (sixfold dihedral rotational symmetry) was imposed in the rM179 oligomer and control reconstruction, along with low C6 symmetry (sixfold rotational symmetry) and without imposing any symmetry to check to see that the particles indeed have these noted symmetries. Final maps were visualized using University of California, San Francisco Chimera software (36). The resolution of the reconstructions was determined to be at 18.7 Å using the Fourier shell correlation (37) (Fig. S7D). Contour levels were chosen for the surface views to enclose 100% of the protein volume calculated from the relative molecular weights of rM179 and rM166, and using an average protein partial-specific volume of 0.73 cm3/g (38).
Mineralization Experiments.
Stock solutions of recombinant murine amelogenins rM179 and rM166 were mixed with CaCl2 with Na2HPO4 solution to establish the final protein concentration 0.2 mg/mL, 1 mM phosphate, and 1.67 mM Ca2+. Solution pH was adjusted to 8.0 by adding NaOH. Because of its small volume, we were not able to maintain constant pH during the mineralization reaction. Because the mineral formation is accompanied by drop in the pH (for example, see ref. 8), we chose to establish the starting pH higher than that found in vivo, to compensate for this pH drop. The droplets of the solutions were mounted on lacey carbon grids and incubated at 37 °C in a humidity chamber for 2 h. The samples were taken from the humidity chambers at 10, 30, and 120 min after the beginning of the reaction, blotted, and immediately plunge-frozen into ethane slush cooled by liquid nitrogen (LN) using Vitrobot automated vitrification station (FEI). The samples were transferred into an LN cryoholder (Gatan) and studied under FEI Tecnai F20 cryo-TEM equipped with field-emission gun at 200 kV. The data were recorded using a Gatan 4k × 4k CCD camera in a low-dose mode. The images were taken at nominal magnification 50,000 (the calibrated magnification is 70,093) under low-dose condition at 20 e/Å2 to minimize radiation damages to the samples. To enhance the image contrast, underfocusing in the range of 1–3 μm was used to record the images. FTIR spectroscopy of amelogenin in solution was performed as previously described (39).
Supplementary Material
Acknowledgments.
Support for this work was provided by National Institutes of Health Grants R01DE016376 (to H.C.M.) and R01DE016703 (to E.B.) and by the Commonwealth of Pennsylvania Grant SAP 4100031302 (to J.F.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.J.D. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106228108/-/DCSupplemental.
References
- 1.Beniash E, Simmer JP, Margolis HC. The effect of recombinant mouse amelogenins on the formation and organization of hydroxyapatite crystals in vitro. J Struct Biol. 2005;149:182–190. doi: 10.1016/j.jsb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Margolis HC, Beniash E, Fowler CE. Role of macromolecular assembly of enamel matrix proteins in enamel formation. J Dent Res. 2006;85:775–793. doi: 10.1177/154405910608500902. [DOI] [PubMed] [Google Scholar]
- 3.Du C, Falini G, Fermani S, Abbott C, Moradian-Oldak J. Supramolecular assembly of amelogenin nanospheres into birefringent microribbons. Science. 2005;307:1450–1454. doi: 10.1126/science.1105675. [DOI] [PubMed] [Google Scholar]
- 4.Chai H, Lee JJW, Constantino PJ, Lucas PW, Lawn BR. Remarkable resilience of teeth. Proc Natl Acad Sci USA. 2009;106:7289–7293. doi: 10.1073/pnas.0902466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowenstam HA, Weiner S. On Biomineralization. Oxford: Oxford Univ Press; 1989. pp. 175–176. [Google Scholar]
- 6.Gibson CW, et al. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 7.Paine ML, et al. Enamel biomineralization defects result from alterations to amelogenin self-assembly. J Struct Biol. 2000;132:191–200. doi: 10.1006/jsbi.2000.4324. [DOI] [PubMed] [Google Scholar]
- 8.Kwak S-Y, et al. Role of 20-kDa amelogenin (P148) phosphorylation in calcium phosphate formation in vitro. J Biol Chem. 2009;284:18972–18979. doi: 10.1074/jbc.M109.020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang XD, et al. How amelogenin orchestrates the organization of hierarchical elongated microstructures of apatite. J Phys Chem B. 2010;114:2293–2300. doi: 10.1021/jp910219s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delak K, et al. The tooth enamel protein, porcine amelogenin, is an intrinsically disordered protein with an extended molecular configuration in the monomeric form. Biochemistry. 2009;48:2272–2281. doi: 10.1021/bi802175a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lakshminarayanan R, Fan D, Du C, Moradian-Oldak J. The role of secondary structure in the entropically driven amelogenin self-assembly. Biophys J. 2007;93:3664–3674. doi: 10.1529/biophysj.107.113936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiedemann-Bidlack FB, Beniash E, Yamakoshi Y, Simmer JP, Margolis HC. pH triggered self-assembly of native and recombinant amelogenins under physiological pH and temperature in vitro. J Struct Biol. 2007;160:57–69. doi: 10.1016/j.jsb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fincham AG, et al. Evidence for amelogenin nanospheres as functional components of secretory-stage enamel matrix. J Struct Biol. 1995;115:50–59. doi: 10.1006/jsbi.1995.1029. [DOI] [PubMed] [Google Scholar]
