Abstract
Pumilio and FBF homology (PUF) proteins represent highly promising candidates for engineering sequence-specific RNA recognition, but were only known to recognize G, A and U, significantly limiting applications. Two groups (Filipovska et al., 2011; Dong et al., 2011) have now reported the discovery of the cytosine-recognition code for PUF proteins.
RNA-binding proteins (RBPs) play essential roles in regulating every step of RNA maturation and function. Typically, they recognize specific RNA structural features and sequences through a small number of very common RNA-binding modules (Lunde et al., 2007) and are often complemented by other enzymatic or structural domains that perform additional functional roles. It would be very useful to engineer RBPs with desired RNA-binding specificities for investigating RNA biology and for potential biomedical applications, but this task has proven to be very challenging (Mackay et al., 2011). The only exception has been the PUF domain, which has been successfully manipulated to target single-stranded RNA in a sequence specific manner. Thus, this domain represents the most promising candidate for the routine generation of designer RBPs with required RNA-binding specificity, were it not for one limitation: PUF domains have not been observed to recognize cytosine so far.
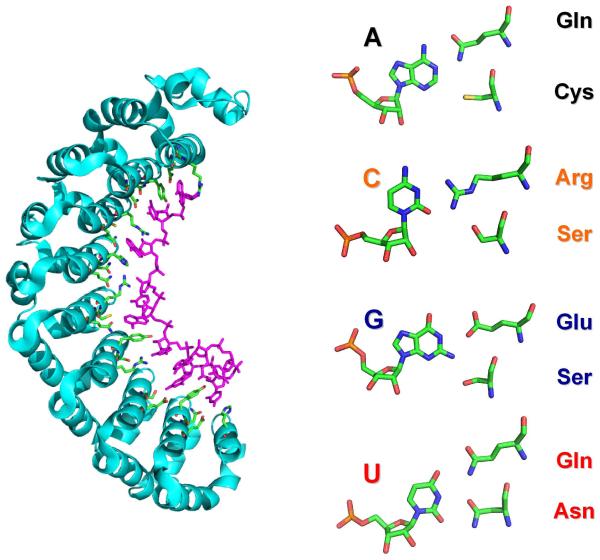
PUF proteins are named after their founding members PUMILIO and fem-3 binding factor (FBF). They regulate gene expression by binding to specific sequences in the 3′-UTR of their target mRNAs and promote mRNA degradation and translational repression by recruiting effector proteins to the targeted sequence. All PUF domains contain multiple repeats, typically 8, of the same sequence-specific RNA-binding module, a structurally conserved 36-amino acid repeat. The single-stranded RNA (ssRNA) runs antiparallel to the protein and binds to the inner concave surface generated by the multiple PUF repeats (Figure 1) (Wang et al., 2002). Remarkably, each PUF repeat recognizes one nucleotide using three well-conserved amino acids in a base-specific manner. The amino acid side chain at position 13 in the repeat forms stacking interactions with the aromatic ring of the RNA base, and the Watson-Crick edge of the base is recognized by a specific combination of two amino acids (at positions 12 and 16). Thus, a single recognition code was generated: cysteine and glutamine bind adenine, asparagine and glutamine bind uracil, and serine and glutamate bind guanine (Figure 1). The simplicity of this recognition code allowed switching the specificity of individual repeats from one nucleotide to the other by just mutating only the two amino acids that make specific contacts with the RNA base edge (Cheong et al., 2006). Many of the resulting mutant PUF proteins can bind to their cognate RNAs as tightly and specifically as the wild-type protein they were derived from.
FIGURE1.
RNA recognition by PUF proteins.
Shown here is the crystal structure of an engineered PUF domain in complex with target RNA containing cytosine (2YJY.pdb; Dong et al., 2011). On the left, the RNA recognition code of PUF repeats is illustrated.
Once proof of principle was provided that specificity could be engineered, different effector domains were attached to the designed PUF domains to engineer new functions. This technology was used to characterize the localization and trafficking of mitochondrial (mtRNA) in single cells (Ozawa et al., 2007), and to engineer artificial splicing factors with desired sequence specificity and activity by combining a designed PUF domain with different splicing modulation domains (activator or repressor) (Wang et al., 2009). These artificial splicing factors were shown to modulate splicing of an endogenous human gene, BCL-X.
