Abstract
Phylogenetic analyses of genes with demonstrated involvement in evolutionary transitions can be an important means of resolving conflicting hypotheses about evolutionary history or process. In sunflower, two genes have previously been shown to have experienced selective sweeps during its early domestication. In the present study, we identified a third candidate early domestication gene and conducted haplotype analyses of all three genes to address a recent, controversial hypothesis about the origin of cultivated sunflower. Although the scientific consensus had long been that sunflower was domesticated once in eastern North America, the discovery of pre-Columbian sunflower remains at archaeological sites in Mexico led to the proposal of a second domestication center in southern Mexico. Previous molecular studies with neutral markers were consistent with the former hypothesis. However, only two indigenous Mexican cultivars were included in these studies, and their provenance and genetic purity have been questioned. Therefore, we sequenced regions of the three candidate domestication genes containing SNPs diagnostic for domestication from large, newly collected samples of Mexican sunflower landraces and Mexican wild populations from a broad geographic range. The new germplasm also was genotyped for 12 microsatellite loci. Our evidence from multiple evolutionarily important loci and from neutral markers supports a single domestication event for extant cultivated sunflower in eastern North America.
Keywords: agriculture, Helianthus annuus, phylogeography
Many genetic changes essential for crop evolution have been identified, and these alleles provide useful tools for inferring how, where, and when early societies transformed wild plants into agricultural staples (1, 2). Examining sequence diversity of these alleles in cultivated lineages, wild progenitors, and archaeological specimens can reveal critical information about the rate, timing, geography, and order of the domestication process (3). For instance, sequences of domestication loci obtained from archaeological maize cobs indicated that although domestication alleles for two traits became fixed early, fixation of domestication alleles for a third trait occurred several thousand years later (4). Patterns of sequence diversity around domestication alleles have yielded estimates of the strength of selection during domestication for several loci in rice and maize (5–7). In addition, the geographic distributions of domestication alleles in extant cultivated and wild germplasm have been used to assess how crops and crop alleles have spread from domestication centers and reveal whether convergent traits in independent lineages evolved from the same or unique suites of mutations (5, 8–11).
Here, we examine three genes that experienced selective sweeps during early domestication of sunflower, Helianthus annuus, to address a recent controversy about the number and location of sunflower domestication centers (12–17). Molecular, archaeological, and linguistic evidence has long supported the presence of a domestication center in eastern North America (ENA) (16). However, pre-Columbian archaeological finds at several sites in southern Mexico have suggested the possibility of a second independent domestication center and challenged the view that domesticated sunflower reached Mexico only after introduction by Spanish traders (14, 15, 17, 18). The archaeological evidence suggests two possible alternative explanations: an independent origin of domesticated sunflower in Mexico or dispersal to Mexico through trade routes established before Spanish colonization. The identification and significance of the archaeological remains, the historic record, and modern linguistic evidence have been debated heavily (16, 17, 19–26). Previous molecular studies using neutral nuclear and chloroplast DNA markers have indicated that extant Mexican landraces are descended from the same lineage as ENA landraces, suggesting that if an independent Mexican lineage evolved, it did not contribute to modern cultivated sunflower germplasm (27, 28). However, the provenance and genetic purity of the two Mexican landrace samples included in those studies have been questioned. In addition, few Mexican wild H. annuus populations were included in these analyses.
We have made new, extensive collections of Mexican wild sunflowers and domesticated cultivars grown by indigenous Mexican populations. Sequencing genes demonstrated to have experienced selective sweeps during domestication in ENA provided an opportunity to test whether extant Mexican landraces descended from the same or a unique lineage. In particular, if domesticated sunflower reached Mexico through trade routes from the North, we predicted that ENA and Mexican landraces would share the same haplotypes at these genes. If two independent domestication events occurred in isolation, it is highly unlikely these parallel events would have selected for the same causative mutations arising on the same haplotype backgrounds at multiple loci (8–10). In addition, the presence of the domesticated haplotype in ENA wild populations and its absence from wild Mexican populations would exclude the possibility of a parallel selective sweep from equivalent standing variation.
