Abstract
The circadian clock is closely associated with energy metabolism. The liver clock can rapidly adapt to a new feeding cycle within a few days, whereas the lung clock is gradually entrained over one week. However, the mechanism underlying tissue-specific clock resetting is not fully understood. To characterize the rapid response to feeding cues in the liver clock, we examined the effects of a single time-delayed feeding on circadian rhythms in the liver and lungs of Per2::Luc reporter knockin mice. After adapting to a night-time restricted feeding schedule, the mice were fed according to a 4, 8, or 13 h delayed schedule on the last day. The phase of the liver clock was delayed in all groups with delayed feeding, whereas the lung clock remained unaffected. We then examined the acute response of clock and metabolism-related genes in the liver using focused DNA-microarrays. Clock mutant mice were bred under constant light to attenuate the endogenous circadian rhythm, and gene expression profiles were determined during 24 h of fasting followed by 8 h of feeding. Per2 and Dec1 were significantly increased within 1 h of feeding. Real-time RT-PCR analysis revealed a similarly acute response in hepatic clock gene expression caused by feeding wild type mice after an overnight fast. In addition to Per2 and Dec1, the expression of Per1 increased, and that of Rev-erbα decreased in the liver within 1 h of feeding after fasting, whereas none of these clock genes were affected in the lung. Moreover, an intraperitoneal injection of glucose combined with amino acids, but not either alone, reproduced a similar hepatic response. Our findings show that multiple clock genes respond to nutritional cues within 1 h in the liver but not in the lung.
Introduction
The mammalian circadian clock consists of a central pacemaker in the suprachiasmatic nucleus (SCN) of the hypothalamus and various oscillators in most peripheral tissues [1]. The molecular oscillator of the circadian clock is thought to depend on a negative transcriptional feedback loop of core clock genes such as Per1, Per2, Cry1, Cry2, Clock and Bmal1 [2]. In addition to these genes, several other clock genes such as Rev-erbα, Dbp, Dec1 and Dec2, reinforce the molecular oscillator of the transcriptional circuit [3], [4], [5]. The circadian clock is not only robust but it is also flexible enough to adapt to surrounding circumstances. Light-dark (LD) cycles comprise a critical cue for the central clock in the SCN, whereas cyclic feeding behavior is the predominant cue for many peripheral tissue clocks [6], [7]. Restricted daytime feeding obviously entrains the circadian clocks of many peripheral tissues in nocturnal animals and even in extra-SCN brain regions, even though SCN activity is locked to LD cues [8], [9], [10]. Although the phases of circadian gene expression are similarly shifted in most peripheral tissues after one week of daytime feeding, food-induced phase resetting proceeds faster in the liver than in the kidney, heart, pancreas or lung [8], [9]. Two days of restricted feeding efficiently shifts the phase of the liver clock in mice and rats [9], [11] and even a 30-min feeding stimulus induces rapid Per2 and Dec1 gene expression within 1 h in the rat liver and shifts the circadian phases of clock gene expression on the following day [12]. However, the molecular profile underlying the variable sensitivity of tissues to feeding cues and the nutrients required to affect the peripheral clocks remain obscure.
Findings from behavioral and cell culture experiments suggest that an increase in the glucose level is involved in feeding-induced entrainment [13], [14]. A 100% glucose diet causes food anticipatory activity (FAA) rhythms in mice and rats, whereas feeding on glucose alone does not entrain the mouse liver clock [11], [14]. In contrast, the oral intake of sugars plus proteins can entrain the liver clock in mice [11], indicating that a balanced diet is required for proper entrainment of this clock.
Enteral nutrition at restricted times entrains the circadian rhythm of blood cortisol in humans [15]. In contrast, total parenteral feeding despite a restricted duration abolishes the adrenocortical rhythm, although the blood urea level, which is signalled by the time of feeding, remains at the same level as that for oral feeding [16]. Moreover, jejunal resection attenuates the daily rhythm of corticosterone in rat blood [17]. These results suggest that nutritional digestion in the gastrointestinal region is critical to entrain peripheral clocks to feeding. However, this is controversial because total parenteral nutrition could entrain the clocks of the SCN and liver in rats [18].
Although feeding cues obviously entrain many peripheral clocks, studies of the molecular mechanism underlying the food entrainment of each peripheral clock are scant. The present study compares the response of the lung and liver clocks in mice to clarify the molecular profile underlying the rapid response of the liver clock to nutritional cues.
