Abstract
Symbiotic interactions between ascidians (sea-squirts) and microbes are poorly understood. Here we characterized the cyanobacteria in the tissues of 8 distinct didemnid taxa from shallow-water marine habitats in the Bahamas Islands by sequencing a fragment of the cyanobacterial 16S rRNA gene and the entire 16S–23S rRNA internal transcribed spacer region (ITS) and by examining symbiont morphology with transmission electron (TEM) and confocal microscopy (CM). As described previously for other species, Trididemnum spp. mostly contained symbionts associated with the Prochloron-Synechocystis group. However, sequence analysis of the symbionts in Lissoclinum revealed two unique clades. The first contained a novel cyanobacterial clade, while the second clade was closely associated with Acaryochloris marina. CM revealed the presence of chlorophyll d (chl d) and phycobiliproteins (PBPs) within these symbiont cells, as is characteristic of Acaryochloris species. The presence of symbionts was also observed by TEM inside the tunic of both the adult and larvae of L. fragile, indicating vertical transmission to progeny. Based on molecular phylogenetic and microscopic analyses, Candidatus Acaryochloris bahamiensis nov. sp. is proposed for this symbiotic cyanobacterium. Our results support the hypothesis that photosymbiont communities in ascidians are structured by host phylogeny, but in some cases, also by sampling location.
Introduction
Symbioses between cyanobacteria and marine invertebrates are common, especially in sponges (reviewed in [1]) and ascidians (reviewed in [2]). However, little is known about the nature of these symbioses and much remains to be described. In particular, few studies have employed molecular approaches to more accurately assess bacterial diversity and host-specificity [3], [4], [5], [6], [7], [8]. The majority of ascidian-microbe studies have focused on species within the ascidian family Didemnidae (Aplousobranchia), which often establish symbiotic relationships with unicellular cyanobacteria from the genera Prochloron (Prochlorales) and Synechocystis (Chroococcales). The type species of these genera are Prochloron didemni, first found in Didemnum spp. from Baja California [9], [10], and Synechocystis trididemni, found in the Caribbean ascidian Trididemnum cyanophorum [11]. The cell morphology of both cyanobacterial species is very similar [11], and molecular phylogenetic analyses revealed that they had evolved from a common cyanobacterial ancestor [12], [13].
More recently, a new oxygenic photoautotroph was found in the tropical didemnid Lissoclinum patella [14]. This photosymbiont was tentatively named Acaryochloris marina and presented a set of unique characteristics, the most remarkable being that it uses chlorophyll d (chl d) as a predominant photosynthetic pigment [14], [15]. Chl d is a minor photosynthetic pigment that was found in association with red macroalgae and though to be an artifact [16]. Since then, the presence of Acaryochloris has also been reported in other ascidian species. Kühl et al. [17] reported Acaryochloris-like cells growing on biofilms beneath the didemnid ascidians L. patella, Diplosoma similis and D. virens. Martínez-García et al. [8] also observed small patches of Acaryochloris-like cells on the basal tunic layer of the Mediterranean ascidian Cystodytes dellechiajei (Polycitoridae).
Although the role of photosymbionts in most symbiotic relationships is unknown, the few studies that have investigated ascidian-cyanobacterial symbioses proposed a mutualistic relationship (reviewed in [18] for Prochloron symbiosis), with direct transmission of symbionts between adult generations through the larva [19], [20], [21], [22], [23]. Vertical transmission allows the maintenance of the symbiotic relationship and ensures that offspring have immediate access to the microbes necessary for their survival [18], [24]. This strategy is believed to give hosts a competitive edge from an early stage, and it is normally associated with obligate symbioses.
In the Caribbean, the colonial ascidian Trididemnum solidum Van Name 1902 is known to overgrow and kill corals [25]. This species is distributed in patches and releases larvae throughout the year, the majority settling within 15 min [26]. Both the larvae and the adult of T. solidum are associated with cyanobacteria of the genus Synechocystis [12], [27], [28]. Concentrations of chl a are much higher in the larvae than in the adults, suggesting that the ascidian is highly dependent on its photosymbionts [28], [29]. Trididemnum cyanophorum Lafargue & Duclaux 1979 was first described together with its symbiont Synechocystis trididemni, which was present in high densities both in the tunic of the adult and of the larvae, suggesting an obligate symbiosis [11].
The cosmopolitan didemnid Lissoclinum fragile (Van Name, 1902) is also found in the Caribbean and is known to carry symbiotic cyanobacteria [30], [31]. Monniot [30] described the symbiont of L. fragile as an alga located in the cloacal cavities of the colonies, in the tunic pouches around the abdomen of each zooid, in the mantle surrounding the gonads, and in the surface layer of the larvae [30]. In contrast, Kott et al. [32] and Cox [33] reported that the symbiont of L. fragile was a species of Prochloron, usually found in patches on the surface of the colonies.
The aim of this study was to assess the diversity of the cyanobacterial community inhabiting didemnid ascidians from the Bahamas Islands. We established the genetic identity and diversity of both the ascidian hosts and their photosymbionts in order to better understand the degree of host-specificity. To achieve this goal, we determined host phylogeny by sequencing a fragment of the mitochondrial gene cytochrome oxidase I (COI) that is commonly used to determine species boundaries and diversity among ascidian taxa [34], [35], [36], [37], [38]. In order to identify and describe the photosymbiont diversity from within ascidian tissues, we sequenced a fragment of the 16S rRNA gene and the entire 16S–23S rRNA internal transcribed spacer region (16S–23S ITS). We also examined the morphology of the photosymbionts by transmission electron microscopy (TEM). Finally, we used confocal microscopy (CM) to investigate the presence of chlorophyll d (chl d) and phycobiliproteins (PBPs) in some of our ascidian samples.
Materials and Methods
Ascidian samples and identification
Ascidian samples were collected from mangroves of Sweeting's Cay, and coral reefs of Little San Salvador and Plana Cay (Bahamas) by SCUBA diving in 2008 and 2010 (Table 1). Collection of samples was performed with the permission of the Government of the Bahamas (to JRP). Pictures of each sampled colony were taken in situ before fixation in absolute ethanol (Figure 1). A piece of each colony was anaesthetized by cold exposure as described elsewhere [39], and fixed in formaldehyde for examination of zooids in a relaxed state. Spicules were obtained from small pieces of the tunic (≈5 mm2) previously boiled in commercial bleach until complete oxidation of the tissue. Spicules were then washed several times in water, dehydrated in absolute ethanol, and sputter-coated with gold. All spicule samples were observed with a Hitachi H1200 scanning electronic microscope available at the Scientific and Technical Services of the University of Barcelona (Figure 1). Samples were identified based on Lafargue & Duclaux [11], Kott [40], and Monniot [30], [41].
Table 1. Ascidian species analyzed in this study.
| Species | Date | Code | Location | GPS position | Acc. Num. COI |
| Trididemnum cyanophorum | 30-May-08 | SC 2-1 | Sweeting's Cay | 26°38′35″N; 77°57′44″W | JF506187 |
| Lissoclinum aff. fragile A | 4-Jun-08 | LSS 1-7 | Little San Salvador | 24°35′9″N; 75°58′26″W | JF506183 |
| Lissoclinum fragile A | 7-Jun-08 | WPC 1-1 | West Plana Cay | 22°36′15″N; 73°37′39″W | JF506185 |
| Lissoclinum fragile B | 7-Jun-08 | WPC 3-6 | West Plana Cay | 22°35′50″N; 73°37′45″W | JF506184 |
| Lissoclinum fragile C | 8-Jun-08 | EPC 1-2 | East Plana Cay | 22°36′23″N; 73°33′37″W | JF506181 |
| Lissoclinum fragile D | 8-Jun-08 | EPC 1-5 | East Plana Cay | 22°36′23″N; 73°33′37″W | JF506180 |
| Lissoclinum aff. fragile B | 9-Jun-08 | LSS 1-2 | Little San Salvador | 24°35′7″N; 75°58′25″W | JF506182 |
| Trididemnum solidum | 9-Jun-08 | LSS 2-1 | Little San Salvador | 24°35′6″N; 75°58′20″W | JF506186 |
Species name, sampling date, code, location within Bahamas, GPS position, and GenBank accession numbers for the cytochrome oxidase I (COI) gene.
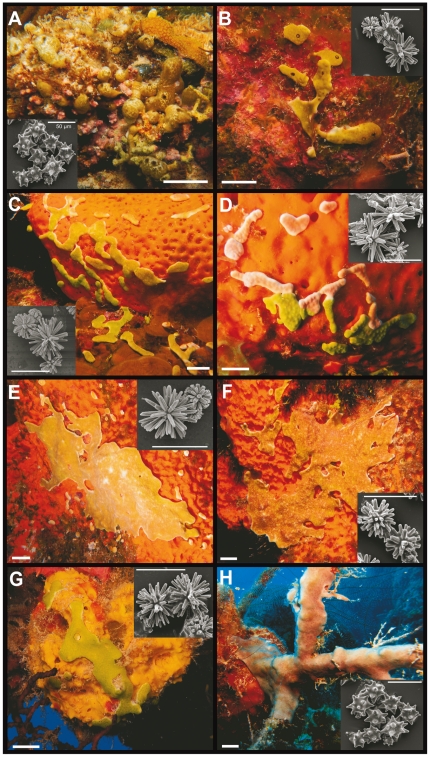
Figure 1. Images of Bahamian ascidians and their spicule types.
