Abstract
Sequence analyses of fungal genomes have revealed that the potential of fungi to produce secondary metabolites is greatly underestimated. In fact, most gene clusters coding for the biosynthesis of antibiotics, toxins, or pigments are silent under standard laboratory conditions. Hence, it is one of the major challenges in microbiology to uncover the mechanisms required for pathway activation. Recently, we discovered that intimate physical interaction of the important model fungus Aspergillus nidulans with the soil-dwelling bacterium Streptomyces rapamycinicus specifically activated silent fungal secondary metabolism genes, resulting in the production of the archetypal polyketide orsellinic acid and its derivatives. Here, we report that the streptomycete triggers modification of fungal histones. Deletion analysis of 36 of 40 acetyltransferases, including histone acetyltransferases (HATs) of A. nidulans, demonstrated that the Saga/Ada complex containing the HAT GcnE and the AdaB protein is required for induction of the orsellinic acid gene cluster by the bacterium. We also showed that Saga/Ada plays a major role for specific induction of other biosynthesis gene clusters, such as sterigmatocystin, terrequinone, and penicillin. Chromatin immunoprecipitation showed that the Saga/Ada-dependent increase of histone 3 acetylation at lysine 9 and 14 occurs during interaction of fungus and bacterium. Furthermore, the production of secondary metabolites in A. nidulans is accompanied by a global increase in H3K14 acetylation. Increased H3K9 acetylation, however, was only found within gene clusters. This report provides previously undescribed evidence of Saga/Ada dependent histone acetylation triggered by prokaryotes.
Keywords: histone modification, microbial communication, secondary metabolism gene clusters
Microorganisms have an enormous potential to produce all kinds of different low molecular-weight molecules. Many of these molecules have found their way as important compounds to medicine or agriculture, such as the penicillins and avermectins, respectively (1, 2). However, a major question is how many potentially useful compounds are actually overlooked, not only because the majority of microorganisms has not yet been cultivated, but also because the increasing number of genome sequences of fungi and bacteria indicates that the genetic capability of these organisms to produce different compounds has been largely underestimated. The latter conclusion was based on the presence of genes coding for thiotemplate assembly lines, such as polyketide synthases (PKSs) (3) and nonribosomal peptide synthetases (NRPSs) (4) in various microorganisms. For example, the important model fungus Aspergillus nidulans harbors 28 putative PKS and 24 NRPS gene clusters (4), giving this fungus the potential to produce at least 52 different secondary metabolites. Recent data further complicate a serious estimation because communication occurs between gene clusters, which could potentially lead to even more compounds (5). A major hurdle to identifying this untapped reservoir, however, is the observation that most of these gene clusters are silent under standard laboratory conditions (6–8). To address this problem, methods were established to activate silent gene clusters by genetic engineering (6, 7, 9). The most challenging question, however, is the identification of the true function of these compounds in the habitat. Because these functions are largely unknown, it is not possible to develop strategies on a rationale basis for the activation of their biosyntheses. It is highly likely that many of these compounds are produced as chemical signal molecules or for defending the habitat (10, 11). In this context, we recently discovered that the intimate physical interaction of A. nidulans with a distinct soil-dwelling bacterium identified from a collection of 58 species of actinomycetes, i.e., Streptomyces hygroscopicus (renamed S. rapamycinicus) (12) led to activation of a silent polyketide synthase (PKS) gene cluster. This finding indicated that communication between microorganisms can indeed play a key role in activating silent gene clusters (13). The identified gene cluster encodes the production of the archetypal polyketide orsellinic acid, its derivative lecanoric acid, and the cathepsin K inhibitors F-9775A and F-9775B.
Because A. nidulans is a model organism well-suited to study various fundamental biological questions, this opened up the opportunity to shed light on the molecular mechanisms that form the basis of communication between the fungus and the bacterium. Previously, it was reported that chromatin modifications also contribute to the regulation of gene clusters (9, 14, 15). However, none of these studies addressed the question of whether this regulation is important in a natural setting of interacting microorganisms.
Here, our integrative study led to the discovery that the bacterium induces the histone modification via the main histone acetyltransferase (HAT) complex Saga/Ada in A. nidulans. Furthermore, because Saga/Ada is also required for the biosynthesis of penicillin, sterigmatocystin, and terrequinone, we expand the function of this complex to a set of genes not previously known.
Our work provides insight into bacteria-triggered histone modification in a fungus as a potential basis for a crosstalk between different species of microorganisms.
Results and Discussion
A Functional Saga/Ada Complex Is Required for ors Gene Cluster Activation.
Posttranslational modifications of histones are involved in the regulation of secondary metabolism in filamentous fungi (9). Therefore, we analyzed a number of chemical epigenetic modifiers for their impact on orsellinic and lecanoric acid production induced by the interaction of A. nidulans with S. rapamycinicus. The addition of the HAT inhibitor anacardic acid blocked the transcription of the orsA gene (Fig. S1), but not the actin gene analyzed as a control. By contrast, the histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA) activated the orsA gene without the need for coincubation of A. nidulans with S. rapamycinicus (Fig. S1C). These effects were confirmed by HPLC analysis of the fungal culture extracts (Fig. S1 A and B).