Materials and Methods
Animals and Handling
Animals were handled according to the guidelines of the Ministry of Agriculture, Forestry and Fisheries for laboratory animal studies and the study was reviewed and approved by the Animal Care and Use Committee of the National Food Research Institute, National Agriculture and Food Research Organization (NARO), Japan (approval ID; H21-083, 084, 092, H22-001 and 010).
BALB/cAn mice (males, 10–30 weeks) were obtained from the Institute for Animal Reproduction, Charles River Japan. Homozygous of the Clock mutant mice (Jcl:ICR genetic background) were described previously [19]. Per2::Luc knockin mice [20] established by Dr. Joseph Takahashi (Northwestern University, USA) were supplied by the Jackson Laboratory (USA) and bred as homozygotes. All mice were housed under 24±1°C, 55±5% humidity and a 12 h light-dark (LD) photocycle (light period from 08:00 to 20:00) with free access to water and a standard diet (NMF; Oriental Yeast, Japan).
Luminescent Analysis of Explants from Per2::Luc Mice
Liver and lung explants were prepared [11], [21], [22] from male and female Per2::Luc mice (23–50 weeks). The circadian rhythmicity in the liver explants did not significantly differ between the sexes [23], [24]. The mice were sacrificed at ZT3 to record bioluminescence rhythmicity in the liver and the lung. The livers and the lungs were rapidly removed from the mice and placed in cold Hanks' balanced salt solution (HBSS; Sigma, USA) with 10 mM HEPES (Sigma) and penicillin-streptomycin (Sigma). The blocks were cut into pieces and explanted in 35-mm dishes, sealed with Parafilm and incubated at 37°C under a 5% CO2 atmosphere with DMEM containing high glucose (Invitrogen, USA), 10 mM HEPES, penicillin-streptomycin and 0.1 mM luciferin (Promega, USA). Bioluminescence was measured and integrated for 1 min at intervals of 10 min using a dish-type luminometer (Kronos Dio AB-2500; Atto, Japan).
Luminescence data were analyzed as described [11], [22], [25]. The original data were smoothed using an adjusting-averaging method with 100-min running means and then the dataset was detrended by subtracting the 24-h running average from the raw data. Peaks were defined as points at which the bioluminescence was higher than that on both sides of the points and confirmed from waveforms. The peak phase time was determined from the peak that initially appeared between 12 and 36 h of culture.
DNA Microarray Analysis
We performed DNA microarray analysis using the fibrous DNA chip Genopal® (Mitsubishi Rayon, Japan) as described [22], [26], [27]. We synthesized DNA oligonucleotide probes to detect 206 genes related to metabolic processes and installed them in Genopal®, which comprises hollow plastic fibers containing a gel to which the probes can attach [28]. All probes (65-mer), including those for positive and negative controls, were designed to have an annealing temperature (Tm) ranging from 65°C to 75°C throughout the microarray.
We randomly selected 8 of 30 Clock mutant mice (males, 39–62 weeks) and placed them in cages equipped with infrared sensors (AS10D; Melquest, Japan) to record locomotor activity. All of the mice were bred under 12 h LD conditions for two weeks followed by constant light (LL) for three weeks. Behavioral data were visualized using CIF-II software (Melquest) and rhythmicity was evaluated using Metlab software (MathWorks Inc., USA). After breeding under LL for 3 weeks, the mice were sacrificed at 6, 12, 18, and 24 h after starting to fast and at 1, 2, 4 and 8 h after resumed feeding (that is, 25, 26, 28 and 32 h from the start of fasting) by cervical dislocation and then the livers and lungs were immediately collected in RNAlater™ (Ambion, USA). Total RNA was extracted from the excised tissues using the RNeasy Mini kit (Qiagen KK, Japan) and amplified using the MessageAmpII biotin-enhanced amplification kit (Applied Biosystems, USA) according to the manufacturer's instructions. Biotinylated aRNA was disintegrated using fragmentation reagents (Applied Biosystems) and then incubated at 95°C for 7.5 min. Hybridization, washing and fluorescent-labelling proceeded using the Genopal® UE-104 system (Mitsubishi Rayon). Hybridization was performed using a DNA microarray (Genopal®) in hybridization buffer, 0.12 M Tris-HCl/0.12 M NaCl/0.05% Tween-20, and the fragmented biotinylated aRNA at 65°C for 16 h. The hybridized DNA microarray was washed in 1 mL of 0.12 M Tris-HCl/0.12 M NaCl/0.05% Tween-20 at 65°C and in 1 mL of 0.12 M Tris-HCl/0.12 M NaCl. The microarray was then fluorescently labelled with streptavidin-Cy5 (GE Healthcare Bio-Sciences KK, Japan), washed in 0.12 M Tris-HCl/0.12 M NaCl/0.05% Tween-20 at room temperature and in 1 mL of 0.12 M Tris-HCl/0.12 M NaCl.