(A) Trididemnum cyanophorum from Sweeting's Cay (SC 2-1); (B) Lissoclinum aff. fragile from Little San Salvador (LSS 1-7); (C) Lissoclinum fragile from West Plana Cay (WPC 1-1); (D) Lissoclinum fragile from West Plana Cay (WPC 3-6); (E) Lissoclinum fragile from East Plana Cay (EPC 1-2); (F) Lissoclinum fragile from East Plana Cay (EPC 1-5); (G) Lissoclinum aff. fragile from Little San Salvador (LSS 1-2); and (H) Trididemnum solidum from Little San Salvador (LSS 2-1). Scale bar on ascidian photos = 2 cm. Scale bar on spicule photos = 50 µm.
All ascidians sampled for this study belonged to the family Didemnidae (Aplousobranchia). The sample collected in mangroves from Sweeting's Cay was identified as Trididemnum cyanophorum, while a similar species collected in Little San Salvador overgrowing corals was T. solidum (Figure 1, Table 1). The 4 samples collected on the reefs of East and West Plana Cay ranged in color from bright green to pink (i.e. Figure 1D), but examination of their zooid and spicule morphology revealed that they all belonged to the same species, identified as Lissoclinum fragile (Table 1). Samples collected on the pinnacles of Little San Salvador were morphologically similar to the ones collected in Plana Cay, although the pigmentation pattern was slightly different, with a black circle around the common cloacal apertures of the colonies. This feature was not described in the literature and was absent in other L. fragile colonies (Figure 1). To differentiate between these two groups of samples, we named the samples collected in Little San Salvador Lissoclinum aff. fragile (Table 1).
Ascidian DNA extraction and PCR amplification
Samples fixed in absolute ethanol were kept at −20°C until processed. To maximize ascidian DNA yield, several zooids were carefully separated from the tunic under a stereomicroscope. DNA was extracted using the Puregene kit (Qiagen). The primer set LCO1490 and HCO2198 described by Folmer et al. [42], and Tun_forward and Tun_reverse2 described by Stefaniak et al. [43] were used to amplify a fragment of the Cytochrome c Oxidase subunit I (COI) mitochondrial gene. Amplification was performed with 1 µL of each primer (10 µM), 12.5 µL GoTaq™ reaction buffer (Promega), 10 to 20 µg/mL DNA, and PCR water to a total-reaction volume 25 µL. For both primer sets, a single soak at 94°C for 5 min, was followed by 40 amplification cycles (denaturation at 94°C for 30 sec; annealing at 40°C for 30 sec; and extension at 68°C for 2 min), and a final extension at 72°C for 10 min, in a Peltier PTC-200 gradient PCR. PCR products were purified using the Wizard purification kit (Promega). Sequencing reactions were carried out with the BigDye™ terminator v. 3.1 and the same primer set used during the amplification step, and analyzed on an ABI Prism 3100 automated sequencer. All sequences have been deposited in GenBank (accession numbers listed in Table 1).
Cyanobacteria DNA extraction and PCR amplification
Several ascidian colonies per species and sampling site were ground together in absolute ethanol using a sterile pestle. The resulting greenish liquid was decanted into two 20 mL glass scintillation vials and evaporated under vacuum to leave a powdery organic residue. To re-suspend the extract, 1 mL of lysis solution from the Puregene kit (Qiagen) was added to each scintillation vial, vortexed, and incubated at 55°C for 65 min. The remaining solution was placed in two 2 mL sterile tubes and incubated for an additional hour at 55°C. After allowing the tubes to cool down, we continued with the DNA extraction protocol following manufacturer's instructions. The primer set CYA781F [44] and CYA23S1R or Primer 340 [45] was used to amplify the 3′ end of the 16S rRNA gene, the complete 16S–23S ITS region, and a small fragment of the 23S rRNA gene. Total reaction volume was 25 µL with: 12.5 µL of GoTaq mastermix (Promega), 1 µL of each primer (10 µM), 10.5 µL of PCR grade water, and 1 µL of DNA. PCR program was set as follows: A single soak at 85°C for 5 min, 38 amplification cycles (denaturation at 94°C for 1.5 min; annealing at 50°C for 2 min; and extension at 72°C for 3 min), and a final extension at 72°C for 10 min, in a Peltier PTC-200 gradient PCR. PCR products were run in a low-melting-point agarose gel (1%). One single band of around 1400 bp was observed for each sample, except for Trididemnum cyanophorum for which two clear bands were recorded (a ≈1400 bp band, hereafter named A; and a ≈1500 bp band, named B). Bands were processed separately and purified using the Wizard PCR Preps DNA Purification System (Promega).
Purified DNA was cloned in E. coli using the TOPO® TA Cloning® Kit and One Shot® TOP10 competent cells (Invitrogen), according to manufacturer's instructions. Eight separate clones per ascidian sample were screened by PCR using the plasmid primers T7 and M13R and a total reaction volume of 25 µL: 1 µL of each primer (10 µM), 12.5 µL of GoTaq mastermix (Promega), 10.5 µL of PCR grade water, and 1 µL of each clone, and the following cycle parameters: a single soak at 95°C for 10 min, followed by 30 amplification cycles (95°C for 30 s; 55°C for 30 s and 72°C for 1.5 min), and a final step at 72°C for 2 min. PCR amplicons were run in a low-melting-point agarose gel (1%) to confirm insert size before sequencing using BigDye TM terminator v. 3.1 on an ABI Prism 3100 automated sequencer. Because of the length of some of our sequences (up to 1445 bp), direct sequencing with primers T7 and M13R did not always result in a complete sequence, so we used the primer U1098F [46] to close any remaining gap. All sequences have been deposited in GenBank (accession numbers JF506188 to JF506255).
Phylogenetic analysis
Sequences were aligned using Clustal X [47] with a gap opening penalty of 24 and a gap extension penalty of 4 [48]. To build each phylogenetic tree, additional sequences were retrieved from the Barcode of Life Data System [49] and GenBank (see accession numbers and codes in Figures 2, 3, and 4). Neighbor-joining (NJ) and maximum parsimony analyses were conducted in MEGA 4 [50]. For NJ analyses, the Tajima-Nei model of nucleotide substitution was used and data were re-sampled using 10,000 bootstrap replicates. For MP analyses, the Close-Neighbor-Interchange (CNI) branch-swapping algorithm was implemented and data were re-sampled using 1,000 bootstrap replicates [51]. JModeltest [52] was used to select the best model of DNA substitution for maximum likelihood (ML) and Bayesian inference (BI) analyses according to the Akaike information criterion (AIC). To analyze the COI and 16S rRNA sequences, the GTR+I+G [53] model with substitution rates varying among sites according to an invariant and gamma distribution was used. For sequences including the ITS region, we used the F81+G model [54] with substitution rates varying among sites. ML analyses were performed with Treefinder v. October 2008 [55] and the GTR+I+G substitution model. Data were re-sampled using 1000 bootstrap replicates. For Bayesian inference, MrBayes 3.1.2 [56] was run to obtain a majority-rule consensus tree and posterior probabilities of branch nodes, implementing the corresponding likelihood model for each gene fragment. To analyze the 16S rRNA gene fragment, and the same fragment plus the full ITS region, the Monte Carlo Markov Chain length was set to 16 and 4 million generations, respectively, with sampling every 100th generation. After 15,013,000 generations (16S rRNA fragment) and 3,550,000 (16S–23S ITS region), the average standard deviation of split frequencies between two independent chains reached less than 0.01. For the ascidian sequences, the Monte Carlo Markov Chain length was set to 1,000,000 with sampling every 100th generation. The average standard deviation of split frequencies between two independent chains reached less than 0.01 after 404,000 generations. For all analyses, the first 25% of the resulting trees were discarded as burn-in.
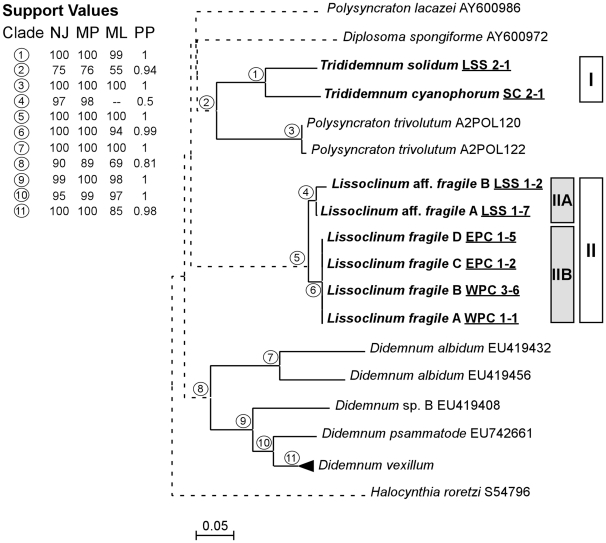
Figure 2. Host phylogeny with partial COI gene sequences.