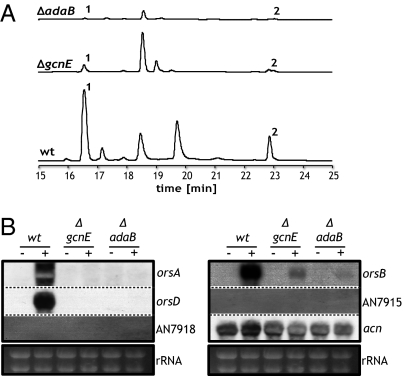
Based on these inhibitor studies, it was likely that acetylation of histones or other proteins, such as transcription factors, is required for the induction of the ors gene cluster by the streptomycete. By genome mining, we identified 40 genes encoding putative acetyltransferases. Deletion of 36 of these genes was successful; deletion of the remaining four appeared to be lethal (Table S1). Importantly, one of the deletion mutants showed a dramatically decreased ability to induce the ors genes after coincubation with the streptomycete. This mutant lacks the gcnE gene (16) that encodes a HAT orthologous to the Saccharomyces cerevisiae GCN5 (17, 18), also named KAT2 (19) (Table S1). GCN5 and its orthologs are members of several multisubunit coactivator complexes, such as SAGA, ADA, and NuA4, involved in histone acetylation and chromatin restructuring [reviewed in Baker and Grant (20)]. Gcn5 acetylates various lysine residues of histones H3 and H2B. Thereby, Gcn5 is involved in global acetylation, but also facilitates gene transcription via specific targeted acetylation (21). In S. cerevisiae, SAGA is recruited to promoter regions by interaction of its Tra1 subunit with transcription factors (22) such as Gal4 and Gcn4. Genome-wide expression profiling suggested a role for SAGA in the regulation of stress-related genes responding to environmental stress, such as metabolic starvation, DNA damage, and heat (23). Depending on the organism, Saga/Ada consists of about 20 subunits. In A. nidulans, it was shown that GcnE and AdaB are essential core subunits (16). To provide evidence that GcnE is part of the Saga/Ada complex, we also deleted the adaB gene (Fig. S2). The gcnE and adaB deletion strains generated here were viable, indicating that the complex is not required for essential cellular functions. Furthermore, both mutants exhibited the previously reported phenotype (16), i.e., strongly reduced formation of spores and a slightly reduced growth rate. As demonstrated by HPLC analysis of the culture supernatants, both mutants showed significantly reduced production of orsellinic and lecanoric acid when cocultured with S. rapamycinicus (Fig. 1A). Consistently, in the gcnE and adaB mutant strains, there were hardly any transcripts of the ors genes analyzed, i.e., orsA, orsB, and orsD (Fig. 1B). The reintegration of the gcnE gene into the ΔgcnE mutant strain fully restored the wild-type phenotype, including expression of ors cluster genes and orsellinic acid production during cocultivation of A. nidulans with S. rapamycinicus (Fig. S3A). These data proved that GcnE is responsible for the described phenotypes and confirmed the function of the Saga/Ada complex for induction of the ors gene cluster. By contrast, the actin gene used for normalization of expression did not show a major differential regulation in both the gcnE and adaB mutant strains, suggesting that the Saga/Ada complex specifically regulates expression of the ors genes.
Fig. 1.
Orsellinic acid and lecanoric acid biosynthesis in gcnE and adaB deletion strains of A. nidulans during cocultivation with S. rapamycinicus. (A) HPLC profiles of ethyl acetate extracts of ΔgcnE and ΔadaB and the wild type cocultivated with S. rapamycinicus. 1, orsellinic acid; 2, lecanoric acid. (B) Northern blot analysis of ors cluster genes in wild-type and deletion strains incubated without (−) or with (+) S. rapamycinicus. The genes AN7915 and AN7918 lie next to the ors gene cluster. The β-actin–encoding gene acn, which is not induced by S. rapamycinicus and not affected by Saga/Ada, served as an internal control.
To further analyze the global meaning of GcnE on gene expression, we performed full genome microarray analysis. We showed that GcnE plays a major role in the fast fungal response to the streptomycete (SI Results and Discussion and Fig. S4).
Saga/Ada Complex Is a Specific Regulator of Secondary Metabolism of A. nidulans.