Hybridization signals were acquired using a DNA microarray reader and multibeam excitation technology (Yokogawa Electric Co., Japan). The DNA microarrays were scanned at multiple exposure durations of 0.1, 0.4, 1.0, 4.0, and 30 s. Intensity values with the optimal exposure condition for each spot were selected according to saturation. The relative amount of each transcript was normalized to the amount of 18S ribosomal RNA in the same sample.
Quantitative RT-PCR
BALB/c mice fasted overnight from ZT9 resumed feeding from the next ZT 1, or were intraperitoneally injected with 0.3 mL of nutrients, 30% glucose (Glc; 1.7 M; 3.0–3.5 g/kg body weight), a mixture of 12 amino acids (12AA; 50× MEM amino acids solution (PromoCell GmbH, Germany) comprising L-isomers of 10 mM F, 10 mM H, 20 mM I, 20 mM K, 20 mM L, 5 mM M, 30 mM R, 20 mM T, 20 mM V, 2.5 mM W, 10 mM Y and 5 mM Cystine), Glc with 3 amino acids (3AA; L-isomers of F, M, and T; the concentrations of all of these were the same as in the mixture containing 12AA), Glc with 6 amino acids (6AA; L-isomers of K, H, I, L, R, and V; the concentrations of all of these were the same as in the mixture containing 12AA), Glc with 9 amino acids (9AA; mixture of 3AA and 6AA comprising L-isomers of F, K, H, I, M, L, R, T, and V) or PBS (control) at ZT1. The final osmolarity of each solution was not adjusted. The mice were then sacrificed by cervical dislocation at ZT2 for one-time sampling, or at ZT2, 5, 8, and 11 for multi-time sampling. The livers and lungs were immediately collected in RNAlater and total RNA was extracted described above. Reverse transcription proceeded using Superscript II reverse transcriptase (Invitrogen) with random hexamers according to the manufacturer's protocol. Gene-specific primers (Table 1) and Thunderbird SYBR qPCR Mix (Toyobo, Japan) were included in the real-time PCR protocol. The reaction proceeded at 95°C for 15 s, 60°C for 31 s and for 40 cycles. Amounts of PCR products were monitored using an ABI Prism 7000 sequence detection system and analyzed using ABI Prism 7000 SDS software (Applied Biosystems). All primer pairs spanned exon junctions. The relative amounts of each transcript were normalized to the amount of GAPDH transcript in the same cDNA.
Table 1. Primer pairs used for real-time RT-PCR.
| Gene names | Forward (5′-3′) | Reverse (5′-3′) |
| mPer1 | gcttcgtggacttgacacct | tgctttagatcggcagtggt |
| mPer2 | caacacagacgacagcatca | tcctggtcctccttcaacac |
| mDec1 | ccatgggttaggtgagccatg | tgtctgcacttagtagagtctgag |
| mDec2 | ctacacacactctcagactgg | gtttctgtcctgtaatctgtgg |
| mRev-erbα | ccctggactccaataacaaca | tgccattggagctgtcact |
| mDbp | gcattccaggccatgagact | ccagtacttctcatccttctgt |
| mGck | gtgaggtcggcatgattgt | tccaccagctccacattct |
| mGapdh | catggccttccgtgttccta | cctgcttcaccaccttcttga |
Statistical Analysis
All values are expressed as means ± SEM. Differences in expression levels and peak times were statistically evaluated using Student's t-test for single comparisons, and one-way ANOVA with Dunnett's post-hoc test for multiple comparisons using Prism 5 software (GraphPad Software; USA).