Two strong candidate early domestication genes in sunflower have been described. A genomic screen found that sequence diversity in the gene c4973, a chorismate synthase homolog involved in aromatic amino acid synthesis, is significantly reduced in North American landraces relative to its sequence diversity in neutral loci (29). HaFT1, a homolog of the floral inducer FLOWERING LOCUS T, shows a similar signature of a selective sweep during early domestication (30). Although this evidence indicates that the genomic regions containing these genes reflect a history of recent selection, we describe these genes as candidate domestication genes because additional work is required to distinguish whether they or other tightly linked loci carry the specific mutations responsible for domestication traits and targeted by selection. Notably, however, several lines of genetic and functional evidence have provided strong support for a frameshift mutation in the domesticated allele of HaFT1 as the likely causative mutation that responded to selection through its effects on flowering time or head diameter (30). Here, we identify a third candidate early domestication gene and show that patterns of diversity in these genes, as well as 12 neutral markers, in Mexican landraces and wild H. annuus are consistent with all extant landraces originating from a single domestication center in eastern North America.
Results
Selection on Gibberellin 2-Oxidase Homolog HaGA2ox During Early Domestication.
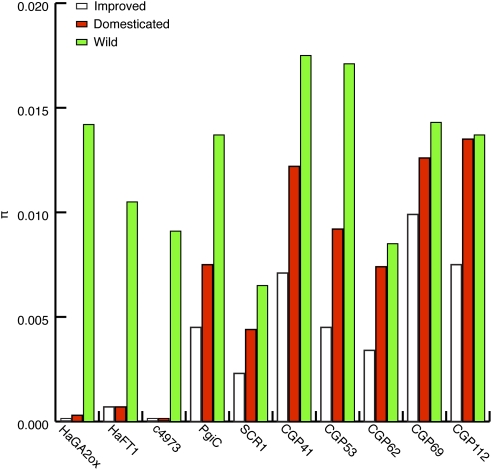
An 885-bp portion of the gibberellin 2-oxidase homolog HaGA2ox was examined as part of an ongoing study of a genetic diversity panel from the United States and Canada (31). This panel included 18 individuals from improved modern crop lines, 19 individuals from Native American landraces, and 23 individuals from a geographically diverse sample of wild sunflower populations (Table S1). Six Helianthus argophyllus individuals also were sequenced to obtain divergence estimates. Average pairwise nucleotide diversity (π) in HaGA2ox was much reduced in domesticated and improved lines relative to neutral loci, similar to previous observations for HaFT1 and c4973 (Fig. 1). Only two haplotypes—distinguished from each other by two SNPs—segregated in Native American landraces (Fig. 2A). Improved lines were fixed for the major landrace haplotype, and this haplotype was present only at minor frequency in wild populations, indicative of a selective sweep (Table 1). All coding substitutions along the branch distinguishing the domesticated and wild haplotypes were synonymous changes. A maximum-likelihood Hudson–Kreitman–Aguadé (HKA) test, which compares the likelihood of a neutral evolutionary model of polymorphism and divergence with the likelihood of a model in which a candidate gene is under selection, was conducted for HaGA2ox and seven previously sequenced putative neutral loci (31, 32). Although HaGA2ox was found to be evolving neutrally in wild populations (P = 0.9748), there were significant signatures of a selective sweep in the landrace (P = 0.0032) and improved pools (P = 0.0002).
Fig. 1.
Average pairwise nucleotide diversity of three candidate domestication genes (HaFT1, HaGA2ox, and c4973) is significantly reduced relative to seven neutral loci during early domestication.
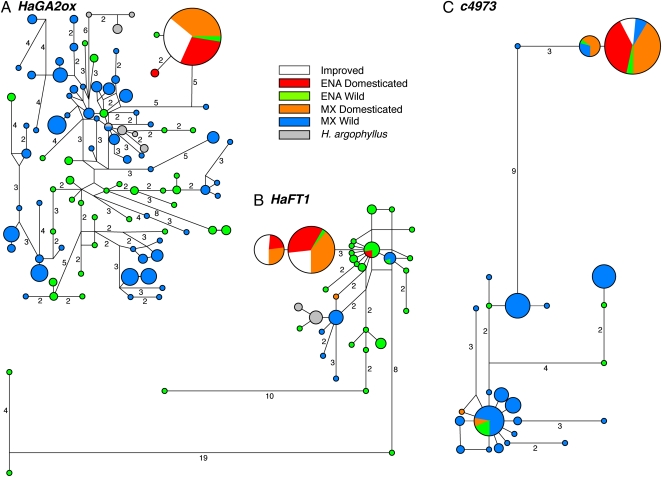
Fig. 2.
