Abstract
We investigated the effect of methionine sulfoximine (MetSox), a potent inhibitor of glutamine synthetase, on Mycobacterium tuberculosis. M. tuberculosis encodes four glutamine synthetases, of which MetSox targets the type I enzyme encoded by glnA1. Trancriptional profiling revealed that glutamate synthetase (gltB) and a type II glutamine synthetase (glnA3) were induced after exposure to MetSox. In addition, we observed a high rate (10−5) of spontaneous resistance to MetSox. All resistant strains had a single-nucleotide deletion in the 5′ region of glnA1, and Western analysis revealed that GlnA1 expression was increased in resistant as compared with sensitive strains. These data show that M. tuberculosis can respond to the effect of MetSox inhibition either by up-regulation of GlnA3 or by GlnA1. The high frequency of resistance suggests that MetSox and other compounds specifically targeting GlnA1 are not likely to become successful anti-mycobacterial agents.
Introduction
Nitrogen assimilation is a critical metabolic pathway in Mycobacterium tuberculosis, as several of the key enzymes have been identified as being essential for growth or for virulence1,5–8,11 Glutamine synthetase (GS) plays a key role in nitrogen assimilation, as it catalyzes the ATP-dependent reaction between glutamate and ammonia to generate glutamine. M. tuberculosis has four GSs, although only GlnA1, the major GS, appears to be required for in vitro growth.6 Methionine sulfoximine (MetSox) is a potent inhibitor of GS.9 MetSox is phosphorylated by GS in the presence of ATP, and it irreversibly binds to the active site, thereby preventing the entry of the substrate glutamate. MetSox inhibits the growth of M. tuberculosis in axenic culture and during intracellular infection of macrophages.4 In addition, we recently demonstrated that MetSox-mediated inhibition of GS activity, in combination with L-glutamine (L-gln) supplementation, was sufficient to allow the isolation of strains with deletions in GlnE, an adenylyl transferase which post-transcriptionally regulates GS activity.1,8
Novel drugs are urgently required for treating M. tuberculosis infections; therefore, we wanted to assess whether GlnA1 inhibition was a rational approach for treatment. In order to evaluate this, we investigated the effect of MetSox treatment on M. tuberculosis, measuring growth inhibition, profiling drug-exposure signatures, and defining resistance mutations.
Materials and Methods
Bacterial strains, growth media, and antibiotics
M. tuberculosis H37Rv (ATCC 25618) was grown in Middlebrook 7H9 medium plus 10% (vol/vol) oleic acid-albumin-dextrose-catalase (OADC) supplement (Becton Dickinson) and 0.05% (wt/vol) Tween 80 or on Middlebrook 7H10 agar plus 10% (vol/vol) OADC. Methionine sulfoximine, L-glutamine, and D-glutamine were used as described.
Transcriptomics
M. tuberculosis H37Rv was grown to mid-log phase (day 4, OD580 approx 0.4) and exposed to 200 μM methionine sulfoximine for 4 and 8 hours alongside a carrier control (100 μl H2O). Mycobacterial RNA was extracted using the GTC/Trizol method,12 DNase-treated and purified using RNeasy columns (Qiagen). A M. tuberculosis whole genome microarray, generated by the Bacterial Microarray Group at St. George's (ArrayExpress accession number A-BUGS-23; http://bugs.sgul.ac.uk/A-BUGS-23), was hybridized as described12,13 using M. tuberculosis genomic DNA as a common reference. Three biological replicates of RNA derived from MetSox-treated and control cultures were hybridized in duplicate. Comparative spot intensities from the images were calculated using Imagene 5.5 (BioDiscovery) and imported into GeneSpring GX 7.3 (Agilent Technologies) for further analysis. These data were normalized to the 50th percentile of all genes detected to be present on the array and filtered to include only genes flagged to be present on 80% of the arrays. Genes with a fold change >1.5 relative to the carrier control were classed as differentially expressed. The Hypergeometric probability (p-value <0.05) was used to identify functional categories of genes significantly over-represented. Differentially expressed gene lists together with fully annotated microarray data have been deposited in BμG@Sbase (accession number: E-BUGS-112; http://bugs.sgul.ac.uk/E-BUGS-112) and ArrayExpress (accession number: E-BUGS-112).