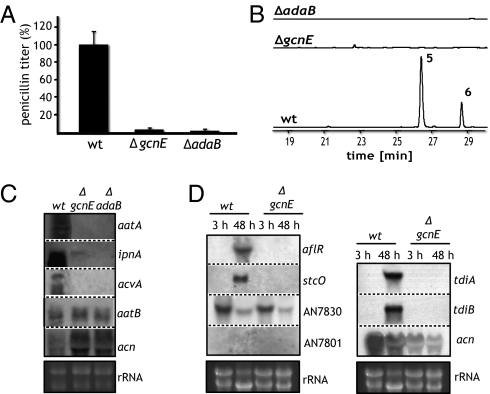
To address the question of whether the Saga/Ada complex is also required for the regulation of other secondary metabolite gene clusters in A. nidulans, we incubated the wild-type and deletion strains in Aspergillus minimal medium (AMM) for 72 h. Within this time period, the fungus started to produce compounds like sterigmatocystin and terrequinone A (24). HPLC profiles of whole-culture extracts indicated that both the gcnE and adaB mutant strains were impaired in the production of sterigmatocystin and terrequinone A (Fig. 2B). Northern blot analysis confirmed this observation, because in the ΔgcnE mutant, mRNA of sterigmatocystin and terrequinone biosynthesis genes was not detected (Fig. 2D). The specific role played by the Saga/Ada complex for the biosynthesis of these compounds was further supported by the observation that the mRNA levels of genes not belonging to these clusters, such as the genes AN7830 and AN7801, were not affected by a nonfunctional Saga/Ada complex (Fig. 2D).
Fig. 2.
Saga/Ada activity is required for the production of penicillin, sterigmatocystin, and terrequinone A in A. nidulans. Calculated penicillin titers (A) and HPLC profiles of ethyl acetate extracts (B) of wild type, ΔgcnE, and ΔadaB. The wild-type penicillin titer was set to 100%. 5, sterigmatocystin; 6, terrequinone A. Northern blot of penicillin biosynthesis genes acvA, ipnA, aatA, and aatB (C) and of selected sterigmatocystin and terrequinone cluster genes (D) in wild-type and deletion strains. Strains were cultivated in fermentation medium for 48 h (C) and under nonproducing (3 h of cultivation in AMM) and producing (48 h of cultivation in AMM) conditions (D). aflR and stcO are members of the sterigmatocystin cluster; the genes AN7830 and AN7801 are located directly adjacent to the cluster. tdiA and tdiB represent central genes of the terrequinone gene cluster. The actin gene acn served as a control for a gene not affected by Saga/Ada.
Another well-studied secondary metabolite produced by A. nidulans is penicillin. Four genes, namely acvA, ipnA, aatA, and aatB, encode the enzymes required for penicillin's biosynthesis (25, 26). Whereas acvA, ipnA, and aatA are located in a cluster on chromosome VI, aatB is separately located on chromosome I. As measured by a bioassay, the Saga/Ada mutant strains ΔgcnE and ΔadaB produced only ∼3% of this antibiotic compared with the wild type (Fig. 2A). This finding was reflected by a strongly reduced expression of the biosynthesis genes aatA, ipnA, and acvA (Fig. 2C). Interestingly, no difference for aatB expression was observed between the wild-type and the Saga/Ada-deficient strains. Consistent with the data on aatB, the actin gene is also not part of the cluster and thus not regulated by Saga/Ada (Fig. 2C), further confirming the specificity of the complex for distinct gene clusters. Penicillin production was restored to that of the wild type in the complemented gcnEc strain (Fig. S3C).
We also analyzed whether the laeA gene, encoding a potential methyltransferase involved in the expression of several secondary metabolism gene clusters (27), influences activation of the ors gene cluster. As shown in Fig. S5, the laeA gene was neither expressed during bacterial–fungal cocultivation nor influenced by Saga/Ada during sterigmatocystin production. Importantly, deletion of the laeA gene did not affect either the ors gene cluster expression or orsellinic acid and lecanoric acid production, thus making an involvement of LaeA in the bacteria-induced activation of the ors gene cluster unlikely.
Saga/Ada Complex Catalyzes Gene Cluster-Specific Acetylation.