Sequences obtained in this study are highlighted (bold lettering). The Stolidobranchia species Botryllus schlosseri was used as an outgroup taxa. Labels on terminal nodes of reference sequences indicate the ascidian species and GenBank accession number or code in the Barcode of Life database. Labels on terminal nodes of sequences from this study include species name and sample code as in Table 1 (underlined: SC: Sweeting's Cay; LSS: Little San Salvador; WPC: West Plana Cay, and EPC: East Plana Cay). Bars and labels highlight the two clades of interest (I and II). Samples corresponding to Lissoclinum aff. fragile and L. fragile are further highlighted with a grey bar (IIA and IIB, respectively). Tree topology was obtained from neighbor-joining (NJ) analysis. Individual bootstrap values from NJ, maximum parsimony (MP) and maximum likelihood (ML) analyses and posterior probabilities (PP) from Bayesian inference are located in the upper-left box and correspond to circle numbers on tree nodes. Solid lines indicate bootstrap support greater than 50% from at least 2 of the 4 phylogenetic criteria, and dashed lines indicate weakly supported branches. The Didemnum vexillum cluster includes GenBank sequences: EU419439, -57, EU742661, -68, -69, -75. Scale bar represents 0.05 substitutions per site.
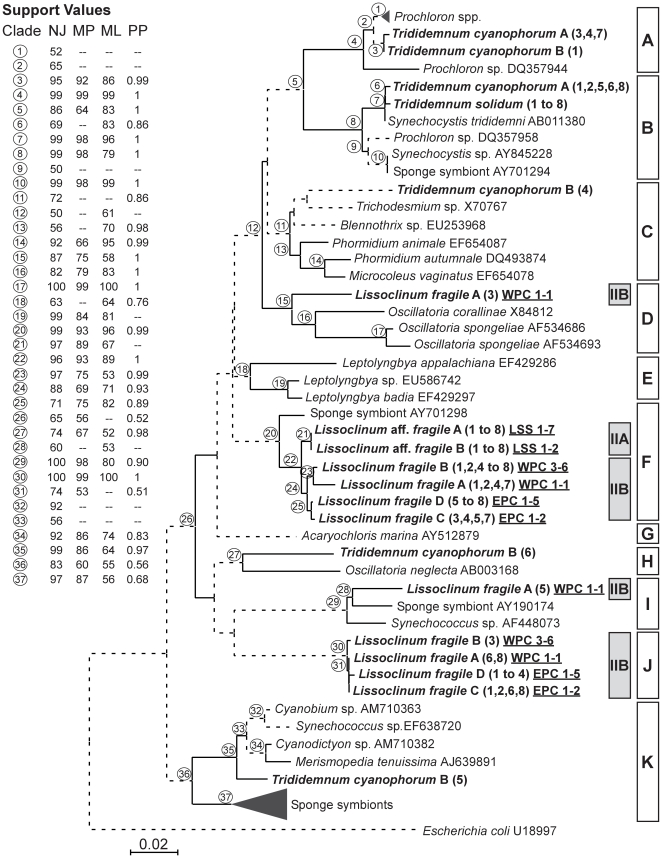
Figure 3. Phylogeny of partial 16S rRNA gene sequences from cyanobacteria isolated from Bahamian ascidians.
Sequences obtained in this study are highlighted (bold lettering). Labels on terminal nodes of reference sequences indicate other cyanobacteria species and GenBank accession numbers. Labels on terminal nodes of sequences from this study include sample name, clone sequenced (in parenthesis), and sample code for the Lissoclinum species (underlined: LSS: Little San Salvador; WPC: West Plana Cay, and EPC: East Plana Cay). Tree topology was obtained from neighbor-joining (NJ) analysis. Individual bootstrap values from NJ, maximum parsimony (MP) and maximum likelihood (ML) analyses and posterior probabilities (PP) from Bayesian inference are located in the upper-left box and correspond to circle numbers on tree nodes. Solid lines indicate bootstrap support greater than 50% from at least 2 of the 4 phylogenetic criteria, and dashed lines indicate weakly supported branches. Bars and labels highlight the 11 major cyanobacterial groups detected in this study (A to K). Samples corresponding to Lissoclinum aff. fragile and L. fragile are further highlighted with a grey bar (IIA and IIB, respectively). The Prochloron spp. clade includes GenBank sequences: DQ357949, -50, -53, -57, -62, -66, -67, DQ385852, and X63141. The sponge symbiont clade includes GenBank sequences: AY701287, -1288, -1290, -1296, -1299, -1301, -1302, -1304, -1306, -1311, EU307448, -455, -458, -477, -488, -489, -492, -509, EF121775, -89, -98. Scale bar represents 0.02 substitutions per site.
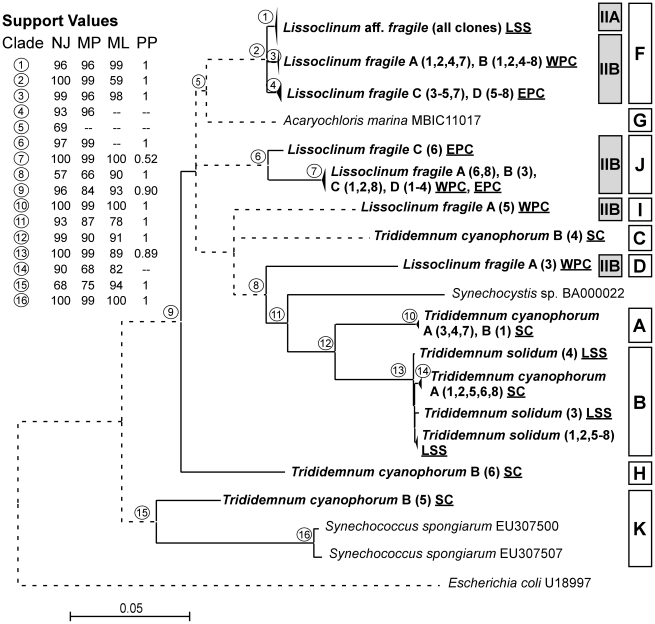
Figure 4. Phylogeny of 16S–23S ITS gene sequences from cyanobacteria isolated from Bahamian ascidians.
Sequences obtained in this study are highlighted (bold lettering). Labels on terminal nodes of sequences include sample name and clone sequenced (in parenthesis). Labels on terminal nodes of sequences from this study include sample name, clone sequenced (in parenthesis), and sample location (underlined: SC: Sweeting's Cay, LSS: Little San Salvador; WPC: West Plana Cay, and EPC: East Plana Cay). Bars as in Figure 3. Tree topology corresponds to the consensus obtained from neighbor-joining (NJ) analysis. Individual bootstrap values from NJ, maximum parsimony (MP) and maximum likelihood (ML) analyses and posterior probabilities (PP) from Bayesian inference are located in the upper-left box and correspond to circle numbers on tree nodes. Solid lines indicate bootstrap support greater than 50% from at least 2 of the 4 phylogenetic criteria, and dashed lines indicate weakly supported branches. Scale bar represents 0.05 substitutions per site.
Transmission electron microscopy
The ultrastructure of the most common symbionts in the ascidian tunic was examined by transmission electron microscopy (TEM). Small pieces (about 2 mm3) of Lissoclinum fragile, Lissoclinum aff. fragile, Trididemnum cyanophorum, and T. solidum were cut from each colony, and fixed in 2.5% glutaraldehyde 2% paraformaldehyde using filtered seawater as buffer. Only samples of L. fragile contained mature larvae, which were carefully separated from adults and observed separately. Samples were incubated in the fixative mixture overnight at 4°C, and then washed several times in filtered seawater. To construct resin blocks, samples were dehydrated in a graded ethanol series and embedded in Spurr's resin at room temperature. Semi-thin (5 microns) and ultrathin sections (ca. 60 nm) were cut with a Reichert Ultracut microtome. Ultrathin sections were stained with uranyl acetate and lead citrate for ultrastructural observation [57]. TEM observations were performed at the Microscopy Unit of the Scientific and Technical Services of the University of Barcelona on a JEOL JEM-1010 (Tokyo, Japan) coupled with a Bioscan 972 camera (Gatan, Germany).