Growth curves
Liquid cultures were diluted to give a starting optical density at 580 nm (OD580) of 0.01 in 4 ml of medium in 12 mm tubes. Each tube contained a magnetic stirrer and was incubated at 37°C on a Wheaton Biostir. OD580 readings were taken periodically.
GlnA1 amplification
Primers GlnA1F (5′-GTAAAGGAGCATTCTGTGACGGAA-3′) and GlnA1R (5′-ACGTCGTAGTACAGCGCGAATT-3′) were used to amplify glnA1 and the upstream region for sequencing (product size 1,448 bp).
Western analysis of GlnA1
Cell extracts were prepared from liquid cultures. Cells were harvested by centrifugation, washed twice in 10 mM Tris (pH 8.0), resuspended in 1 ml of 10 mM Tris (pH 8.0), and added to lysing matrix B tubes (QBiogene). Cells were disrupted using the Fastprep (QBiogene) set at speed 6.0 for 30 seconds. Samples were centrifuged for two min, and the supernatant was recovered and filter sterilized. Protein was quantified using a BCA kit (Pierce), and 15 μg of total protein was loaded onto 12% polyacrylamide gels and transferred to PVDF membranes (Invitrogen) for Western blot analysis. Membranes were probed with rat anti-GlnA1 antibody from Ida Rosenkrands (Statens Serum Institut, Denmark). The primary antibody was detected using horseradish peroxidase goat-anti-rat (Sigma), and activity was detected using an ECL kit (GE Healthcare).
Results
The effects of MetSox on M. tuberculosis growth and survival
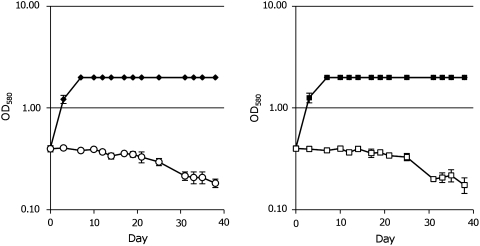
We determined the effects of MetSox on the growth of M. tuberculosis by measuring both changes in optical density and counting viable bacilli. First, we determined that the MIC99 of MetSox for M. tuberculosis on solid medium was 50 μM. When treated with 200 μM, MetSox cultures showed complete inhibition of growth, and, in fact, the OD580 slowly decreased from 0.4 to 0.18 (Fig. 1A). Inhibition of growth was completely relieved by the addition of 3 mM L-glutamine (Fig. 1B), which was expected as MetSox treatment should have the equivalent phenotypic effects as the deletion of glnA1, encoding glutamine synthestase, that is, glutamine auxotrophy. We titrated the minimum amount of L-gln required to overcome inhibition by MetSox: cells were able to grow in medium containing 300 μM, but not 30 μM L-gln. Interestingly, 3 mM D-gln was not able to rescue the cells from MetSox-dependent killing, suggesting that the bacteria lack any isomerase able to convert D-gln to L-gln.
FIG. 1.
Inhibition of Mycobacterium tuberculosis H37Rv growth by MetSox. M. tuberculosis was cultured in (A) medium only (solid diamond) or medium plus 200 μM MetSox (open circle) and (B) medium plus 2 mM MetSox (open box) or 200 μM MetSox and 3 mM L-gln (filled box). Data represent the average and standard deviations of four independent cultures.