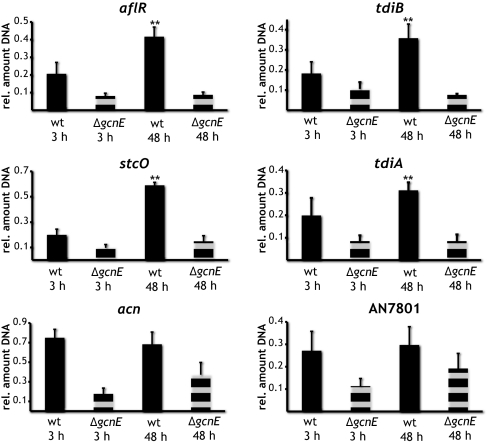
Prominent targets of histone acetylation by the Saga/Ada complex are lysine K9 and K14 of histone 3 (H3). Recently, Reyes-Dominguez et al. (28) showed that acetylation of these H3 residues in the promoter region of the sterigmatocystin biosynthesis regulatory gene aflR was increased when the fungus was shifted to sterigmatocystin production conditions. However, a high level of acetylation did not necessarily lead to a high expression of cluster genes. It thus remained elusive whether these histone marks play an important role in secondary metabolite regulation and whether this acetylation is catalyzed by the Saga/Ada complex. Therefore, we determined the histone acetylation of both sterigmatocystin and terrequinone cluster genes by ChIP and used antibodies targeted against both K9 and K14 of H3. In the wild-type strain, for all promoter regions of genes residing inside the aflatoxin or terrequinone gene cluster (i.e., aflR, stcO, tdiA, and tdiB), we detected increased acetylation at both lysines of H3 after 48 h compared with the 3-h incubation (Fig. 3). This increased acetylation coincided with the expression of the genes after 48 h (Fig. 2D). Promoters of genes not confined to the clusters, such as acn and AN7801, did not show any change in acetylation of H3 (Fig. 3). These results indicate a cluster-specific acetylation of H3 in response to secondary metabolite production conditions. Furthermore, in the Saga/Ada deletion strain, at both time points, i.e., 3 h and 48 h, we observed a significant reduction in K9/K14 acetylation of H3 compared with the acetylation level in the wild-type strain (Fig. 3). Hence, Saga/Ada is responsible for these histone marks, which are likely required for secondary metabolite gene cluster activation. In addition, Saga/Ada also affected promoter acetylation of genes whose regulation did not change and that are not organized in clusters, as seen for the genes acn and AN7801. Similar results were obtained for proline utilization genes. Deletion of gcnE drastically reduced the degree of acetylation, but not the level of gene activation (16). Despite reduced H3K9K14 acetylation levels in the proline utilization genes, the loss of nucleosome positioning associated with the gene activation event was identical between the wild-type and the gcnE mutant. It was proposed that the strength of interaction between the activator and its target in the transcription complex elicits the extent to which activation will depend on the global acetylation function (29). We suggest the same is true for targeted acetylation by the Saga/Ada complex reported here.
Fig. 3.
Saga/Ada-mediated increase of H3 acetylation of sterigmatocystin and terrequinone cluster genes. Wild-type and ΔgcnE strains were incubated under nonproducing (3-h incubation) and producing (48-h incubation) conditions. ChIP was carried out by probing acetylated K9 and K14 at H3 followed by qRT-PCR analysis of the promoter regions of the genes aflR, stcO, tdiA, tdiB, AN7801, and acn. Data are given as the amount of DNA obtained from acetylated H3 relative to the total amount of H3 protein. SDs of the mean of three biological replicates are indicated. The missing increase of the DNA amounts of both the sterigmatocystin cluster genes aflR and stcO and the terrequinone cluster genes tdiA and tdiB under production conditions in ΔgcnE indicates the need for Saga/Ada for the expression of these two gene clusters. Statistical significance of data are given by the P value (*P < 0.05; **P < 0.01).
Next, we addressed the question of which lysine of H3, K9 or K14, is more important for the cluster-specific gene activation. Previously, it was reported that K14 of H3 is an activation mark for light-induced gene expression in the filamentous fungus Neurospora crassa (30). Therefore, by ChIP analysis we studied this modification for the promoter of the sterigmatocystin regulatory gene aflR (Fig. S6A). We found a highly increased acetylation level under sterigmatocystin production conditions that was dependent on the Saga/Ada component GcnE. Surprisingly, we observed a significant increase of K14 acetylation for genes transcriptionally not regulated by Saga/Ada, i.e., acn and AN7801. By contrast, ChIP experiments with K9-specific antibodies only revealed an augmentation in precipitated DNA for promoter regions residing inside the secondary metabolite gene cluster. As seen in Fig. S6B, and consistent with the results obtained with the combined H3K9K14-specific antibody (Fig. 3), the cluster genes aflR and stcO showed a strong increase of K9 acetylation after 48 h that was absent in the gcnE deletion strain. This increase was not observed in promoters of genes that are not part of the clusters, such as acn and AN7801. Taken together, our data indicate that the production of secondary metabolites in A. nidulans is associated with a global increase in H3K14 acetylation. Specificity for cluster genes, however, depends on H3K9 acetylation.
S. rapamycinicus-Induced Activation of the ors Cluster in A. nidulans Is Mediated by Targeted Histone Acetylation Dependent on Saga/Ada.
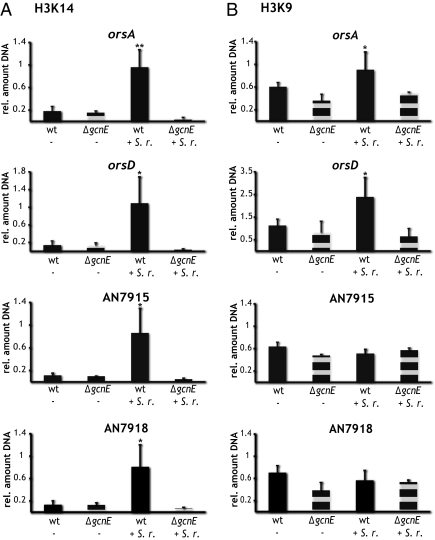
Our data pointed to the question of whether the acetylation level of H3 is decisive for induction of the A. nidulans ors gene cluster by S. rapamycinicus. We approached this question by performing ChIP experiments with antibodies targeted against either K9 or K14. Two promoter regions of genes located next to the ors cluster and of genes within the ors cluster were analyzed (Fig. 4). The acetylation pattern reflected the data seen for the sterigmatocystin gene cluster. Both acetylation marks (K9 and K14 of H3) increased during the bacterial–fungal interaction, but were reduced in the gcnE deletion strain, indicating the requirement of a functional Saga/Ada complex for (i) the acetylation triggered by the streptomycete and (ii) for acetylation of K9 and K14 (Fig. 4).