Results
Single Feeding Delays Circadian Clock in the Liver but not in the Lung
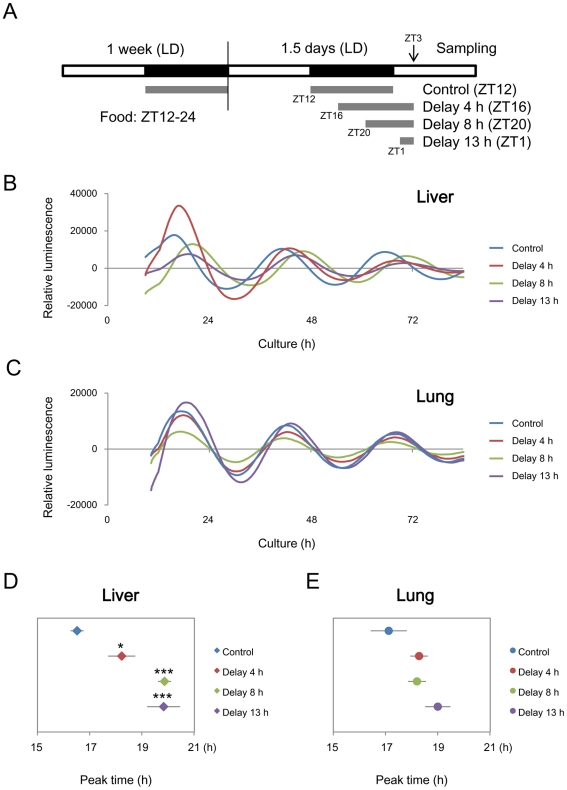
To understand the response of peripheral clocks induced by feeding cues, we examined the effects of a single delayed feeding on the circadian clocks in the liver and lungs. Per2::Luc knock-in mice [20] received a night-time restricted feeding schedule (ZT12-24) for one week, and then the feeding start time was delayed on the last day by 0 (control), 4, 8 and 13 h; that is, feeding from ZT12, 16, 20 and on the next day at ZT1 (Figure 1A). Figure S1 shows representative results of temporal consumption suggesting that the mice consumed similar amounts or more food during feeding that was delayed by 4 h and 8 h to the controls, and that the mice with feeding delayed by 13 h consumed less food than the controls. The liver and lung explants were isolated from the mice at ZT3 and then circadian luminescence was recorded (Figure 1B and 1C). Peak time statistically differed in the liver among groups (one-way ANOVA, p<0.001, n = 5–7; Figure 1D). The peak times of the groups given delayed feeding differed from control values (p<0.05, Dunnett's test). In contrast, the peak time was not shifted in the lungs of all four groups (one-way ANOVA, p>0.05, n = 5–7; Figure 1E). These results indicated that a single feeding obviously delays the circadian clock in the liver, but not in the lungs.
Figure 1. Single feeding delays phase of liver, but not lung clock.
After a night-time restricted feeding schedule (ZT12-24) for one week, mice were divided into 4 groups and then delayed feeding was applied as follows. Mice in the 4, 8 and 13 h delayed groups were fed from ZT16, 20, and the next ZT1, respectively. Control mice were fed from ZT12 to ZT24. White and black boxes indicate day and night respectively; gray lines show feeding times (A). Representative circadian profiles of reporter luminescence in explants of livers (B) and lungs (C) isolated from mPer2::Luc knockin mice. Peak times of circadian oscillation between 12 and 36 h in culture are shown for liver (D) and lung (E). Values are shown as means ± SEM (n = 7 for ZT12 and ZT16, and n = 5 for ZT20 and ZT1; *p<0.05, ***p<0.001; vs. control; Dunnett's test).
Acute Response of Clock Genes to Feeding in the Liver of Clock Mutant Mice
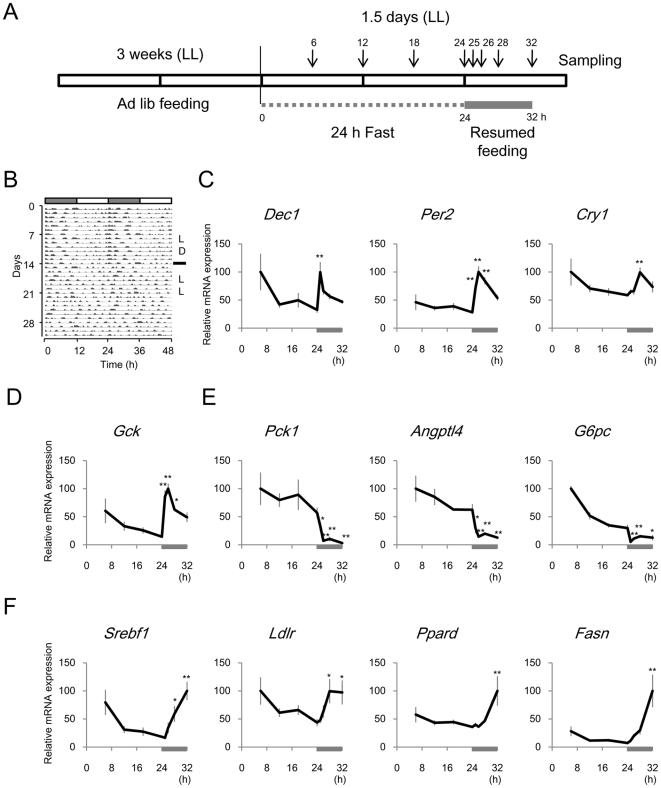