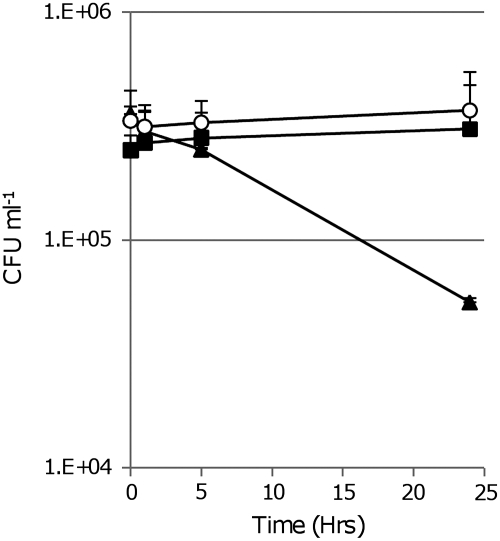
Inhibition of growth can result from either bacteriostatic or bactericidal effects. The decrease in OD suggested that the bacteria were lysing, and, thus, MetSox exposure is lethal. In order to assess this, we cultured M. tuberculosis in liquid medium containing 200 μM MetSox and assayed viability by plating serial dilutions (Fig. 2). In the presence of 200 μM MetSox, the numbers of viable bacteria decreased by almost two logs (3.57 × 105 to 9.07 × 103) in 24 hours. In contrast, the M. tuberculosis control (no MetSox) and rescued cultures (MetSox plus L-gln) showed a 2-fold increase in viability, due to cell multiplication. These data show that MetSox has a rapid effect on cell viability (within 24 hours), although cells do not immediately lyse, as the OD remains the same over the short time period assayed.
FIG. 2.
MetSox-mediated killing of M. tuberculosis. M. tuberculosis H37Rv was cultured in medium only (solid square); 200 μM MetSox (solid triangle); 200 μM MetSox, 3 mM L-Gln (open circle). Viability was measured by counting CFUs. Data represent the average and standard deviations for three independent cultures.
The M. tuberculosis transcriptional signature to MetSox exposure
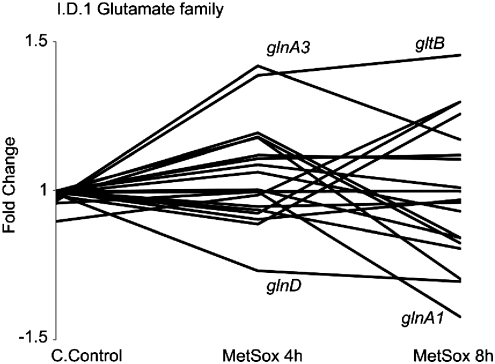
The M. tuberculosis response to the early effects of glutamine synthetase inhibition was determined by treating log phase axenic cultures with 200 μM MetSox and profiling the global gene expression patterns 4 and 8 hours after exposure. The transcriptional response to MetSox was limited, with no genes >3-fold differentially regulated relative to the carrier control at 4 or 8 hours. 60 genes were >1.5-fold differentially expressed after 4 hours of MetSox exposure, and the same with regard to 72 genes after 8 hours. The functional category 1.D.1 Glutamate Family, including genes involved in the metabolism of glutamine/glutamate,2 was significantly enriched in the subset of genes induced after MetSox exposure at 4 or 8 hours (Fig. 3). Specifically, gltB coding for glutamate synthase and glnA3 encoding glutamine synthetase 3 were induced reflecting the changing pool of nitrogen intermediates available on inhibition of glnA1. Rv1879 directly downstream of glnA3 was also up-regulated by MetSox treatment. The induction of glnA3 may reveal a compensatory function for this alternative glutamine synthetase or highlight different mechanisms of regulatory control, with glnA3 transcriptionally induced in response to loss of GS function; whereas glnA1 is regulated primarily at the post-translational level. Interestingly, genes in the functional category I.B.7 encompassing miscellaneous oxidoreductases and oxygenases2 were also significantly over-represented in the subset of genes induced by MetSox treatment. This may constitute the weak inhibition of gshA, encoding a glutamate-cysteine ligase, that disrupts mycobacterial redox potential.6
FIG. 3.