Fig. 4.
H3K9 and H3K14 acetylation during interaction of S. rapamycinicus and A. nidulans. ChIP experiment of wild type and ΔgcnE incubated without (−) or with (+) S. rapamycinicus (S. r.). Promoter regions of orsA, orsD, AN7915, and AN7918 were analyzed by qRT-PCR. SDs of two (H3K14) and three (H3K9) biological replicates are shown. Data are given as the amount of DNA obtained from acetylated H3 protein relative to the total amount of H3 protein. Equal amounts of DNA in the wild-type and ΔgcnE samples under inducing conditions show the Saga/Ada-mediated H3K14 (A) and H3K9 (B) acetylation of the ors genes during cocultivation of A. nidulans and S. rapamycinicus. Statistical significance of data are given by the P value (*P < 0.05; **P < 0.01).
In accordance with the data obtained for the sterigmatocystin gene cluster, the increase in K14 acetylation during fungus/streptomycete coincubation was detected not only in the ors genes, but also in the genes AN7915 and 7918, which are located close to the ors cluster. By contrast, and as also observed for the sterigmatocystin genes (Fig. S6), the K9 acetylation was restricted to genes belonging to the ors cluster, whereas the promoters of AN7915 and AN7918 did not show a difference in this acetylation level (Fig. 4).
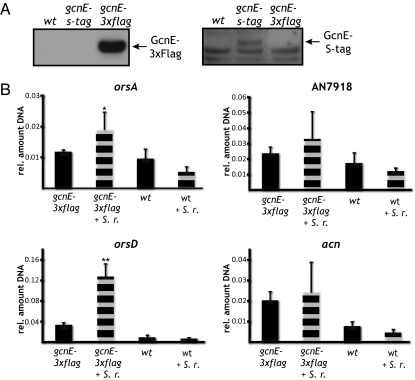
To unequivocally prove that Saga/Ada directly catalyzes the cluster-specific acetylation, we generated a tagged version of GcnE and performed ChIP analysis against this GcnE-3xFLAG protein. As shown in Fig. 5, GcnE-3xFLAG was specifically recruited to the ors cluster during cocultivation of A. nidulans with S. rapamycinicus, fully confirming the previous data obtained with the anti-H3K9/K14 antibodies. The observation that all activation events of the four different secondary metabolite gene clusters investigated here depend on H3 acetylation by Saga/Ada components GcnE/AdaB, but only the ors cluster is activated by bacterial–fungal interaction, deserves some additional comments. These results strengthen the hypothesis that chromatin-level regulation of secondary metabolite gene clusters represents an upper-hierarchical level of regulation, whereas each cluster requires an additional, more-specific signal to be fully activated (31). For some clusters, such as the penicillin or the sterigmatocystin biosynthesis gene cluster, several of these additional signals and regulators have been identified (25, 32). However, it is also conceivable that for some gene clusters, such as the ors locus, full induction only requires chromatin reprogramming induced by the bacterium, which leads to subsequent activation by the general transcription machinery (Fig. 6).
Fig. 5.
GcnE is localized to the ors gene cluster promoters during cocultivation of A. nidulans with S. rapamycinicus. (A) Western blot detection of GcnE-tagged proteins. Protein lysates of wild type, gcnE-S-tag, and gcnE-3xflag coincubated with S. rapamycinicus were analyzed with either monoclonal anti-FLAG antibody or polyclonal anti–S-tag antibody. The anti-FLAG antibody showed high specificity for GcnE-3xFLAG, whereas the anti–S-tag antibody showed lower specificity, resulting in further bands in strains not producing S-tagged GcnE. (B) ChIP analysis of wild type and gcnE-3xflag incubated with or without S. rapamycinicus using an anti–GcnE-3xFLAG monoclonal antibody followed by qRT-PCR analysis of the orsA, orsD, AN7818, and acn promoter regions. Data are given as the amount of DNA obtained from FLAG-tagged GcnE protein relative to the amount of total DNA obtained from input protein. SDs of the mean of three biological replicates are indicated. Statistical significance of data obtained for gcnE-3xflag and gcnE-3xflag + S. rapamycinicus for orsA and orsD promoter was given by calculating the P value (*P < 0.05; **P < 0.01).