Median-joining haplotype networks for three candidate early domestication genes, HaGA2ox (A), HaFT1 (B), and c4973 (C). ENA domesticated and ENA wild are the ENA landraces and American and Canadian wild populations, respectively. MX domesticated and MX wild are the Mexican landraces and Mexican wild populations, respectively. The number of substitutions along a branch is shown for branches with more than one substitution.
Table 1.
Domesticated and wild allele frequencies in eastern North American landraces (ENA domesticated), American and Canadian wild populations (ENA wild), Mexican landraces (MX domesticated), and Mexican wild populations (MX wild)
| Gene | ENA domesticated | ENA wild | MX domesticated | MX wild |
| HaFT1 | ||||
| Domesticated allele | 95% (36) | 5% (2) | 98% (41) | 0% (0) |
| Wild allele | 5% (2) | 95% (38) | 2% (1) | 100% (152) |
| (38 total) | (40 total) | (42 total) | (152 total) | |
| HaGa2ox | ||||
| Domesticated haplotype 1 | 95% (36) | 7% (3) | 100% (48) | 0% (0) |
| Other domesticated haplotypes | 5% (2) | 2% (1) | 0% (0) | 0% (0) |
| Wild haplotypes | 0% (0) | 91% (42) | 0% (0) | 100% (152) |
| (38 total) | (46 total) | (48 total) | (152 total) | |
| c4973 | ||||
| Domesticated haplotype 1 | 100% (44) | 29% (4) | 76% (47) | 7% (8) |
| Domesticated haplotype 2 | 0% (0) | 7% (1) | 18% (11) | 4% (5) |
| Wild haplotype 1 | 0% (0) | 43% (6) | 5% (3) | 20% (23) |
| Other wild haplotypes | 0% (0) | 21% (3) | 1% (1) | 69% (79) |
| (44 total) | (14 total) | (62 total) | (115 total) | |
The number of haplotype sequences per allele class is shown in parentheses.
Description of Collections.
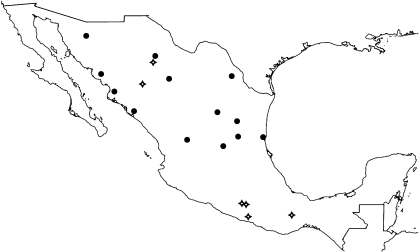
Mexican landraces and wild populations used in this study were collected between 2001 and 2005 (Table S2). Wild specimens were obtained from habitats in all states of Mexico north of Veracruz. Detailed collection procedures and complete morphological descriptions of Mexican landrace achenes can be found in ref. 33. In addition to the genetic affinities of sunflower, we were interested in learning more about how this diverse crop was used by Native Americans in Mexico. Accordingly, we interviewed numerous representatives of indigenous peoples in their homes and fields. Cultivars used in this study were collected from the Tarahumara in Chihuahua, the Nahua in Guerrero, the Tzeltal in Chiapas, and the Mixe in Oaxaca. One domesticated collection was obtained from Mennonites living in Chihuahua who had emigrated recently from Canada and likely brought their sunflower germplasm with them.
Domestication Alleles in Mexican Landraces and Wild Populations.
We next examined sequence diversity of HaGA2ox, HaFT1, and c4973 in 21–31 individuals from the 16 Mexican landrace collections and 57–76 individuals from 15 geographically diverse Mexican wild populations (Table S2 and Fig. 3). For HaGA2ox and c4973, we fully sequenced the same portions previously sequenced from ENA accessions. Although we sequenced the HaFT1 haplotype from all Mexican landraces, we genotyped the HaFT1 allele in wild populations because the putative causal frameshift mutation in the domesticated allele of HaFT1 creates an easily diagnosed restriction site polymorphism.
Fig. 3.
Collection locations of sunflower domesticates (diamonds) and wild populations (circles) in Mexico.