MetSox-mediated changes in the regulation of glutamate metabolism. M. tuberculosis was exposed to 200 μM MetSox for 4 or 8 hours; global gene expression profiles were determined by competitive hybridization of RNA to genomic DNA using whole genome microarrays. The relative levels of expression for each gene were determined and normalized to the control (untreated) population. The gene expression pattern of the 1.D.1 glutamate metabolism family of genes2 is shown; four key genes are marked.
Identification and characterization of MetSox resistant mutants
During the course of these experiments, we noted the appearance of spontaneous resistant mutants. When bacilli from liquid cultures containing MetSox were plated onto 200 μM MetSox, we routinely observed low numbers of viable colonies. Cells plated from the viability experiment were used to estimate the frequency of resistance; from the initial 2.7 × 105 inoculum, 32 colonies were isolated on MetSox medium, giving a resistance frequency of 10−5. Twelve strains were further analyzed. Resistance was confirmed by streaking these strains onto MetSox-containing solid medium. All twelve strains grew on plates containing 100, 200, and 400 μM MetSox, confirming that these strains were truly resistant to MetSox. Wild-type M. tuberculosis did not grow when streaked onto plates containing MetSox unless L-gln was added.
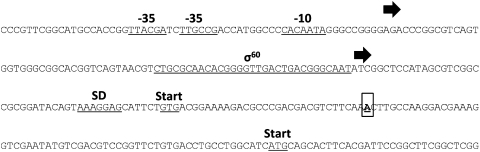
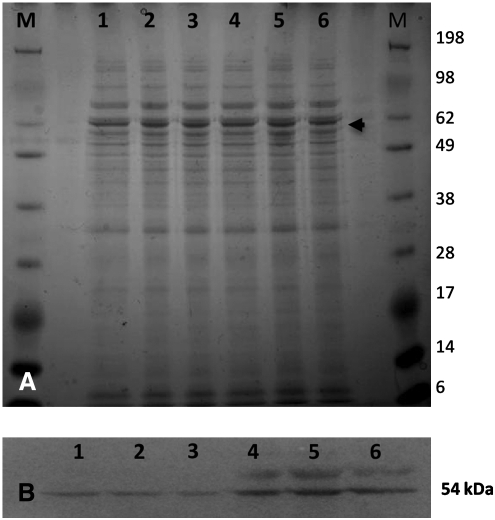
Since the MetSoxR strains isolated were not auxotrophic for L-gln, it indicated that a functional GS was still present. Given the high frequency of spontaneous mutation, we decided to determine the mechanism of resistance to MetSox. We focused on the major type I GS, GlnA1, as it seemed the most likely target of MetSox action and, thus, might harbor resistance-conferring mutations. We sequenced the glnA1 gene from eight resistant strains. We discovered a single nucleotide deletion located in the proposed coding region of glnA1 at +33 relative to the translational start site (genome position 2487647) in all the resistant strains analyzed (Fig. 4). This deletion would potentially lead to a frameshift mutation inactivating the protein. However, sequence analysis revealed a second potential translation start site, which would produce a 447 amino acid protein lacking the first 31 amino acids of the predicted full length GlnA1. Thus, we hypothesized that either the strains were producing a truncated protein with full GS activity or the translation start site of the protein is misannotated in the Tuberculist database (http://genolist.pasteur.fr/TubercuList/index.html) (Fig. 4). In order to examine these two possibilities, we investigated GlnA1 expression in the wild-type and MetSoxR strains. Cell-free extracts were generated from aerated liquid cultures and Western analysis carried out with an antibody generated to M. tuberculosis GlnA1. These data indicated that all MetSoxR strains expressed GlnA1 of the same apparent size as the wild-type strain. However, they all showed an increased level of expression of GlnA1, in comparison to the wild-type strain (Fig. 5). An unknown protein band at a higher molecular weight than GlnA1 was also present in the MetSoxR cell extracts.
FIG. 4.