Confocal microscopy
Based on plylogenetic results, further analyses were performed to assess whether chlorophyll d (chl d) and phycobiliproteins (PBPs) were present in the tunic of Lissoclinum aff. fragile using confocal microscopy. Slices of two colonies previously fixed in 4% formaldehyde were mounted on a slide with distilled water. Images were acquired on a Leica TCS SPE confocal microscope equipped with a NPLAN 100× 1.2 water immersion objective. A 635 nm solid state laser was used for fluorescence excitation. Detection ranges for chl d and PBPs were set to 700–750 nm and 640–670 nm, respectively. Laser power and PMT gain and offset were equally set up for both signals. Confocal images were acquired at the maximum resolution of the objective and the stacks were deconvolved with Huygens Essential (Scientific Volume Imaging, B.V.). Deconvolution was calculated with the optimized iteration mode of this software and all image parameters were read from the Leica. lif format. Object counting and measurements were performed on the deconvolved image stacks after 3D surface rendering using Imaris (Bitplane A.G.) and the Imaris MeasurementPro module. Length and width of at least 80 cells per sample was calculated using the software ImageJ 1.41o. Each stack was analyzed setting the fluorescence threshold to the value that resulted in the maximum number of individual objects identified. Quantification was performed on those objects that showed both signals (PBPs and chl d). Final artwork was done with Photoshop CS (Adobe). Chl d and PBPs images were colored in red and green, respectively.
Results and Discussion
Host phylogeny
Partial COI gene sequences were obtained for all the samples, with a final alignment length of 605 bp. As expected, phylogenetic analyses grouped the two Trididemnum sequences studied in a well-supported clade (bootstrap values >99% in all analysis; Figure 2I). The taxonomic status of T. cyanophorum has been argued in the past, with some authors suggesting that it may be a synonym of T. solidum [30]. Here, we have found 12% sequence variability between T. solidum and T. cyanophorum and some morphological differences in terms of color and colony shape (Figure 1), suggesting that both species names are valid despite the lack of morphological differences among zooids. All the sequences of Lissoclinum obtained in this study formed a well-supported clade (bootstrap values = 100% in all analysis; Figure 2II). Moreover, within the Lissoclinum clade, two subgroups with 3% sequence variability were further distinguished. The first subgroup grouped the two samples obtained from Little San Salvador and named Lissoclinum aff. fragile (Figure 2 IIA). The second subgroup was formed by identical sequences for L. fragile from Plana Cay (Figure 2 IIB). All in all, phylogenetic analyses confirmed morphological observations and determined the taxonomic status of species and groups of samples presenting small morphological differences.
Cyanobacteria phylogeny
After alignment, 68 partial 16S rRNA gene sequences ranging between 740 and 756 bp were obtained, 19 of which were unique. All sequences from Lissoclinum aff. fragile were identical and differed from the ones obtained for L. aff, fragile B by one mutation. Ten unique sequences were retrieved from L. fragile, 3 of which were only encountered in one clone. All sequences obtained from Trididemnum solidum were identical, while 6 unique sequences were retrieved from T. cyanophorum, 4 of which were only sequenced once. The same sequence was never found in both Lissoclinum and Trididemnum species. Analyses conducted using the 16S rRNA fragment described above plus the complete ITS region (269–647 bp) plus a small fragment of the 23S rRNA gene (44 bp), hereafter called the16S–23S ITS region, yielded 56 unique sequences. Identical sequences were observed among specimens of L. aff. fragile A and B (6 clones total), and within clones of L. aff. fragile B (2 clones), L. fragile B (2 clones) and T. cyanophorum A (2 clones).
Blast searches in GenBank with the 16S rRNA fragment showed that the best match for Trididemnum solidum and Trididemnum cyanophorum symbionts were uncultured Prochloron and Synechocystis sequences (>97% max. identity; >93% coverage in all cases). Three other sequences obtained for T. cyanophorum B (clones 4, 5, 6) shared similarities with a range of uncultured cyanobacteria, including Oscillatoriales and Chroococcales. All the sequences from Lissoclinum aff. fragile and most of the sequences from L. fragile (19) shared up to 95% identity with Acaryochloris sp. (100% coverage). The remaining sequences from L. fragile (11 in total) moderately resembled Leptolyngbia antactica (93% max. identity; 100% coverage). Finally, two sequences of L. fragile from West Plana Cay (A, clones 3 and 5) shared 94% and 97% identities with Phormidium sp. (100% coverage) and Synechococcus sp. sequences (100% coverage), respectively.
Phylogenetic analyses based on a fragment of the 16S rRNA gene (Figure 3), and the 16S–23S ITS region (Figure 4), revealed a total of 11 groups, 10 of which were equivalent among analyses. The first one (named A in Figure 3 and 4) included 4 sequences obtained from T. cyanophorum and had strong bootstrap support (>99%). Phylogenetic analyses conducted with the 16S rRNA fragment showed that these sequences from T. cyanophorum grouped with 9 sequences of Prochloron spp. retrieved from GenBank. Five sequences from T. cyanophorum and all sequences from T. solidum (8) were grouped with two species of Synechocystis and one sequence of Prochloron in clade B (Figure 3 and 4). High similarity of the cyanobacterial sequences obtained here for T. solidum and T. cyanophorum with Synechocystis trididemni is not surprising (clade B), as this cyanobacterium was first described in T. cyanophorum [11], and has also been found in other Trididemnum species [3]. Along with Synechocystis, some Trididemnun species can also establish symbiotic relationships with Prochloron (e.g. T. miniatum [58], T. paracyclops [3]). For T. cyanophorum, symbionts of both Prochloron and Synechocystis appeared to coexist. This phenomenon has also been reported for other ascidians, and is especially common in Trididemnum species [32], [59], [60].
The third clade (named C) from the 16S rRNA analyses was formed by different cyanobacterial species belonging to the Chroococcales, together with one sequence obtained here for T. cyanophorum B (clone 4). The position of the sequence from T. cyanophorum B (4) within this group was not resolved. Further analyses, including those of the 16S–23S ITS region, did not yield clearer results, probably due to the limited number of available cyanobacterial ITS sequences in GenBank (Figure 4). Other sequences of Chroococcales were found in the 16S rRNA clade I, which grouped clone 5 from L. fragile A, an uncultured cyanobacterium sequence obtained from the Australian sponge Cymbastela sp., and a cultured strain of Synechococcus sp. This clade had a bootstrap support >80% in all analyses, however, it did not form a monophyletic group with other Synechococcus sequences retrieved from GenBank (located in clade K). Clade K was entirely formed by Chroococcales, including many sponge symbionts, some free-living species, and clone sequence 5 from T. cyanophorum B (Figure 3 and 4). Cyanobacteria are common members of sponge-associated microbial communities. In particular, Synechococcus species have been widely reported as the major photosymbionts inhabiting sponges [48], [61], [62], [63]. Some of these symbionts were reported to be host-specific, others were found in different sponge species, and some varied according to location [48]. The relative integration of these symbionts varies depending on the sponge species, ranging from obligate symbiosis to a commensal existence, with cyanobacterial cells interspersed among sponge cells [64], [65]. In this study, only one sequence related to the sponge symbiont Synechococcus was found, indicating that this cyanobacterial genus may only form facultative associations in ascidians. Moreover, the sequence obtained herein appeared more closely related to free-living Synechococcus, and may just have been captured from the water column by the ascidian.
In the 16S rRNA analyses, one sequence (clone 5) obtained from L. fragile A formed clade D with cyanobacterial species belonging to the Oscillatoriales (Figure 3). In contrast, the 16S–23S ITS sequence obtained for L. fragile A (5) (Figure 4) held a basal position within the Prochloron-Synechocystis group (clades A and B). Other sequences of Oscillatoriales appeared in clade H, which, for the 16S rRNA analysis, only included 2 sequences: one obtained from T. cyanophorum B (clone 6) and a sequence from Oscillatoria neglecta. Although the existence of this clade appeared to be supported by bootstrap analysis, its connection with the other clades remained undetermined, especially considering that other Oscillatoriales were grouped in clade D (Figure 3). The 16S–23S ITS analysis (Figure 4) showed that the T. cyanophorum B (clone 6) sequence appeared basal to all clades except clade K (bootstrap support >84% in all analyses). Thus, based on 16S rRNA and 16S–23S ITS results, the identity and phylogenetic position of L. fragile A (clone 5) and T. cyanophorum B (clone 6) within the Oscillatoriales could not be resolved.
The last clade formed by sequences of Oscillatoriales was found only in the 16S rRNA analysis and grouped three sequences of Leptolyngbya spp. retrieved from GenBank (clade E, Figure 3). Although Blast searches returned Leptolyngbya species as best match for 11 of our 16S rRNA sequences from L. fragile, none of them were included in this clade based on phylogenetic analyses. Several studies have reported the occurrence of Oscillatoriales associated with ascidians [9], [33], [66], [67], [68], [69], [70], [71]. However, most of these studies include only short references to these symbionts [9], [67], [68], a few are accompanied by electron microscopical observations [33], [66], [69], [70], [71], and even fewer provided a name or a sequence to identify the symbiont [66], [70]. Thus, to date, the degree of host-specificity and strength of association between ascidians and Oscillatoriales remains unresolved.