As would be expected if the extant ENA and Mexican landraces are descended from a single domestication event, the Mexican landraces were fixed for the same major domesticated haplotype for HaGA2ox (Fig. 2A and Table 1). Although present at minor frequency in ENA wild populations, this haplotype was not observed in wild Mexican populations.
Nearly the same results were observed for HaFT1. All but one haplotype sequenced from Mexican landraces contained the frameshift allele (Fig. 2B and Fig. S1). Although the frameshift-containing domesticated allele does segregate at minor frequency in wild populations from Oklahoma, this allele was not found in any Mexican wild individuals, an observation inconsistent with a parallel sweep from standing variation in Mexico (Table 1). We predicted that the single Mexican wild haplotype present in the Mexican landraces is the result of a recent hybridization event. If true, then nearby genes also should segregate for one American domesticate-like allele and one Mexican wild-like allele, because adequate time will not have passed to break down linkage disequilibrium between alleles in closely linked genes. We sequenced portions of HaFT2 and HaFT3, which are within 2.7 cM of HaFT1 (30), from this individual and observed this predicted pattern. For both genes, one allele exactly matched the major allele in improved lines and ENA landraces. The other allele at each locus was similar to alleles in ENA wild populations but contained one or more unique SNPs, likely indicative of Mexican derivation.
Although the results of HaFT1 and HaGA2ox sequencing were completely consistent with a single origin of extant domesticated sunflower, the results for c4973 did not distinguish unambiguously between the single- and multiple-origin scenarios. As for the other two genes, the haplotype distribution is disjunct, with most wild sequences forming one cluster and most domesticated sequences forming a second cluster (Fig. 2C). Two haplotypes, which differ from each other by only a single SNP, were at high frequency in Mexican landraces. One of these two haplotypes was fixed in our sample that included previously collected data from a small set of ENA domesticates (29) and additional sequencing we conducted on the landraces in our own diversity panel (Fig. 2C and Table 1). Wild haplotypes also were observed to segregate at minor frequency in the Mexican landraces, possibly indicating recent hybridization as observed for HaFT1. Notably, both domesticated haplotypes were found in wild populations from both regions, meaning a parallel selective sweep from standing variation in Mexico cannot be excluded. The domesticated alleles were present at higher frequency in American wild populations, however (Table 1), and these alleles were found in populations from the eastern and northern United States. Thus, ENA is more likely to be the geographic source of these domesticated alleles.
Ancestry Assignment with Neutral Markers.
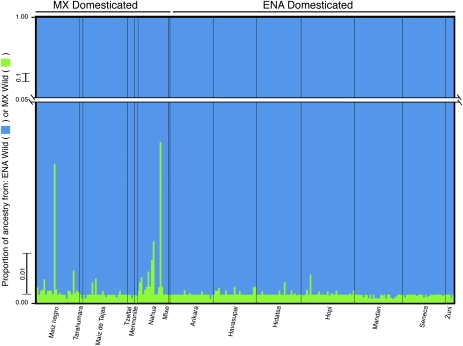
Under certain evolutionary scenarios, incongruities may arise between domestication gene trees and lineage histories because a domestication allele that contributes to desirable phenotypes may be bred from one lineage into an independent lineage through hybridization and selective breeding (5, 6, 11). Therefore, to determine ancestry and corroborate our findings with neutral markers, we genotyped our new, more thorough collections from Mexico for 12 microsatellite loci that had been scored previously on a large set of ENA cultivars and wild populations (27). As expected, domesticated lines contained a subset of the diversity present in wild populations, with the exception of two cultivar-specific alleles present in both ENA and Mexican domesticates (Table S3). In addition, although several alleles present in ENA wild populations but not in Mexican wild populations segregated in domesticates from either or both regions, only one allele observed in one Mexican domesticated individual was found solely in wild individuals from Mexico (Table S3). The estimation of the ancestral provenance of ENA and Mexican landrace samples—as performed in the program Structure through analysis of multilocus genotypes (34)—yielded a clear, unequivocal result. All Mexican and ENA cultivars clustered with the ENA wild samples and not with the Mexican wild samples (Fig. 4), consistent with the results from the three selected loci. For the Mexican landraces, the proportion of genotype with ancestry assigned to the ENA-wild group was >0.96 in all cases.
Fig. 4.