Identification of a single base deletion in the upstream region of glnA1 in MetSox resistant strains. The upstream region of glnA1 is indicated. The upstream region and coding sequence of the resistant isolates were determined. The single nucleotide (A) that was deleted in all the MetSoxR strains is boxed and indicated in bold. The two identified transcriptional start sites are denoted with curved arrows; potential translational start sites, promoter regions (−10 and −35), the σ60 binding site, and Shine-Dalgarno (SD) sites are all indicated.
FIG. 5.
Expression of GlnA1 in MetSox resistant strains. Cell-free extracts were prepared from wild-type and MetSoxR strains grown under aerated conditions for 1 week. 15 μg of total protein was analyzed by (A) SDS-PAGE and (B) Western blotting. (A) Gels were stained with SimplyBlue (Invitrogen). Protein molecular marker sizes are displayed; the arrow indicates the approximate location of GlnA1 (57 kDa). (B) Western blotting was carried out with a GlnA1-specific antibody. Lanes 1–3: wild-type H37Rv; Lanes 4–6: three independent MetSoxR strains.
Discussion
We investigated the role of the potent GS inhibitor MetSox on both the transcriptome and growth of M. tuberculosis. Global expression analyses suggest that M. tuberculosis responds to MetSox exposure and the inhibition of GS activity by increasing transcription of several genes involved in nitrogen assimilation. Of these, GlnA3 is one of the four GS encoded by M. tuberculosis (GlnA1-4) and, similar to GlnA1, is involved in the synthesis of L-gln.6 However, GlnA3 is a heat-labile type II GS that is not post-translationally controlled by adenylylation, in contrast to the heat stable type I GS, GlnA1, controlled by adenlylyation.3,10 It is, therefore, plausible that GlnA3 expression increases in response to a decrease in the glutamine pool due to GlnA1 inactivation. Interestingly, expression of GltB, which encodes glutamate synthesis and is part of the GOGAT pathway, was increased. This suggests that GlnA3 is replacing the role of GlnA1; but, since activity cannot be regulated post-transcriptionally, similar to GlnA1, glutamine is being converted back to glutamate by GltB to regulate the internal nitrogen pool. A high frequency of spontaneous resistance was observed after MetSox exposure. All resistant strains had a single bp deletion downstream of the annotated translational start site. However, a second start site was located 93 bases from the original. Western analysis revealed that GlnA1 was still translated, leading us to conclude that the second translational start site is active and that the resistance-conferring deletion is in the upstream region either within the GlnA1 promoter or within a regulatory site. Thus, the single-nucleotide deletion conferring MetSox-resistance likely disrupts the regulation of glutamine synthetase, thereby resulting in increased GlnA1 expression.
Since MetSox resistance was easily generated by over-expression of the target GlnA1, it is likely that other inhibitors targeting the same enzyme would also result in resistance. Thus, we suggest that GlnA1 is not a suitable antimycobacterial drug target in the search for novel therapeutic agents.
Acknowledgments
This work was funded by the European Union Project LSHP-CT-2005-018923. The authors would like to thank Ida Rosenkrands for providing the GlnA1 antibody, Jasvir Dhillon for assistance with the transcriptional analysis, and Tanjore Balganesh for helpful discussions. PB would like to thank The Wellcome Trust for funding the Bacterial Microarray Group at St. George's (grant no. 062511). M. tuberculosis H37Rv reference DNA was provided by Colorado State University (HHSN266200400091C; NIH, NIAID N01-AI-40091).
Disclosure Statement
No competing financial interests exist.