The 16S rRNA clade F (Figure 3) grouped most of the sequences obtained from L. fragile A, B, C, and D, all the sequences from L. aff. fragile, and a single sequence obtained from the Bahamian sponge Pseudoaxinella flava by [63]. The best matches of this group according to blast searches were Acaryochloris species. Based on 16S rRNA analyses, Acaryohcloris marina (clade G in Figure 3) appeared at the base of clades A, B, C, D, E and F, while the 16S–23S ITS analyses showed that A. marina was related only to clade F (Figure 4). Acaryochloris marina is a recently discovered oxygenic photoautotroph that uses chl d as the predominant photosynthetic pigment [17], [72], [73]. A closer analysis of the Acaryochloris-like sequences (clade F) compared to A. marina (clade G) reported a sequence divergence >5%, while sequence variation within the Acaryochloris-like clade obtained here was <1%.
Within clade F, sequences were grouped according to host and sampling location. The first sub-clade contained all sequences of L. aff. fragile from Little San Salvador (Figures 3 and 4, IIA). Sequences for L. fragile formed two groups according to sampling site: West Plana Cay (samples A and B), and East Plana Cay (samples C and D, 7 kilometers away from West Plana Cay, and 330 km away from Little San Salvador). Thus, even adjacent populations had distinct cyanobacterial symbionts. All the remaining sequences of L. fragile A, B, C, and D formed a well-supported clade (J) by themselves, without any clear correlation with location (bootstrap support >97% in all analyses except ML for 16S–23S ITS; Figure 3 and 4). Therefore, as previously reported for sponges [48], some ascidian photosymbionts are host-specific, while a few may depend on environmental parameters associated with a given location.
The phylogenetic position of clade J within the cyanobacteria could not be resolved with analyses of the 16S rRNA fragment or the 16S–23S ITS region. Moreover, although the closest blast match with an identified cyanobacterium was Leptolyngbia antactica, none of the phylogenetic analyses related clade J with other Oscillatoriales (clades D, E, and H; Figure 3 and 4). Based on currently available information, we are unable to identify or provide a taxonomic position for sequences in clade J.
Cyanobacteria morphology
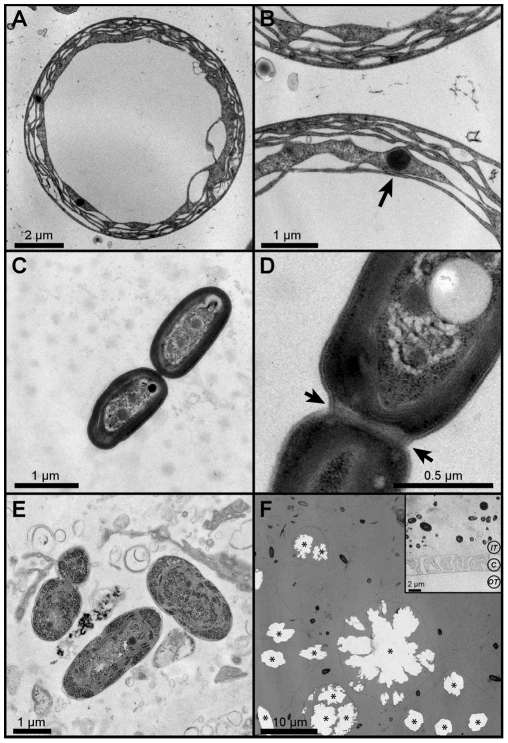
Photosymbionts of Trididemnum solidum and T. cyanophorum were embedded in the adult tunic, but were never observed to be in direct contact with ascidian cells (Figure 5A). The symbionts observed in T. solidum and T. cyanophorum symbionts were morphologically identical to those reported from previous studies of Synechocystis trididemni [11]. Specifically, S. trididemni was reported to be round, with a diameter of 8 to 11 µm, and 5 to 7 thylakoids around the periphery of the cell. The diameter of Synechocystis observed herein fell within the range indicated above, however they had a few less thylakoids per cell (3 to 5 instead of 5 to 7). As described for S. trididemni, some of these thylakoids were also irregular and developed bladders or vesicles close to the nucleoplasma [11]. Polyedric bodies were also observed in close contact with the thylacoides (Figure 5B, [11]). Thus, morphological observations of the ultrastructure of photosymbionts of T. solidum and T. cyanophorum confirmed our phylogenetic results, indicating that Synechocystis was the major symbiont in these ascidian species.
Figure 5. Transmission electron microscopical images of photosymbionts from Trididemnum cyanophorum and Lissoclinum aff. fragile.
(A) Synechocystis sp. symbiont in Trididemnum cyanophorum; (B) Polyedric bodies located in the periphery of the Synechocystis cell; (C) Acaryochloris bahamiensis nv. sp. after division in the tunic of Lissoclinum fragile; (D) Detail showing the peptoglycan layer between two cells of Acaryochloris bahamiensis nv. sp. during the final stage of division; (E) Other cyanobacterial symbionts observed in the tunic of L. fragile; (F) Spicules and cyanobacteria distribution inside the tunic of L. fragile (asterisks indicate spicule location). The inset shows cyanobacteria in the tunic of the larvae (C: Cuticle; IT: Inner tunic; OT: Outer tunic).
A very different type of cyanobacterium was observed in L. fragile and L. aff. fragile samples (Figure 5). As hinted by the phylogenetic analyses, the overall structure of the photosymbionts (i.e. cell shape, size, and major features) was consistent with previous research describing the ultrastructure of Acaryochloris species [14], [74], [75], [76]. A consensus on the number and arrangement of the thylakoids within Acaryochloris cells does not exist in the literature. The first studies on A. marina reported more than 7 thylakoids surrounding the cytoplasm [14], [74], [75], while subsequent studies reported a lower number of thylakoids evenly spaced along the periphery of the cell [76], [77]. The Acaryochloris-like cells observed herein better fit this last description, and presented 5 to 7 thylakoids evenly spaced along the periphery of the cell (Figure 5C). Several symbiont cells were also observed undergoing division, and followed the major division steps described by Marquart et al. [75]. Notably, the photosymbiont cells observed herein also remained connected by a common peptidoglycan layer after cell division (Figure 5D). Another cyanobacterial type containing abundant glycogen granules was observed in the tunic around the zooid abdomen (Figure 5E). Cyanobacteria were consistently found outside the sheaths that surround the calcareous spicules of the tunic (Figure 5F).
Acaryochloris-like cells were also found inside the tunic of the larva isolated from L. fragile, indicating that the symbionts were transmitted to the progeny (Figure 5E inset). Vertical transmission of photosymbionts to larvae has often been observed in ascidians and is assumed to be essential for host survival [18], [78]. To date, three transmission modes have been described. The first mode involves the formation of a tunic extension at the posterior end of the larval trunk, just above the tail insertion called rastrum [40]. The rastrum has been described in most Diplosoma species with photosymbionts in the cloacal cavities [22], [40], [68], [78], [79], [80], [81], [82], [83], [84]. The second mode has been observed in some Didemnum, Trididemnum and Lissoclinum species and is associated with the adhesion of the photosymbionts to either the posterior end of the larval trunk or around the entire larva except for the sensory and adhesive organs [19], [21], [40], [78], [85], [86]. These ascidian species also harbor their photosymbionts in the cloacal cavities and, as in the first mode, symbionts are acquired when larvae pass through these cloacal cavities.
A more recently described transmission mode applies to ascidian species that harbor photosymbionts within their tunic. This process was described for Trididemnum miniatum and is thought to involve host cells acting as a vehicle for transporting symbionts from the tunic of the adult to that of the larvae [20]. Although no larvae were obtained from T. solidum and T. cyanophorum, the process described for T. miniatum or a similar mechanism may apply to the Trididemnum species analyzed in the present study. Our observations for L. fragile suggest that this species may also acquire their symbionts by an active transport mechanism during the formation of the deeper layer of the tunic, or inner tunic. In support of this hypothesis, the cyanobacteria were observed in the inner tunic separated by a folded cuticle from the outer tunic (Figure 5E inset). Photosymbionts are sufficiently abundant in the inner tunic to confer obvious pigment to the tadpole larve, except for the regions around the adhesive and sensory organs. Further research is needed to assess the exact process involved in the transfer of photosymbionts to the larvae and to determine whether ascidian cells are implicated in this process.