However, naturally occurring PUF domains have not been observed to recognize cytosine, leaving designers without a recognition code for ‘C’ and therefore limiting the potential target sites. In a significant advance, two groups (Filipovska et al., 2011; Dong et al., 2011) now report using directed evolution methods to select for PUF repeat variants that specifically recognize cytosine. Both studies use the yeast three-hybrid system to link the interaction between PUF domains and RNA to a life-death growth selection. A DNA library based on the PUM1 PUF domain containing randomized amino acids at positions 12 and 16 was combined with an RNA, in which the targeted base was cytosine, so that only variants capable of binding to C would survive the selection. In the study by Filipovska et al., five unique PUF mutants selectively interacted with RNAs containing cytosine, but not with those containing adenine, guanine or uracil RNA bases; all had an arginine at position 16 and an amino acid with a small or nucleophilic side chain at position 12 (Gly, Ala, Ser, Thr or Cys). The result of study by Dong et al. was more focused: almost all the selected clones contained an SXXXR sequence, perhaps a consequence of more stringent selection conditions and tighter binding, and Y/H/R were found to be the best stacking residues in the cognate repeat to achieve specific binding of cytosine. Once the selected proteins were overexpressed and purified, the directed evolution experiments were proven to have been successful; specificity was re-engineered without loss of binding affinity. Dong et al. then went further by engineering a new splicing factor that recognize C-containing target sequences, and demonstrated the specific modulation of alternative splicing of the VEGF-A mRNA, a key regulator of angiogenesis.
The crystal structure of the mutated PUF (SYXXR) protein revealed how cytosine is recognized (Dong et al., 2011). The essential arginine contacts the O2 and N3 positions of the cytosine, while the serine forms a hydrogen bond with an amino group of the arginine side chain to position it for recognizing C (Figure 1). The cytosine base has to move away slightly from the RNA-binding surface to accommodate the longer arginine side chain. The specific interactions with cytosine are consistent with previous structure-based analysis of protein-RNA interactions, which had already indicated that the guanidinium group of arginine is most frequently used to simultaneously interact with Cyt-N3 and O2.
An additional step was undertaken by Filipovska et al., who engineered PUFs with 16 RNA-binding repeats to achieve higher level of RNA sequence discrimination than would be possible with the 8 nucleotides that are typically recognized by naturally occurring PUF proteins. The extended PUF bound its cognate 16 nucleotide RNA target in yeast and activated transcription more efficiently than a canonical 8-repeat PUF protein with its cognate RNA. It will now be interesting to establish the structure of this extended PUF protein to examine how it interacts with its extended RNA target. PUF domains adopt a crescent shape (Figure 1), so the doubling the number of repeats may not be feasible without distorting the superstructure of the PUF repeats and affecting their RNA binding ability. It may also increase the possibility of some bases flipping out from the RNA-binding surface, leading to reduced RNA-binding specificity. Nonetheless, this study suggests that it is feasible to use extended PUF repeats to target unique RNA sequences, which would be difficult to achieve with eight-repeat PUF.
About 20 years ago, Yen Choo and Aaron Klug reported the application of engineered zing finger proteins to bind to a desired DNA sequence and alter gene transcription (Choo et al., 1994). Even if a sequence-specific recognition code for zinc fingers does not quite exist, this study led to many successful applications of engineered zinc finger proteins and nucleases, to the point where we can now simply order a designed zinc finger protein with any targeting DNA sequence specificity from a chemical supplier. PUF proteins are even more easily adaptable than zinc fingers; the discovery of the cytosine-recognition code clearly provides a straightforward two amino acid tool-kit to design RNA recognition. With the caveat that recognition of sequences longer than 8 nucleotides may still be challenging, it should now be possible to simply dial in an RNA sequence to generate a PUF protein with any desired specificity. Applications in research on RNA metabolism are limited only by our ingenuity and perhaps even biomedical applications of PUF proteins will be possible.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Cheong C, Hall TM. Proc. Natl. Acad. Sci. USA. 2006;103:13635–14639. doi: 10.1073/pnas.0606294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo Y, Sanchezgarcia I, Klug A. Nature. 1994;372:642–645. doi: 10.1038/372642a0. [DOI] [PubMed] [Google Scholar]
- Dong S, Wang Y, Cassidy-Amstutz C, Lu G, Bigler R, Jezyk MR, Li C, Hall TM, Wang Z. J. Biol. Chem. 2011 doi: 10.1074/jbc.M111.244889. Advanced online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipovska A, Razif MFM, Nygård KKA, Rackham O. Nat. Chem. Biol. 2011 doi: 10.1038/nchembio.577. Advanced online publication. [DOI] [PubMed] [Google Scholar]
- Lunde BM, Moore C, Varani G. Nat. Rev. Mol. Cell. Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay JP, Font J, Segal DJ. Nat. Struct. Mol. Biol. 2011;18:256–261. doi: 10.1038/nsmb.2005. [DOI] [PubMed] [Google Scholar]
- Ozawa T, Natori Y, Sato M, Umezawa Y. Nature Methods. 2007;4:413–419. doi: 10.1038/nmeth1030. [DOI] [PubMed] [Google Scholar]
- Wang Y, Cheong G, Hall TM, Wang Z. Nature Methods. 2009;6:825–830. doi: 10.1038/nmeth.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McLachlan J, Zamore PD, Hall TM. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]