References
- 1.Carroll P. Pashley C.A. Parish T. Functional analysis of GlnE, an essential adenylyl transferase in Mycobacterium tuberculosis. J. Bacteriol. 2008;190:4894–4902. doi: 10.1128/JB.00166-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cole S.T. Brosch R. Parkhill J. Garnier T. Churcher C. Harris D. Gordon S.V. Eiglmeier K. Gas S. Barry C.E. Tekaia F. Badcock K. Basham D. Brown D. Chillingworth T. Connor R. Davies R. Devlin K. Feltwell T. Gentles S. Hamlin N. Holroyd S. Hornby T. Jagels K. Krogh A. McLean J. Moule S. Murphy L. Oliver K. Osborne J. Quail M.A. Rajandream M.A. Rogers J. Rutter S. Seeger K. Skelton J. Squares R. Squares S. Sulston J.E. Taylor K. Whitehead S. Barrell B.G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 3.Darrow R.A. Knotts R.R. Two forms of glutamine synthetase in free-living root-nodule bacteria. Biochem. Biophys. Res. Commun. 1977;78:554–559. doi: 10.1016/0006-291x(77)90214-5. [DOI] [PubMed] [Google Scholar]
- 4.Harth G. Horwitz M.A. Expression and efficient export of enzymatically active Mycobacterium tuberculosis glutamine synthetase in Mycobacterium smegmatis and evidence that the information for export is contained within the protein. J. Biol. Chem. 1997;272:22728–22735. doi: 10.1074/jbc.272.36.22728. [DOI] [PubMed] [Google Scholar]
- 5.Harth G. Horwitz M.A. An inhibitor of exported Mycobacterium tuberculosis glutamine synthetase selectively blocks the growth of pathogenic mycobacteria in axenic culture and in human monocytes: extracellular proteins as potential novel drug targets. J. Exp. Med. 1999;189:1425–1435. doi: 10.1084/jem.189.9.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harth G. Maslesa-Galic S. Tullius M.V. Horwitz M.A. All four Mycobacterium tuberculosis glnA genes encode glutamine synthetase activities but only GlnA1 is abundantly expressed and essential for bacterial homeostasis. Mol. Microbiol. 2005;58:1157–1172. doi: 10.1111/j.1365-2958.2005.04899.x. [DOI] [PubMed] [Google Scholar]
- 7.Lee S. Jeon B.Y. Bardarov S. Chen M. Morris S.L. Jacobs W.R. Protection elicited by two glutamine auxotrophs of Mycobacterium tuberculosis and in vivo growth phenotypes of the four unique glutamine synthetase mutants in a murine model. Infect. Immun. 2006;74:6491–6495. doi: 10.1128/IAI.00531-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parish T. Stoker N.G. glnE is an essential gene in Mycobacterium tuberculosis. J. Bacteriol. 2000;182:5715–5720. doi: 10.1128/jb.182.20.5715-5720.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ronzio R.A. Meister A. Phosphorylation of methionine sulfoximine by glutamine synthetase. Proc. Natl. Acad. Sci. U.S.A. 1968;59:164–170. doi: 10.1073/pnas.59.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart G.R. Wernisch L. Stabler R. Mangan J.A. Hinds J. Laing K.G. Butcher P.D. Young D.B. The heat shock response of Mycobacterium tuberculosis: linking gene expression, immunology and pathogenesis. Comp. Funct. Genomics. 2002;3:348–351. doi: 10.1002/cfg.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart G.R. Wernisch L. Stabler R. Mangan J.A. Hinds J. Laing K.G. Young D.B. Butcher P.D. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology. 2002;148:3129–3138. doi: 10.1099/00221287-148-10-3129. [DOI] [PubMed] [Google Scholar]
- 12.Waddell S.J. Butcher P.D. Use of DNA arrays to study transcriptional responses to anitmycobacterial compounds. Methods Mol. Biol. 2010;642:75–91. doi: 10.1007/978-1-60327-279-7_6. [DOI] [PubMed] [Google Scholar]
- 13.Waddell S.J. Stabler R.A. Laing K.G. Reynolds K. L., R.C. Besra G.S. The use of microarray analysis to determine the gene expression profiles of Mycobacterium tuberculosis in response to anti-bacterial compounds. Tubercule. 2004;84:263–274. doi: 10.1016/j.tube.2003.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]