To characterize which genes are associated with rapid response of the liver clock induced by feeding, we performed DNA microarray analysis using a focused DNA chip containing 206 metabolic genes including 18 clock genes (Materials and Methods and Table S1). We minimized the endogenous circadian rhythm of gene expression in the liver by breeding Clock mutant mice under constant light (LL). In fact, the behavioural rhythms of the mice were attenuated after three weeks (Figure 2B). Moreover, Clock mutant mice are entrained normally by feeding cues [29], [30]. We examined the gene expression profile under 24 h of fasting and for 8 h after feeding was resumed (Figure 2A). The endogenous rhythm of clock gene expression was quite weak under 24 h fasting (Figure 2C and Table S1). Although the expression of most clock genes was not significantly affected within 8 h after feeding was resumed (one-way ANOVA, p>0.05), Per2, and Dec1 were transiently induced within 1 h after resuming feeding and Cry1 was slightly increased after 4 h (p<0.01; Figure 2C). With respect to other genes, Gck (glucokinase) was transiently induced within 1 h after resuming feeding as well as Per2 and Dec1 (p<0.01; Figure 2D). None of the genes on the DNA chip was acutely induced within 1 h after feeding like Gck (Table S1). Several genes related to glucose production such as Pck1 (phosphoenolpyruvate carboxykinase 1), Angptl4 (angiopoietin-like 4) and G6pc (glucose-6-phosphatase) were acutely decreased within 1 h (Figure 2E), whereas those associated with fatty acid synthesis such as Ppard (peroxisome proliferator activator receptor delta), Srebf1 (sterol regulatory element binding transcription factor 1), Ldlr (low density lipoprotein receptor) and Fasn (fatty acid synthase) gradually increased for 8 h after feeding was resumed (Figure 2F).
Figure 2. Rapid response of gene expression after resumed feeding in liver of Clock mutant mice.
(A) Experimental design. Clock mutant mice were bred under constant light (LL) for 3 weeks, fasted for 24 h and then fed for 8 h. (B) Representative actogram from mice under 12-h light-dark (LD) cycle for 2 weeks that were switched to LL. (C–F) Expression profiles of mRNAs for clock genes (C), genes related to glucose metabolism (D–E), and fatty acid synthesis (F). Gray bars in both panels, duration of resumed feeding. Data are shown as means ± SEM (n = 5 for 25 h, n = 4 for 12, 18, 24 and 26 h, and n = 3 for 6, 28, and 32 h; *p<0.05, **p<0.01; 24 h vs. 25–32 h; Dunnett's test).
Acute Response of Clock Genes to Feeding in Wild Type Mice
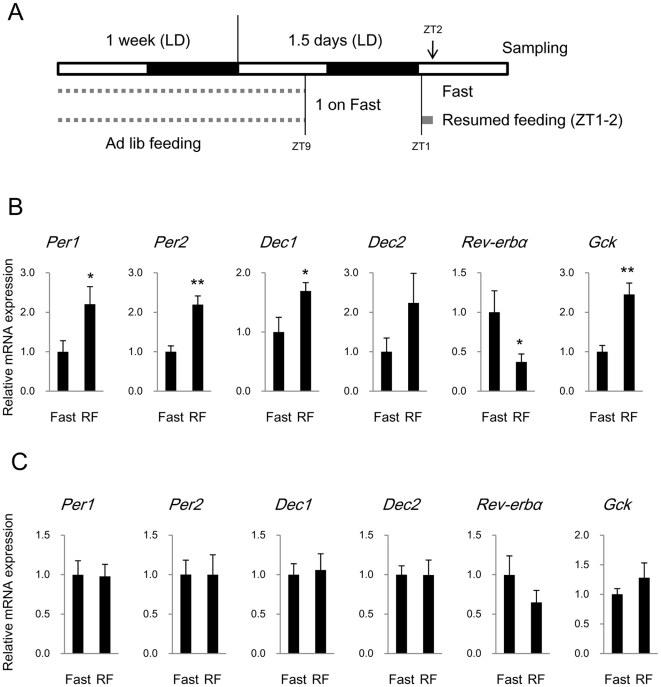