Chlorophyll d in Lissoclinum aff. fragile
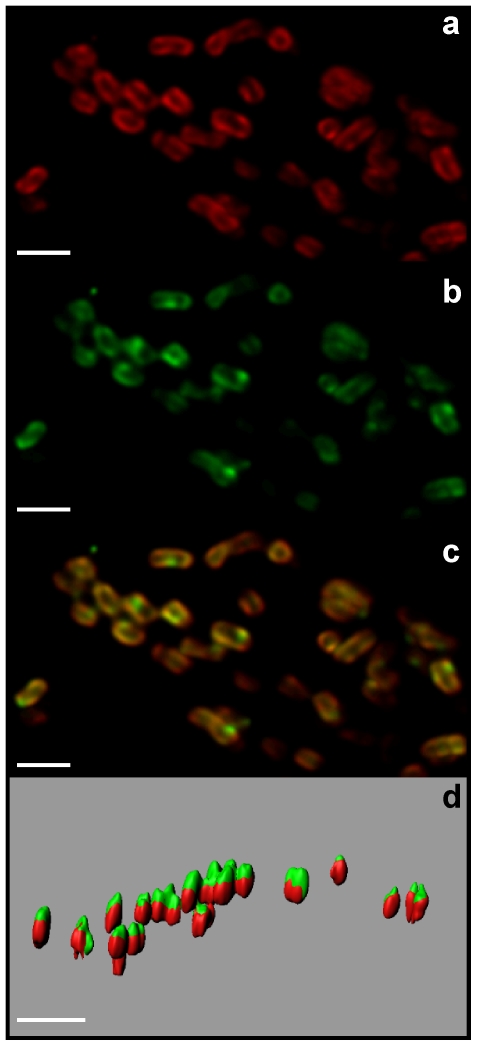
Confocal microscopic examination revealed the presence of numerous cells containing both chlorophyll d (chl d) and phycobiliproteins (PBPs) in the tunic of Lissoclinum aff. fragile (Figure 6). Chl d is a predominant photosynthetic pigment in Acaryochloris species, a genus discovered to live in close association with ascidians [8], [17], [72], [73], [87]. Measurements performed on the deconvolved image stacks of these Acaryochloris-like cells revealed an average length of 1.75 µm (±0.38; SD), width of 1.13 µm (±0.24; SD), and volume of 2.32 µm3 (±1.07; SD). The diameter and length of the observed cells were within the range reported in the literature for Acaryochloris spp. (1.5–1.7 µm diameter and 1.8–2.1 µm in length [75], 1–1.5 µm in diameter and 1.5–3 µm in length [15]).
Figure 6. Confocal microscopical images and model reconstruction of Acaryochloris-like cells in Lissoclinum aff. fragile.
Cells were excited with a 635 nm red laser and fluorescence was detected between 700–750 nm for chlorophyll d (a) and 640–670 nm for phycobiliproteins (b). (c) Composite image from combining the chlorophyll d (red) and the phycobiliproteins (green) channels. (d) 3D surface rendering after image deconvolution showing the emplacement of chlorophyll d and phycobiliproteins within the Acaryochloris-like cells. Scale bar = 2 µm.
To date, Acaryochloris spp. have only been found growing on biofilms beneath didemnid ascidians [17], [72], [73] or forming small aggregations at the base of the ascidian tunic [8]. However, in this study, Acaryochloris-like cells were abundant (Figure 6) and appeared scattered through the tunic. Three-dimensional reconstruction of these cells also revealed that chl d and PBPs co-localized in the center of each Acaryochloris-like cell, however, they were compartmentalized, with chl d mainly present on one side and PBPs on the other (Figure 6d). A particular orientation of chl d and PBPs within the photosymbiont cell and the host tunic may be necessary to ensure optimal sunlight absorption.
Taken together with phylogentic analyses and TEM observations, our results indicate that L. fragile harbors photosymbionts from the Acaryochloris group within its tunic. Based on unique 16S rRNA and 16S–23S ITS sequences, we propose here to name Candidatus Acaryochloris bahamiensis nov. sp. for this photosymbiont. The importance of A. bahamiensis nov. sp. to the survival of its host remains to be assessed. However, vertical transfer of A. bahamiensis nov. sp. to the larvae suggests a close association between host and symbiont, and that this relationship is important to host survival. This is the first report of Acaryochloris forming a symbiotic relationship with an ascidian and living within its tunic.
Conclusion
In conclusion, using molecular and electron microscopical techniques, we have shown that the ascidians examined in this study harbor a considerable diversity of photosymbionts. The primary taxa of symbionts found were Synechocystis in the tunic of Trididemnum solidum and T. cyanophorum, and Acaryochloris-like symbionts in Lissoclinum fragile and L. aff. fragile. Host identity strongly correlated with the identity of the photosymbionts found in the tunic, although in some cases (e.g. Lissoclinum fragile) differences could be related to sampling location. Analyses of 16S rRNA and 16S–23S ITS sequences from the symbionts in two varieties of Lissoclinum fragile revealed two major clades (F and J). Clade J could not be associated with any known cyanobacterium, while clade F was related to Acaryochloris. Ultrastructural examination confirmed the similarity of clade F symbionts with other species of Acaryochloris. These symbionts were also observed in the inner tunic of the larvae, suggesting that specialized mechanisms of vertical transmission exist. Analyses using CM revealed the presence of chl d and PBP, further reinforcing the classification of these photosymbionts as Acaryochloris spp. Based on these results and unique 16S rRNA and 16S–23S ITS sequences, we propose the name Candidatus Acaryochloris bahamiensis nov. sp. for this photosymbiont. Substantial research is still required to determine the diversity, host-specificity, and function of microbial symbionts in ascidians.
Acknowledgments
We thank Dr. Patrick M. Erwin for his advice on primer selection, and Dr. Min Chen for kindly sharing his knowledge on the photosystems of Acaryochloris sp. We also thank the government of the Bahamas Islands for permission to perform research in their territorial waters.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was funded by the Marie Curie International Reintegration Grant FP7-PEOPLE-2010-RG 277038 (within the 7th European Community Framework Program), by a Systematics Research grant from the Linnean Society and the Systematics Association, and by the Spanish Government project CTM2010-17755. Use of the UNOLS Research Vessel Walton Smith was funded by a grant from the US National Science Foundation (OCE 0550468 to JRP). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Webster NS, Taylor MW. Marine sponges and their microbial symbionts: love and other relationships. Environmental Microbiology. 2011 doi: 10.1111/j.1462-2920.2011.02460.x. doi: 10.1111/j.1462-2920.2011.02460.x. [DOI] [PubMed] [Google Scholar]
- 2.Hirose E, Neilan BA, Scmidt EW, Murakami A. Enigmatic life and evolution of Prochloron and related cyanobacteria inhabiting colonial ascidians. In: Gault PM, Marler HJ, editors. Handbook on cyanobacteria. Nova Science Publishers, Inc; 2009. pp. 161–189. [Google Scholar]
- 3.Münchhoff J, Hirose E, Maruyama T, Sunairi M, Burns BP, et al. Host specificity and phylogeography of the prochlorophyte Prochloron sp., an obligate symbiont in didemnid ascidians. Environmental Microbiology. 2007;9:890–899. doi: 10.1111/j.1462-2920.2006.01209.x. [DOI] [PubMed] [Google Scholar]
- 4.Tait E, Carman M, Sievert SM. Phylogenetic divrsity of bacteria associated with ascidians in Eel Pond (Woods Hole, Massachusetts, USA). Journal of Experimental Marine Biology and Ecology. 2007;342:138–146. [Google Scholar]
- 5.Yokobori S, Kurabayashi A, Neilan BA, Maruyama T, Hirose E. Multiple origins of the ascidian-Prochloron symbiosis: molecular phylogeny of photosymbiotic and non-symbiotic colonial ascidians inferred from 18S rDNA sequences. Molecular Phylogenetics and Evolution. 2006;40:8–19. doi: 10.1016/j.ympev.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 6.Martínez-García M, Díaz-Valdés M, Wanner G, Ramos-Esplà A, Antón J. Microbial community associated with the colonial ascidian Cystodytes dellechiajei. Environmental Microbiology. 2007;9:521–534. doi: 10.1111/j.1462-2920.2006.01170.x. [DOI] [PubMed] [Google Scholar]
- 7.Martínez-García M, Díaz-Valdés M, Antón J. Diversity of pufM genes, involved in aerobic anoxygenic photosynthesis, in the bacterial communities associated with colonial ascidians. FEMS Microbiology Ecology. 2010;71:387–398. doi: 10.1111/j.1574-6941.2009.00816.x. [DOI] [PubMed] [Google Scholar]
- 8.Martínez-García M, Koblízek M, Lopez-Legentil S, Antón J. Epibiosis of oxygenic phototrophs containing chlorophylls a, b, c, and d on the colonial ascidian Cystodytes dellechiajei. Microbial Ecology. 2011;61:13–19. doi: 10.1007/s00248-010-9694-6. [DOI] [PubMed] [Google Scholar]
- 9.Lewin RA, Cheng L. Associations of microscopic algae with didemnid ascidians. Phycologia. 1975;14:149–152. [Google Scholar]
- 10.Lewin RA. Prochloron, type genus of the Prochlorophyta. Phycologia. 1977;16:217. [Google Scholar]
- 11.Lafargue F, Duclaux G. Premier example, en Atlantique tropical, d'une association symbiotique entre une ascidie Didemnidae et une cyanophycée Chroococcale: Trididemnum cyanophorum nov. sp. et Synechocystis trididemni nov. sp. Annales de l'Institut Océanographique. 1979;55:163–184. [Google Scholar]