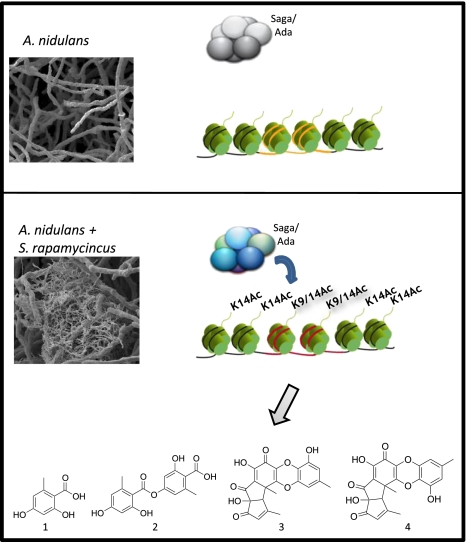
Fig. 6.
Model for histone acetylation-mediated secondary metabolite gene cluster activation in A. nidulans by S. rapamycinicus. Secondary metabolite genes (orange) under noninducing conditions are characterized by deacetylated histone H3. The intimate contact between A. nidulans and S. rapamycinicus leads to an increased acetylation of histone H3 catalyzed by the Saga/Ada complex. The modification of H3K9 is specific for the secondary metabolite gene clusters (red), whereas H3K14 acetylation is not specifically targeted. Hence, Saga/Ada triggers the expression of the ors genes and the formation of orsellinic acid (1), lecanoric acid (2), F-9775A (3), and F-9775B (4).
As shown here, upon direct physical contact, the bacterium induces targeted histone acetylation that requires the Saga/Ada complex. This posttranslational regulation results in the specific response of the fungus, including the production of orsellinic acid and its derivatives (Fig. 6). Based on the importance of GcnE for the production of these substances and the impact of the Saga/Ada complex on bacteria-induced secondary metabolite production, we thus anticipate a key role of GcnE in the fungal reaction to the microbial surrounding. It is intriguing that bacteria-mediated posttranslational modifications are increasingly recognized as key strategies used by pathogens to modulate factors of mammalian hosts critical for infection (33). As shown here, such mechanisms might have been evolved very early during evolution of communication systems between microorganisms of different kingdoms.
In conclusion, we show that bacteria are able to trigger alterations of histone modification in fungi. This reprogramming leads to specific activation of the biosynthesis of distinct secondary metabolites derived from orsellinic acid. Our conclusion was based on HAT inhibitor studies and the deletion of most of the acetyltransferase genes encoded by A. nidulans, including HATs. Among these genes, we identified the gcnE gene required for the induction of the ors gene cluster by HPLC, liquid chromatography–mass spectrometry (LS-MS), and Northern blot analysis. Because deletion of the adaB gene gave the same phenotype as gcnE deletion, we concluded that the Saga/Ada complex is the regulatory target of bacteria-induced histone modification. Impressively, Saga/Ada is also required for production of other natural products, such as penicillin, sterigmatocystin, and terrequinone A. The production of these compounds and the mRNA levels of their biosynthesis genes were massively down-regulated in both the gcnE and adaB mutant strains. Thus, besides of one of the proposed functions of Saga/Ada in stress regulation, we expanded the function of the Saga/Ada complex to the induction of secondary metabolism gene clusters. It remains to be elucidated whether this induction caused by the bacterium represents a type of stress signal or a communication signal. We applied extensive ChIP experiments to successfully determine the histone modifications catalyzed by the Saga/Ada complex and associated with the bacterial control of orsellinic acid production. Our data suggests that specificity for activation of cluster genes depends on H3K9 acetylation. Several distinct interactions between fungi and bacteria have been identified thus far, and only a few of them have been investigated in detail, such as the symbiosis of bacteria and fungi in lichens and the symbiosis of intracellular bacteria in zygomycetes (34, 35). Here, by the finding that the bacteria-induced secondary metabolite production in A. nidulans is mediated by histone modification via the Saga/Ada complex, we postulate a possible mechanism by which fungi integrate stimuli from interacting microbes.
Materials and Methods
Strains.
A. nidulans and bacterial strains used in this study are listed in Table S2. Deletion strains were obtained by transformation (36) using the argB gene of A. nidulans as a selectable marker. Deletion cassettes were produced as described in Szewczyk (37). Strains with tagged gene variants were obtained using the pabaA1 gene of A. nidulans as a selectable marker.
Media, Cultivation Conditions, and Penicillin Bioassay.