Estimated ancestry of domesticated H. annuus from Mexico (MX domesticated) and North America (ENA domesticated). Each vertical bar represents an individual’s genome and is colored in proportion to the estimated ancestry from MX wild (green) or ENA wild (blue). Vertical black lines delimit individuals into groups by landrace, as reported below each group.
Discussion
Taken together, the findings from three candidate domestication loci and additional neutral markers indicate that domesticated sunflowers grown in Mexico today are descended from the same cultivated genetic lineage as ENA domesticated sunflower. All ENA and Mexican landraces shared the same divergent HaGA2ox haplotypes that, although observed in ENA wild populations, never were present in Mexican wild populations. Although the signature of a selective sweep on HaGA2ox could also be explained by selection acting on a closely linked locus, this enzyme is an attractive candidate for the actual target of selection. Gibberellin 2-oxidases convert gibberellins from active to inactive forms and in doing so act to regulate seed development, delay germination, and promote dwarfism (35–38). Reduced seed dormancy and increased plant biomass have been selected for strongly during domestication of many crops (39), and this pressure may have driven evolutionary changes affecting HaGA2ox expression or activity. We are planning future genetic studies to look at the functional evolution of this gene during sunflower domestication.
HaFT1 exhibited the same pattern as HaGA2ox, except for a single Mexican cultivar that was heterozygous for a Mexican wild allele because of a recent hybridization event. Since c4973 domesticated alleles segregate in both ENA and Mexican wild populations, the possibility of a parallel selective sweep on the same allele cannot be excluded. Nevertheless, the higher frequency of domesticated alleles in ENA wild populations suggest that the eastern United States is the more likely geographic source. The low frequency of these alleles in Mexican wild populations and the low frequency of wild alleles in Mexican landraces may have resulted from recent hybridization, as seen with HaFT1. An intriguing alternative explanation for the latter pattern may be that the selective sweep at c4973 was incomplete when cultivated sunflower was dispersed southward to Mexico. If true, then this result would further support an early introduction of cultivated sunflower to Mexico and indicates that, as more domestication alleles and their functions are identified, it may be possible to time selective events as pre- and postdispersal, allowing trait evolution to be ordered relative to this chronological benchmark. Finally, the ambiguous findings with c4973 could also have resulted from altered selection pressures on the domesticated haplotype in Mexico or because c4973 is not an actual domestication gene but is tightly linked to one.
Although these three genes and neutral markers exhibit patterns of variation in concordance with a single-origin scenario for extant cultivated sunflower, the possibility of multiple origins cannot be discounted fully. For instance, despite thorough searching (by R.A.B. and D.L.L.), it is possible that we were unable to locate modern domesticates descended from an independent Mexican lineage. Also, although the data presented herein are more consistent with a single origin and dispersal before the arrival of Spanish explorers (40), our findings do not invalidate the possibility that a separate center of sunflower domestication in Mexico did indeed occur during the pre-Columbian period. An ancient lineage may not be represented in modern germplasm because no descendants of the hypothesized independent Mexican lineage have survived. Although current evidence from archaeological deposits in ENA and Mesoamerica indicate that ancient Mesoamericans cultivated sunflower before the advent of the Spanish in the region, more discoveries will be needed before a full understanding of the trajectory of sunflower crop development in Mesoamerica and ENA emerges.
Lack of modern descendants from a possible independent lineage raises the question of how descendants of that lineage may have gone extinct. It has been suggested that colonial Spanish Christians may have tried to eliminate sunflower because of its important symbolic role in the religious rituals of the solar-worshipping Aztecs and other Native American cultures (17). Alternatively, native domesticates in Mexico may have been replaced much more recently by a substantial influx of seed imports made possible by the North American Free Trade Agreement.
Although the majority of the Mexican archaeological sunflower discoveries were destroyed for radiocarbon dating, archaeological studies at sites throughout Mexico likely will yield additional samples. Furthermore, new sequencing technologies have made ancient DNA studies increasingly successful means of revealing genealogical and phenotypic characteristics of extinct lineages (4, 8, 41–46). Studies of these and newly identified domestication genes in DNA isolated from archaeological plant remains discovered in ENA and Mexico promise to yield even further insight into the timing and geography of sunflower domestication.