- 14.Aichmayer B, et al. Amelogenin nanoparticles in suspension: Deviations from spherical shape and ph-dependent aggregation. Biomacromolecules. 2010;11:369–376. doi: 10.1021/bm900983b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagano T, et al. Mmp-20 and Klk4 cleavage site preferences for amelogenin sequences. J Dent Res. 2009;88:823–828. doi: 10.1177/0022034509342694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moradian-Oldak J, Leung W, Fincham AG. Temperature and pH-dependent supramolecular self-assembly of amelogenin molecules: A dynamic light-scattering analysis. J Struct Biol. 1998;122:320–327. doi: 10.1006/jsbi.1998.4008. [DOI] [PubMed] [Google Scholar]
- 17.Wiedemann-Bidlack FB, et al. Effects of phosphorylation on the self-assembly of native full-length porcine amelogenin and its regulation of calcium phosphate formation in vitro. J Struct Biol. 2011;173:250–260. doi: 10.1016/j.jsb.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pugach MK, et al. The amelogenin C-terminus is required for enamel development. J Dent Res. 2010;89:165–169. doi: 10.1177/0022034509358392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brookes SJ, Lyngstadaas SP, Robinson C, Shore RC, Kirkham J. Intracellular nanosphere subunit assembly as revealed by amelogenin molecular cross-linking studies. Eur J Oral Sci. 2006;114:280–284. doi: 10.1111/j.1600-0722.2006.00311.x. [DOI] [PubMed] [Google Scholar]
- 20.Fincham AG, Leung W, Tan J, Moradian-Oldak J. Does amelogenin nanosphere assembly proceed through intermediary-sized structures? Connect Tissue Res. 1998;38:237–240. doi: 10.3109/03008209809017042. [DOI] [PubMed] [Google Scholar]
- 21.Fukae M, Yamamoto R, Karakida T, Shimoda S, Tanabe T. Micelle structure of amelogenin in porcine secretory enamel. J Dent Res. 2007;86:758–763. doi: 10.1177/154405910708600814. [DOI] [PubMed] [Google Scholar]
- 22.Dey A, et al. The role of prenucleation clusters in surface-induced calcium phosphate crystallization. Nat Mater. 2010;9:1010–1014. doi: 10.1038/nmat2900. [DOI] [PubMed] [Google Scholar]
- 23.Nudelman F, et al. The role of collagen in bone apatite formation in the presence of hydroxyapatite nucleation inhibitors. Nat Mater. 2010;9:1004–1009. doi: 10.1038/nmat2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Posner AS, Betts F. Synthetic amorphous calcium phosphate and its relation to bone mineral structure. Acc Chem Res. 1975;8:273–281. [Google Scholar]
- 25.Onuma K, Ito A. Cluster growth model for hydroxyapatite. Chem Mater. 1998;10:3346–3351. [Google Scholar]
- 26.Beniash E, Metzler RA, Lam RSK, Gilbert PUPA. Transient amorphous calcium phosphate in forming enamel. J Struct Biol. 2009;166:133–143. doi: 10.1016/j.jsb.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahamid J, Sharir A, Addadi L, Weiner S. Amorphous calcium phosphate is a major component of the forming fin bones of zebrafish: Indications for an amorphous precursor phase. Proc Natl Acad Sci USA. 2008;105:12748–12753. doi: 10.1073/pnas.0803354105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deshpande AS, Fang PA, Simmer JP, Margolis HC, Beniash E. Amelogenin-collagen interactions regulate calcium phosphate mineralization in vitro. J Biol Chem. 2010;285:19277–19287. doi: 10.1074/jbc.M109.079939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gebauer D, Volkel A, Colfen H. Stable prenucleation calcium carbonate clusters. Science. 2008;322:1819–1822. doi: 10.1126/science.1164271. [DOI] [PubMed] [Google Scholar]
- 30.Iijima M, Moradian-Oldak J. Control of octacalcium phosphate and apatite crystal growth by amelogenin matrices. J Mater Chem. 2004;14:2189–2199. [Google Scholar]
- 31.Iijima M, Moriwaki Y, Wen HB, Fincham AG, Moradian-Oldak J. Elongated growth of octacalcium phosphate crystals in recombinant amelogenin gels under controlled ionic flow. J Dent Res. 2002;81:69–73. doi: 10.1177/002203450208100115. [DOI] [PubMed] [Google Scholar]
- 32.Cuisinier FJG, Steuer P, Senger B, Voegel JC, Frank RM. Human amelogenesis—high-resolution electron-microscopy of nanometer-sized particles. Cell Tissue Res. 1993;273:175–182. doi: 10.1007/BF00304624. [DOI] [PubMed] [Google Scholar]
- 33.Banfield JF, Welch SA, Zhang HZ, Ebert TT, Penn RL. Aggregation-based crystal growth and microstructure development in natural iron oxyhydroxide biomineralization products. Science. 2000;289:751–754. doi: 10.1126/science.289.5480.751. [DOI] [PubMed] [Google Scholar]
- 34.Simmer JP, et al. Isolation and characterization of a mouse amelogenin expressed in E. coli. Calcif Tissue Int. 1994;54:312–319. doi: 10.1007/BF00295956. [DOI] [PubMed] [Google Scholar]
- 35.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 36.Goddard TD, Huang CC, Ferrin TE. Software extensions to UCSF Chimera for interactive visualization of large molecular assemblies. Structure. 2005;13:473–482. doi: 10.1016/j.str.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 37.van Heel M, Schatz M, Orlova E. Correlation functions revisited. Ultramicroscopy. 1992;46:307–316. [Google Scholar]
- 38.Harpaz Y, Gerstein M, Chothia C. Volume changes on protein folding. Structure. 1994;2:641–649. doi: 10.1016/s0969-2126(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 39.Elangovan S, Margolis HC, Oppenheim FG, Beniash E. Conformational changes in salivary proline-rich protein 1 upon adsorption to calcium phosphate crystals. Langmuir. 2007;23:11200–11205. doi: 10.1021/la7013978. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.