A. nidulans was incubated in Aspergillus minimal medium (AMM) (38). When required, supplements were added as follows: arginine (final concentration 50 μM), p-aminobenzoic acid (3 μg⋅mL−1), pyridoxine HCl (5 μg⋅mL−1), and biotin (0.6 μg⋅mL−1). Each experiment was started with an AMM overnight culture that was inoculated with 4 × 106 spores mL−1 and incubated at 37 °C. The gcnE and adaB deletion strains were incubated 4 h longer than the wild type to warrant the formation of equal amounts of biomass. The biomass was separated from the medium using Miracloth and inoculated into fresh AMM. For sterigmatocystin and terrequinone production experiments, experimental cultures were grown at 37 °C for 72 h. Samples for RNA isolation were taken after 3 h and 48 h. Cross-linking for ChIP experiments was carried out at the same time points. For coincubation experiments, freshly prepared S. rapamycinicus was added to an A. nidulans culture, as described in Schroeckh et al. (13). If required, SAHA (4 mM) and anacardic acid (100 μM) were added at the same time point. For both RNA isolation and cross-linking for ChIP, mycelia were harvested after 3 h of coincubation. After 24 h of cultivation, the coculture was prepared for HPLC analysis. For penicillin production, fermentation media were applied as previously described (39). Penicillin bioassay was carried out with Bacillus calidolactis C953 as an indicator organism (40). Three biological replicates were analyzed to calculate SDs.
Preparation of Chromosomal DNA, Total RNA, Northern and Southern Blot Analyses, and Quantitative RT-PCR.
Isolation of chromosomal DNA and of total RNA of A. nidulans, Northern and Southern blot analysis, and quantitative RT-PCR (qRT-PCR) were performed as described in Schroeckh et al. (13). Oligonucleotides are listed in Table S3.
Western Blot, ChIP Coupled to qRT-PCR Analysis, Generation of Transformation Cassettes, Microarray, and Analysis of Secondary Metabolites.
Western blot analysis, ChIP coupled to qRT-PCR analysis, generation of deletion cassettes, microarrays, and extraction, HPLC, and LC-MS of secondary metabolites are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank N. Hannwacker, C. Täumer, M. G. Schwinger, and K. Perlet for technical assistance in cultivation of microorganisms and Southern and Northern blot analyses, and A. Perner for MS measurements. We are grateful to the Electron Microscopy Center of the University Hospital Jena for providing electron microscopic photographs. This research was supported by the excellence graduate school Jena School for Microbial Communication (JSMC) and the “Pakt für Forschung und Innovation” of the Bundesministerium für Bildung und Forschung and the Thüringer Ministerium für Bildung, Wissenschaft und Kultur, and the Austrian Science Fund Special Research Area - SFB Project F-3703 and Vienna Science and Technology Fund Project WWTF-LS09042.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103523108/-/DCSupplemental.
References
- 1.Brakhage AA. Molecular regulation of beta-lactam biosynthesis in filamentous fungi. Microbiol Mol Biol Rev. 1998;62:547–585. doi: 10.1128/mmbr.62.3.547-585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walsh CT. Natural insights for chemical biologists. Nat Chem Biol. 2005;1:122–124. doi: 10.1038/nchembio0805-122. [DOI] [PubMed] [Google Scholar]
- 3.Cox RJ. Polyketides, proteins and genes in fungi: Programmed nano-machines begin to reveal their secrets. Org Biomol Chem. 2007;5:2010–2026. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- 4.von Döhren H. A survey of nonribosomal peptide synthetase (NRPS) genes in Aspergillus nidulans. Fungal Genet Biol. 2009;46(Suppl 1):S45–S52. doi: 10.1016/j.fgb.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 5.Bergmann S, et al. Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic nonribosomal peptide synthetase gene cluster. Appl Environ Microbiol. 2010;76:8143–8149. doi: 10.1128/AEM.00683-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergmann S, et al. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- 7.Brakhage AA, Schroeckh V. Fungal secondary metabolites—strategies to activate silent gene clusters. Fungal Genet Biol. 2011;48:15–22. doi: 10.1016/j.fgb.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Chiang YM, et al. Molecular genetic mining of the Aspergillus secondary metabolome: Discovery of the emericellamide biosynthetic pathway. Chem Biol. 2008;15:527–532. doi: 10.1016/j.chembiol.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bok JW, et al. Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goh EB, et al. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc Natl Acad Sci USA. 2002;99:17025–17030. doi: 10.1073/pnas.252607699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yim G, Wang HH, Davies J. Antibiotics as signalling molecules. Philos Trans R Soc Lond B Biol Sci. 2007;362:1195–1200. doi: 10.1098/rstb.2007.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar Y, Goodfellow M. Five new members of the Streptomyces violaceusniger 16S rRNA gene clade: Streptomyces castelarensis sp. nov., comb. nov., Streptomyces himastatinicus sp. nov., Streptomyces mordarskii sp. nov., Streptomyces rapamycinicus sp. nov. and Streptomyces ruanii sp. nov. Int J Syst Evol Microbiol. 2008;58:1369–1378. doi: 10.1099/ijs.0.65408-0. [DOI] [PubMed] [Google Scholar]