Materials and Methods
DNA was extracted from leaf tissue using DNeasy 96 Plant or Plant Mini Kits (Qiagen). Portions of HaFT1, HaGA2ox, and c4973 were amplified by PCR and sequenced using gene-specific primers (Table S4) from Mexican cultivars and wild Mexican individuals (Table S2). HaGA2ox also was sequenced on the entire American and Canadian diversity panel (Table S1). We included previously published HaFT1 (30) and c4973 (29) sequences from American and Canadian H. annuus domesticated and wild populations and H. argophyllus (Table S5). Because c4973 had been sequenced previously only on a small sample of American landraces, we obtained additional sequences from our diversity panel to sample c4973 sequence diversity more thoroughly in North American landraces (Tables S1 and S5). Sequence data for seven putative neutral loci were obtained for a previously published study of this diversity panel (Table S5) (31). HaFT1 PCR products were digested with BstEII to genotype individuals for the frameshift mutation nearly fixed in domesticated haplotypes.
For heterozygous individuals, PCR products were cloned (TOPO TA Cloning; Invitrogen), and multiple clones per individual were sequenced using T7/T3 or M13F/M13R primers. Cloned sequences were compared with each other and with sequencing reads from the initial PCR product to ensure that both haplotypes were captured and to eliminate sequence errors introduced by PCR and cloning. Median-joining haplotype networks were constructed with Network 4.5.16 (Fluxus Technology). For all genes, insertion/deletion polymorphisms were recoded as SNPs, and a complex intronic region of nested insertion/deletion polymorphisms was excluded from analysis of HaGA2ox.
Average pairwise nucleotide diversity (π) was calculated using DnaSP (47). The program MLHKA (32) was used to conduct the maximum-likelihood HKA tests for HaGA2ox. A likelihood-ratio test was conducted to compare the likelihood of a neutral model and the likelihood of a model in which HaGA2ox was under selection. Separate tests were performed for improved, domesticated American landraces and American wild samples to determine the timing of selection.
Landraces and wild population samples from Mexico and ENA were genotyped with 12 microsatellite loci (Tables S1, S2, and S4) that had been scored previously on another panel of domesticated and wild H. annuus (27). PCR amplification, fragment separation by capillary electrophoresis, and fragment length scoring were conducted as previously described (48). Data were combined with genotypes from ref. 27 using one to five overlapping samples to assure that allele assignments in each data set were congruent. The ancestry of H. annuus cultivated individuals and the composition of their genotype was evaluated with a Bayesian clustering method, as implemented in Structure v. 2.3.3 (34, 49). Two ancestral source clusters were identified by ref. 27: (i) a group including all Mexican and Arizonan wild populations (MX Wild) and (ii) a group of central United States populations (ENA Wild). Ten independent simulations were performed using the same parameters and prior assumptions as in ref. 27. Results were formatted with HARVESTER v. 0.6.6 (http://taylor0.biology.ucla.edu/structureHarvester/) and permuted with CLUMPP v. 1.1.2 (50). The final matrix of estimated ancestry coefficients was visualized with DISTRUCT v. 1.1 (51).
Supplementary Material
Acknowledgments
We thank A. Harter for generously providing data from her previous work and for providing guidance with the neutral marker analysis. We thank Z. Lai, Q. Yu, A. Raduski, J. Strasburg, M. Kreitzman, L. Washington, and M. Chapman for technical assistance and scientific input. This work was supported by National Science Foundation Grants DEB0608118 and DBI0905958 (to B.K.B), DBI0421630 and DBI0820451 (to L.H.R.), and BCS0228049 (to R.A.B., D.L.L, and L.H.R) and by National Geographic Society Grant 7030-01 (to D.L.L. and R.A.B).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JF428180–JF428779).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104853108/-/DCSupplemental.