- 12.Shimada A, Yano N, Kanai S, Lewin RA, Maruyama T. Molecular phylogenetic relationship between two symbiotic photo-oxygenic prokaryotes, Prochloron sp. and Synechocystis trididemni. Phycologia. 2003;42:193–197. [Google Scholar]
- 13.Palenik B, Swift H. Cyanobacterial evolution and Prochlorophyte diversity as seen in DNA-dependent RNA polymerase gene sequences. Journal of Phycology. 1996;32:638–646. [Google Scholar]
- 14.Miyashita H, Ikemoto H, Kurano N, Adachi K, Chihara M, et al. Chlorophyll d as a major pigmant. Nature. 1996;383:402. [Google Scholar]
- 15.Miyashita H, Ikemoto H, Kurano N, Miyachi S, Chihara M. Acaryochloris marina gen. et sp. nov. (cyanobacteria), an oxygenic photosynthetic prokaryote containing chl d as a major pigment. Journal of Phycology. 2003;39:1247–1253. [Google Scholar]
- 16.Manning WM, Strain HH. Chlorophyll D, a green pigment of red algae. Journal of Biological Chemistry. 1943;151:1–19. [Google Scholar]
- 17.Kühl M, Chen M, Ralph PJ, Schreiber U, Larkum AWD. A niche for cyanobacteria containing chlorophyll d. Nature. 2005;433:820. doi: 10.1038/433820a. [DOI] [PubMed] [Google Scholar]
- 18.Hirose E, Maruyama T. What are the benefits in the ascidian-Prochloron symbiosis? Endocytobiosis and Cell Research. 2004;15:51–62. [Google Scholar]
- 19.Hirose E, Fukuda T. Vertical transmission of photosymbionts in the colonial ascidian Didemnum molle: the larval tunic prevents symbionts from attaching to the anterior part of larvae. Zoological Science. 2006;23:669–674. doi: 10.2108/zsj.23.669. [DOI] [PubMed] [Google Scholar]
- 20.Hirose E, Hirose M. Morphological process of vertical transmission of photosymbionts in the colonial ascidian Trididemnum miniatum Kott, 1977. Marine Biology. 2007;150:359–367. [Google Scholar]
- 21.Kojima A, Hirose E. Transfer of prokaryotic algal symbionts from a tropical ascidian (Lissoclinum punctatum) colony to its larvae. Zoological Science. 2010;27:124–127. doi: 10.2108/zsj.27.124. [DOI] [PubMed] [Google Scholar]
- 22.Hirose E. Plant rake and algal pouch of the larvae in the tropical ascidian Diplosoma similis: An adaptation for vertical transmission of photosynthetic symbionts Prochloron sp. Zoological Science. 2000;17:233–240. [Google Scholar]
- 23.Groepler W, Schuett C. Bacterial community in the tunic matrix of a colonial ascidian Diplosoma migrans. Helgoland Marine Research. 2003;57:139–143. [Google Scholar]
- 24.Usher KM, Bergman B, Raven JA. Exploring cyanobacterial mutualisms. Annual Review of Ecology, Evolution, and Systematics. 2007;38:255–273. [Google Scholar]
- 25.Bak RPM, Sybesma J, van Duyl FC. The ecology of the tropical compound ascidian Trididemnum solidum. II. Abundance, growth and survival. Marine Ecology Progress Series. 1981;6:43–52. [Google Scholar]
- 26.van Duyl FC, Bak RPM, Sybesma J. The ecology of the tropical compound ascidian Trididemnum solidum. I. Reproductive strategy and larval behavior. Marine Ecology Progress Series. 1981;6:35–42. [Google Scholar]
- 27.Holton RW, Stam WT, Boele-Bos SA. DNA-DNA reassociation studies with DNA from Prochloron (Prochlorophyta) samples of Indo-West Pacific origin. Journal of Phycology. 1990;26:358–361. [Google Scholar]
- 28.Sybesma J, Duyl FC, Bak RPM. The ecology of the tropical compound ascodoan Trididemnum solidum. III. Symbiotic association with unicellular algae. Marine Ecology Progress Series. 1981;6:53–59. [Google Scholar]
- 29.Olson RR. Photoadaptations of the Caribbean colonial ascidian-cyanophyte symbiosis Trididemnum solidum. Biological Bulletin. 1986;170:62–74. [Google Scholar]
- 30.Monniot F. Ascidies littorales de Guadeloupe. VIII. Questions de systématique évolutive posées par les Didemnidae. Bulletin du Muséum national d'Histoire naturelle de Paris. 1984;6(A):885–905. [Google Scholar]
- 31.Tarjuelo I, Posada D, Crandall KA, Pascual M, Turon X. Cryptic species of Clavelina (Ascidiacea) in two different habitats: harbours and rocky littoral zones in the northwestern Mediterranean. Marine Biology. 2001;139:455–462. [Google Scholar]
- 32.Kott P, Parry DL, Cox GC. Prokaryotic symbionts with a range of ascidian hosts. Bulletin of Marine Science. 1984;34:308–312. [Google Scholar]
- 33.Cox GC. Comparison of Prochloron from different hosts. I. Structural and ultrastructural characteristics. New Phytologist. 1986;104:429–445. [Google Scholar]
- 34.Hirose E. Ascid container and cellular networks in the ascidian tunic with special remarks on ascidian phylogeny. Zoological Science. 2001;18:723–731. [Google Scholar]
- 35.López-Legentil S, Turon X. Population genetics, phylogeography and speciation of Cystodytes (Ascidiacea) in the western Mediterranean Sea. Biological Journal of the Linnean Society. 2006;88:203–214. [Google Scholar]
- 36.Tarjuelo I, Posada D, Crandall KA, Pascual M, Turon X. Phylogeography and speciation of colour morphs in the colonial ascidian Pseudodistoma crucigaster. Molecular Ecology. 2004;13:3125–3136. doi: 10.1111/j.1365-294X.2004.02306.x. [DOI] [PubMed] [Google Scholar]
- 37.Hirose E. Tunic cells in Leptoclinides echinatus (Didemnidae, Ascidiacea): An application of scanning electron microscopy for paraffin embedding specimens. Hiyoshi Review of Natural Science. 1992;11:5–8. [Google Scholar]
- 38.López-Legentil S, Turon X. How do morphotypes and chemotypes relate to genotypes? The colonial ascidian Cystodytes (Polycitoridae). Zoologica Scripta. 2005;34:3–14. [Google Scholar]
- 39.Turon X. Estudio de las ascidias de las costas de Cataluña e Islas Baleares. Barcelona: University of Barcelona; 1987. [Google Scholar]
- 40.Kott P. Algal-bearing Didemnid ascidians in the Indo-West-Pacific. Memoirs of the Queensland Museum. 1980;20:1–47. [Google Scholar]
- 41.Monniot F. Ascidies de Nouvelle-Calédonie. XII. Le genre Lissoclinum (Didemnidae) dans le lagon sud. Bulletin du Muséum national d'Histoire naturelle de Paris. 1992;14(A):565–589. [Google Scholar]
- 42.Folmer O, Hoeh W, Black M, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology. 1994;3:294–299. [PubMed] [Google Scholar]
- 43.Stefaniak L, Lambert G, Gittenberger A, Zhang H, Lin S, et al. Genetic conspecificity of the worldwide populations of Didemnum vexillum Kott, 2002. Aquatic Invasions. 2009;4:29–44. [Google Scholar]
- 44.Nübel U, Garcia-Pichel F, Muyzer G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Applied and Environmental Microbiology. 1997;63:3327–3332. doi: 10.1128/aem.63.8.3327-3332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iteman I, Rippka R, Tandeau de Marsac N, Herdman M. Comparison of conserved structural and regulatory domains within divergent 16S rRNA–23S rRNA spacer sequences of cyanobacteria. Microbiology. 2000;146:1275–1286. doi: 10.1099/00221287-146-6-1275. [DOI] [PubMed] [Google Scholar]
- 46.Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16A primers. Journal of Microbiological Methods. 2003;55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 47.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Erwin PM, Thacker RW. Cryptic diversity of the symbiotic cyanobacterium Synechococcus spongiarum among sponge hosts. Molecular Ecology. 2008;17:2937–2947. doi: 10.1111/j.1365-294X.2008.03808.x. [DOI] [PubMed] [Google Scholar]
- 49.Monniot C, Monniot F. Additions to the inventory of Eastern tropical Atlantic ascidians: Arrivals of cosmopolitan species. Bulletin of Marine Science. 1994;54:71–93. [Google Scholar]
- 50.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 51.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 52.Posada D. jModelTest: Phylogenetic model averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 53.Tavaré S. Some probabilistic and statistical problems in the analysis of DNA sequences. In: Miura RM, editor. Some mathematical questions in biology - DNA sequence analysis. Providence, RI: American Mathematics Society; 1986. pp. 57–86. [Google Scholar]
- 54.Felsenstein J. Evolutionary trees from DNA sequences: A maximum likelihood approach. Journal of Molecular Evolution. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 55.Jobb G. Treefinder version of October 2008. 2008. October 2008 ed. Munich, Germany.