- 13.Schroeckh V, et al. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci USA. 2009;106:14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scherlach K, Hertweck C. Triggering cryptic natural product biosynthesis in microorganisms. Org Biomol Chem. 2009;7:1753–1760. doi: 10.1039/b821578b. [DOI] [PubMed] [Google Scholar]
- 15.Williams RB, Henrikson JC, Hoover AR, Lee AE, Cichewicz RH. Epigenetic remodeling of the fungal secondary metabolome. Org Biomol Chem. 2008;6:1895–1897. doi: 10.1039/b804701d. [DOI] [PubMed] [Google Scholar]
- 16.Reyes-Dominguez Y, et al. Nucleosome positioning and histone H3 acetylation are independent processes in the Aspergillus nidulans prnD-prnB bidirectional promoter. Eukaryot Cell. 2008;7:656–663. doi: 10.1128/EC.00184-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Timmers HT, Tora L. SAGA unveiled. Trends Biochem Sci. 2005;30:7–10. doi: 10.1016/j.tibs.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allis CD, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 20.Baker SP, Grant PA. The SAGA continues: Expanding the cellular role of a transcriptional co-activator complex. Oncogene. 2007;26:5329–5340. doi: 10.1038/sj.onc.1210603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo MH, vom Baur E, Struhl K, Allis CD. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol Cell. 2000;6:1309–1320. doi: 10.1016/s1097-2765(00)00129-5. [DOI] [PubMed] [Google Scholar]
- 22.Brown CE, et al. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science. 2001;292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- 23.Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 24.Bouhired S, Weber M, Kempf-Sontag A, Keller NP, Hoffmeister D. Accurate prediction of the Aspergillus nidulans terrequinone gene cluster boundaries using the transcriptional regulator LaeA. Fungal Genet Biol. 2007;44:1134–1145. doi: 10.1016/j.fgb.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 25.Brakhage AA, et al. Aspects on evolution of fungal beta-lactam biosynthesis gene clusters and recruitment of trans-acting factors. Phytochemistry. 2009;70:1801–1811. doi: 10.1016/j.phytochem.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 26.Spröte P, et al. Identification of the novel penicillin biosynthesis gene aatB of Aspergillus nidulans and its putative evolutionary relationship to this fungal secondary metabolism gene cluster. Mol Microbiol. 2008;70:445–461. doi: 10.1111/j.1365-2958.2008.06422.x. [DOI] [PubMed] [Google Scholar]
- 27.Bok JW, Keller NP. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reyes-Dominguez Y, et al. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol Microbiol. 2010;76:1376–1386. doi: 10.1111/j.1365-2958.2010.07051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imoberdorf RM, Topalidou I, Strubin M. A role for gcn5-mediated global histone acetylation in transcriptional regulation. Mol Cell Biol. 2006;26:1610–1616. doi: 10.1128/MCB.26.5.1610-1616.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grimaldi B, et al. The Neurospora crassa White Collar-1 dependent blue light response requires acetylation of histone H3 lysine 14 by NGF-1. Mol Biol Cell. 2006;17:4576–4583. doi: 10.1091/mbc.E06-03-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strauss J, Reyes-Dominguez Y. Regulation of secondary metabolism by chromatin structure and epigenetic codes. Fungal Genet Biol. 2011;48:62–69. doi: 10.1016/j.fgb.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu JH, Keller N. Regulation of secondary metabolism in filamentous fungi. Annu Rev Phytopathol. 2005;43:437–458. doi: 10.1146/annurev.phyto.43.040204.140214. [DOI] [PubMed] [Google Scholar]
- 33.Ribet D, Cossart P. Pathogen-mediated posttranslational modifications: A re-emerging field. Cell. 2010;143:694–702. doi: 10.1016/j.cell.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grube M, Berg G. Microbial consortia of bacteria and fungi with focus on the lichen symbiosis. Fungal Biol Rev. 2009;23:72–85. [Google Scholar]
- 35.Lackner G, Moebius N, Hertweck C. Endofungal bacterium controls its host by an hrp type III secretion system. ISME J. 2011;5:252–261. doi: 10.1038/ismej.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ballance DJ, Turner G. Development of a high-frequency transforming vector for Aspergillus nidulans. Gene. 1985;36:321–331. doi: 10.1016/0378-1119(85)90187-8. [DOI] [PubMed] [Google Scholar]
- 37.Szewczyk E, et al. Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- 38.Brakhage AA, Van den Brulle J. Use of reporter genes to identify recessive trans-acting mutations specifically involved in the regulation of Aspergillus nidulans penicillin biosynthesis genes. J Bacteriol. 1995;177:2781–2788. doi: 10.1128/jb.177.10.2781-2788.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Then Bergh K, Litzka O, Brakhage AA. Identification of a major cis-acting DNA element controlling the bidirectionally transcribed penicillin biosynthesis genes acvA (pcbAB) and ipnA (pcbC) of Aspergillus nidulans. J Bacteriol. 1996;178:3908–3916. doi: 10.1128/jb.178.13.3908-3916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brakhage AA, Browne P, Turner G. Regulation of Aspergillus nidulans penicillin biosynthesis and penicillin biosynthesis genes acvA and ipnA by glucose. J Bacteriol. 1992;174:3789–3799. doi: 10.1128/jb.174.11.3789-3799.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.