References
- 1.Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127:1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Purugganan MD, Fuller DQ. The nature of selection during plant domestication. Nature. 2009;457:843–848. doi: 10.1038/nature07895. [DOI] [PubMed] [Google Scholar]
- 3.Gross BL, Olsen KM. Genetic perspectives on crop domestication. Trends Plant Sci. 2010;15:529–537. doi: 10.1016/j.tplants.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaenicke-Després V, et al. Early allelic selection in maize as revealed by ancient DNA. Science. 2003;302:1206–1208. doi: 10.1126/science.1089056. [DOI] [PubMed] [Google Scholar]
- 5.Zhang L-B, et al. Selection on grain shattering genes and rates of rice domestication. New Phytol. 2009;184:708–720. doi: 10.1111/j.1469-8137.2009.02984.x. [DOI] [PubMed] [Google Scholar]
- 6.Olsen KM, et al. Selection under domestication: Evidence for a sweep in the rice waxy genomic region. Genetics. 2006;173:975–983. doi: 10.1534/genetics.106.056473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, et al. The origin of the naked grains of maize. Nature. 2005;436:714–719. doi: 10.1038/nature03863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmer SA, Moore JD, Clapham AJ, Rose P, Allaby RG. Archaeogenetic evidence of ancient nubian barley evolution from six to two-row indicates local adaptation. PLoS ONE. 2009;4:e6301. doi: 10.1371/journal.pone.0006301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azhaguvel P, Komatsuda T. A phylogenetic analysis based on nucleotide sequence of a marker linked to the brittle rachis locus indicates a diphyletic origin of barley. Ann Bot (Lond) 2007;100:1009–1015. doi: 10.1093/aob/mcm129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komatsuda T, et al. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci USA. 2007;104:1424–1429. doi: 10.1073/pnas.0608580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sweeney MT, et al. Global dissemination of a single mutation conferring white pericarp in rice. PLoS Genet. 2007;3:e133. doi: 10.1371/journal.pgen.0030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heiser CB. The sunflower among the North American Indians. Proc Am Philos Soc. 1951;95:432–448. [Google Scholar]
- 13.Heiser CB. The domesticated sunflower in old Mexico? Genet Resour Crop Evol. 1998;45:447–449. [Google Scholar]
- 14.Pope KO, et al. Origin and environmental setting of ancient agriculture in the lowlands of Mesoamerica. Science. 2001;292:1370–1373. doi: 10.1126/science.292.5520.1370. [DOI] [PubMed] [Google Scholar]
- 15.Lentz DL, Pohl MED, Pope KO, Wyatt AR. Prehistoric sunflower (Helianthus annuus L.) domestication in Mexico. Econ Bot. 2001;55:370–376. [Google Scholar]
- 16.Smith BD. Eastern North America as an independent center of plant domestication. Proc Natl Acad Sci USA. 2006;103:12223–12228. doi: 10.1073/pnas.0604335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lentz DL, Pohl MD, Alvarado JL, Tarighat S, Bye R. Sunflower (Helianthus annuus L.) as a pre-Columbian domesticate in Mexico. Proc Natl Acad Sci USA. 2008;105:6232–6237. doi: 10.1073/pnas.0711760105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bye R, Linares E, Lentz DL. México: Centro de Origen de la Domesticatión del Girasol. TIP Revista Especializada en Ciencias Químico-Biológicas. 2009;12:5–12. [Google Scholar]
- 19.Smith BD. Winnowing the archaeological evidence for domesticated sunflower in pre-Columbian Mesoamerica. Proc Natl Acad Sci USA. 2008;105:E45. doi: 10.1073/pnas.0804434105. author reply E49–E50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rieseberg L, Burke JM. Molecular evidence and the origin of the domesticated sunflower. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0804494105. 105:E46; author reply E49--E50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown CH. A lack of linguistic evidence for domesticated sunflower in pre-Columbian Mesoamerica. Proc Natl Acad Sci USA. 2008;105:E47. doi: 10.1073/pnas.0804505105. author reply E49–E50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heiser CB. How old is the sunflower in Mexico? Proc Natl Acad Sci USA. 2008;105:E48. doi: 10.1073/pnas.0804588105. author reply E49–E50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lentz DL, Pohl MD, Bye R. Reply to Rieseberg and Burke, Heiser, Brown, and Smith: Molecular, linguistic, and archaeological evidence for domesticated sunflower in pre-Columbian Mesoamerica. Proc Natl Acad Sci USA. 2008;105:E49–E50. doi: 10.1073/pnas.0804434105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heiser CB. The sunflower (Helianthus annuus) in Mexico: Further evidence for a North American domestication. Genet Resour Crop Evol. 2008;55:9–13. [Google Scholar]