- 56.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 57.Reynolds ES. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. Journal of Cell Biology. 1963;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hirose E, Hirose M, Neilan BA. Localization of symbiotic cyanobacteria in the colonial ascidian Trididemnum miniatum (Didemnidae, Ascidiacea). Zoological Science. 2006;23:435–442. doi: 10.2108/zsj.23.435. [DOI] [PubMed] [Google Scholar]
- 59.Parry DL. Cyanophytes with R-phycoerythrins in association with seven species of ascidians from the Great Barrier Reef. Phycologia. 1984;23:503–513. [Google Scholar]
- 60.Cox GC, Hiller RG, Larkum AWD. An unusual cyanophyte, containing phycourobilin and symbiotic with ascidians and sponges. Marine Biology. 1985;89:149–163. [Google Scholar]
- 61.Erwin PM, Thacker RW. Incidence and identity of photosynthetic symbionts in Caribbean coral reef sponge assemblages. Journal of the Marine Biological Association of the United Kingdom. 2007;87:1683–1692. [Google Scholar]
- 62.Usher KM, Toze S, Fromont J, Kuo J, Sutton DC. A new species of cyanobacterial symbiont from the marine sponge Chondrilla nucula. Symbiosis. 2004;36:183–192. [Google Scholar]
- 63.Steindler L, Huchon D, Avni A, Ilan M. 16S rRNA phylogeny of sponge-associated cyanobacteria. Applied and Environmental Microbiology. 2005;71:4127–4131. doi: 10.1128/AEM.71.7.4127-4131.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thacker RW. Impacts of shading on sponge-cyanobacteria symbioses: a comparison between host-specific and generalist associations. Integrative and Comparative Biology. 2005;45:369–376. doi: 10.1093/icb/45.2.369. [DOI] [PubMed] [Google Scholar]
- 65.López-Legentil S, Song B, McMurray SE, Pawlik JR. Bleaching and stress in coral reef ecosystems: hsp70 expression by the giant barrel sponge Xestospongia muta. Molecular Ecology. 2008;17:1840–1849. doi: 10.1111/j.1365-294X.2008.03667.x. [DOI] [PubMed] [Google Scholar]
- 66.Turon X, Hernández-Mariné M, Catalan J. A new species of Komvophoron (Cyanophyta, Borziaceae) epibiote on ascidians from the Mediterranean Sea. Algological Studies. 1991;64:249–259. [Google Scholar]
- 67.Duclaux G, Lafargue F, Wahl M. First report of Prochloron in association with the genus Polysyncraton didemnid ascidian (Tunicata). Vie et Milieu. 1988;38:145–148. [Google Scholar]
- 68.Kott P. Didemnid-algal symbioses: host species in the Western Pacific with notes on the symbiosis. Micronesica. 1982;18:95. [Google Scholar]
- 69.Larkum AWD, Cox GC, Hiller RG, Parry DL, Dibbayawan TP. Filamentous cyanophytes containing phycourobilin and in symbiosis with sponges and an ascidian of coral reefs. Marine Biology. 1987;95:1–13. [Google Scholar]
- 70.Hernández-Mariné M, Turon X, Catalan J. A marine Synechocystis (Chroococcales, Cyanophyta) epizoic on didemnid ascidians from the Mediterranean Sea. Phycologia. 1990;29:275–284. [Google Scholar]
- 71.Hirose E, Uchida H, Murakami A. Ultrastructural and microspectrophotometric characterization of multiple species of cyanobacterial photosymbionts coexisting in the colonial ascidian Trididemnum clinides (Tunicata, Ascidiacea, Didemnidae). European Journal of Phycology. 2009;44:365–375. [Google Scholar]
- 72.Kühl M, Larkum AWD. The microenvironment and photosynthetic performance of Prochloron sp. in symbiosis with didemnid ascidians. In: Seckbach J, editor. Cellular origin and life in extreme habitats Vol 3: Symbiosis, mechanisms and model systems. Dordrecht: Kluwer Acad. Publ; 2002. pp. 273–290. [Google Scholar]
- 73.Larkum AWD, Kühl M. Chlorophyll d: the puzzle resolved. TRENDS in Plant Science. 2005;10:355–357. doi: 10.1016/j.tplants.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 74.Hu Q, Marquardt J, Iwasaki I, Miyashita H, Kurano N, et al. Molecular structure, localization and function of biliproteins in the chlorophyll a/d containing oxygenic photosynthetic prokaryote Acaryochloris marina. Biochimica et Biophysica Acta. 1999;1412:250–261. doi: 10.1016/s0005-2728(99)00067-5. [DOI] [PubMed] [Google Scholar]
- 75.Marquardt J, Mörschel E, Rhiel E, Westermann M. Ultrastructure of Acaryochloris marina, an oxyphotobacterium containing mainly chlorophyll d. Archives of Microbiology. 2000;174:181–188. doi: 10.1007/s002030000194. [DOI] [PubMed] [Google Scholar]
- 76.Miller SR, Augustine S, Olson TL, Blankenship RE, Selker J, et al. Discovery of a free-living chlorophyll d-producing cyanobacterium with a hybrid proteobacterial/cyanobacterial small-subunit rRNA gene. Proceedings of the National Academy of Sciences. 2005;102:850–855. doi: 10.1073/pnas.0405667102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Swingley WD, Hohmann-Marriott M, Olson TL, Blankenship RE. Effect of iron on growth and ultrastructure of Acaryochloris marina. Applied and Environmental Microbiology. 2005;71:8606–8010. doi: 10.1128/AEM.71.12.8606-8610.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kott P. The Australian Ascidiacea. Part 4, Aplousobranchia (3), Didemnidae. Memoirs of the Queensland Museum. 2001;47:1–407. [Google Scholar]
- 79.Eldredge L-G. A taxonomic review of Indo-Pacific Didemnid ascidians and descriptions of twenty-three central Pacific species. Micronesica. 1966;2:161–261. [Google Scholar]
- 80.Hirose E, Oka AT, Akahori M. Sexual reproduction of the photosymbiotic ascidian Diplosoma virens in the Ryukyu Archipelago, Japan: vertical transmission, seasonal change, and possible impact of parasitic copepods. Marine Biology. 2005;146:677–682. [Google Scholar]
- 81.Oka AT, Suetsugu M, Hirose E. Two new species of Diplosoma (Ascidiacea: Didemnidae) bearing prokaryotic algae prochloron from Okinawajima (Ryukyu Archipelago, Japan). Zoological Science. 2005;22:367–374. doi: 10.2108/zsj.22.367. [DOI] [PubMed] [Google Scholar]
- 82.Hirose E, Oka AT. A new species of photosymbiotic ascidian from the Ryukyu Archipelago, Japan, with remarks on the stability of stigma number in photosymbiotic Diplosoma species. Zoological Science. 2008;25:1261–1267. doi: 10.2108/zsj.25.1261. [DOI] [PubMed] [Google Scholar]
- 83.Hirose E, Oka AT, Hirose M. Two new species of photosymbiotic ascidians of the genus Diplosoma from the Ryukyu Archipelago, with partial sequences of the COI gene. Zoological Science. 2009;26:362–368. doi: 10.2108/zsj.26.362. [DOI] [PubMed] [Google Scholar]
- 84.Hirose M, Hirose E. DNA barcoding in photosymbiotic species of Diplosoma (Ascidiacea: Didemnidae), with the description of a new species from the southern Ryukyus, Japan. Zoological Science. 2009;26:564–568. doi: 10.2108/zsj.26.564. [DOI] [PubMed] [Google Scholar]
- 85.Hirose E, Adachi R, Kuze K. Sexual reproduction of the Prochloron-bearing ascidians, Trididemnum cyclops and Lissoclinum bistratum, in subtropical waters: seasonality and vertical transmission of photosymbionts. Journal of the Marine Biological Association of the United Kingdom. 2006;86:175–179. [Google Scholar]
- 86.Hirose E, Nakabayashi S. Algal symbionts in the larval tunic lamellae of the colonial ascidian Lissoclinum timorense (Ascidiacea, Didemnidae). Zoological Science. 2008;25:1205–1211. doi: 10.2108/zsj.25.1205. [DOI] [PubMed] [Google Scholar]
- 87.Murakami A, Miyashita H, Iseki M, Adachi K, Mimuro M. Chlorophyll d in an epiphytic cyanobacterium of red algae. Science. 2004;303:1633. doi: 10.1126/science.1095459. [DOI] [PubMed] [Google Scholar]