- 25.Heiser CB. Comment: Sunflowers among Aztecs? Int J Plant Sci. 2008;169:980. [Google Scholar]
- 26.Lentz DL. Reply to Heiser. Int J Plant Sci. 2008;169:980. [Google Scholar]
- 27.Harter AV, et al. Origin of extant domesticated sunflowers in eastern North America. Nature. 2004;430:201–205. doi: 10.1038/nature02710. [DOI] [PubMed] [Google Scholar]
- 28.Wills DM, Burke JM. Chloroplast DNA variation confirms a single origin of domesticated sunflower (Helianthus annuus L.) J Hered. 2006;97:403–408. doi: 10.1093/jhered/esl001. [DOI] [PubMed] [Google Scholar]
- 29.Chapman MA, et al. A genomic scan for selection reveals candidates for genes involved in the evolution of cultivated sunflower (Helianthus annuus) Plant Cell. 2008;20:2931–2945. doi: 10.1105/tpc.108.059808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blackman BK, Strasburg JL, Raduski AR, Michaels SD, Rieseberg LH. The role of recently derived FT paralogs in sunflower domestication. Curr Biol. 2010;20:629–635. doi: 10.1016/j.cub.2010.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blackman BK, et al. Contributions of flowering time genes to sunflower domestication and improvement. Genetics. 2011;187:271–287. doi: 10.1534/genetics.110.121327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wright SI, Charlesworth B. The HKA test revisited: A maximum-likelihood-ratio test of the standard neutral model. Genetics. 2004;168:1071–1076. doi: 10.1534/genetics.104.026500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lentz DL, Bye R, Sanchez-Cordero V. Ecological niche modeling and distribution of wild sunflower (Helianthus annuus L.) in Mexico. Int J Plant Sci. 2008;169:541–549. [Google Scholar]
- 34.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oh E, et al. Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 2006;47:124–139. doi: 10.1111/j.1365-313X.2006.02773.x. [DOI] [PubMed] [Google Scholar]
- 36.Thomas SG, Phillips AL, Hedden P. Molecular cloning and functional expression of gibberellin 2- oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA. 1999;96:4698–4703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schomburg FM, Bizzell CM, Lee DJ, Zeevaart JAD, Amasino RM. Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell. 2003;15:151–163. doi: 10.1105/tpc.005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh DP, Jermakow AM, Swain SM. Gibberellins are required for seed development and pollen tube growth in Arabidopsis. Plant Cell. 2002;14:3133–3147. doi: 10.1105/tpc.003046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hammer K. Das Domestikationssyndrom. Genet Resour Crop Evol. 1984;32:11–34. [Google Scholar]
- 40.Smith BD, Yarnell RA. Initial formation of an indigenous crop complex in eastern North America at 3800 B.P. Proc Natl Acad Sci USA. 2009;106:6561–6566. doi: 10.1073/pnas.0901846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erickson DL, Smith BD, Clarke AC, Sandweiss DH, Tuross N. An Asian origin for a 10,000-year-old domesticated plant in the Americas. Proc Natl Acad Sci USA. 2005;102:18315–18320. doi: 10.1073/pnas.0509279102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lalueza-Fox C, et al. A melanocortin 1 receptor allele suggests varying pigmentation among Neanderthals. Science. 2007;318:1453–1455. doi: 10.1126/science.1147417. [DOI] [PubMed] [Google Scholar]
- 43.Miller W, et al. Sequencing the nuclear genome of the extinct woolly mammoth. Nature. 2008;456:387–390. doi: 10.1038/nature07446. [DOI] [PubMed] [Google Scholar]
- 44.Rasmussen M, et al. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature. 2010;463:757–762. doi: 10.1038/nature08835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larson G, et al. Patterns of East Asian pig domestication, migration, and turnover revealed by modern and ancient DNA. Proc Natl Acad Sci USA. 2010;107:7686–7691. doi: 10.1073/pnas.0912264107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 48.Heesacker A, et al. SSRs and INDELs mined from the sunflower EST database: Abundance, polymorphisms, and cross-taxa utility. Theor Appl Genet. 2008;117:1021–1029. doi: 10.1007/s00122-008-0841-0. [DOI] [PubMed] [Google Scholar]
- 49.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jakobsson M, Rosenberg NA. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- 51.Rosenberg NA. Distruct: aA program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.