We clarified the acute response of clock genes to feeding in wild type BALB/c mice using real-time RT-PCR analysis. The mice were fed at ZT1 after an overnight fast and then sampled at ZT2 (Figure 3A) because the microarray analysis showed that one hour is sufficient to induce the expression of early responsive genes such as Gck, Dec1, and Per2 (Figure 2). Moreover, only 2 h feeding at ZT1 after an overnight fast delayed the liver clock (Figure 1), suggesting that the liver responds to feeding cues within 2 h. The expression of clock genes (Per1, Per2, Dec1, Dec2, and Rev-erbα) and Gck at ZT2 was examined in the liver and lungs. Levels of Per1, Per2, and Dec1 rapidly increased within 1 h of feeding in the liver (p<0.05; Figure 3B), but not in the lung (Figure 3C). Dec2 might also be increased in the liver of some mice, but a significant difference was undetected in this cohort. Rev-erbα was significantly decreased in the liver (p<0.05), but not in the lung, and Gck was increased in the liver (p<0.01), but not significantly altered in the lung. We also analyzed the expression of Bmal1 and Dbp in the liver and found that it was not affected after 1 h of feeding (Figure S2). These results were consistent with the above findings using Clock mutant mice.
Figure 3. Rapid response in clock gene and Gck expression in liver and lung after resumed feeding.
(A) Experimental design. BALB/c mice were fasted overnight and assigned to resumed feeding (RF). Mice in RF group resumed feeding for 1 h and those in Fast group (control) continued to fast. All animals were sampled at ZT2. (B and C) Expression of mRNAs for clock genes and Gck in liver (B) and lung (C) is shown as means ± SEM relative to control group (n = 8, *p<0.05, ** p<0.01; t-test).
Intraperitoneal Injection of Nutrients Induces Acute Resetting of Clock in Liver
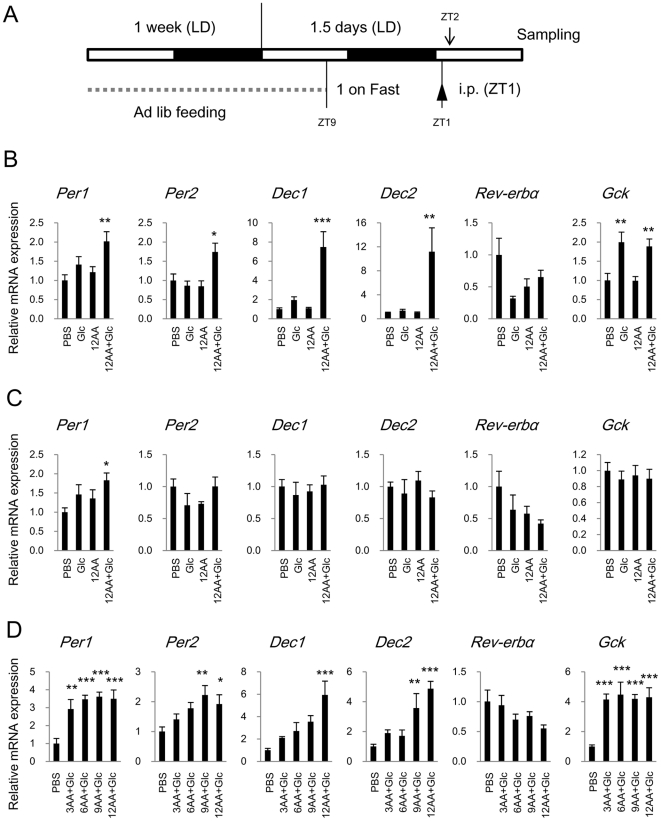
Short term feeding within 1 h induced a rapid response of clock genes in the mouse liver. We examined whether an intraperitoneal injection of nutrients also induces such a response in the liver to determine whether or not intestinal digestion is necessary and which nutrient is required for the rapid response. We used the same time course that was applied in the previous experiment. That is, mice were injected at ZT1 after an overnight fast with 30% glucose (Glc), a mixture of 12 amino acids including all essential amino acids (12AA; L-isomers of F, H, I, K, L, M, R, T, V, W, Y and Cystine), or a mixture of both (12AA+Glc) and then the liver and lungs were sampled at ZT2 (Figure 4A). We found that 12AA+Glc induced Per1, Per2, Dec1 and Dec2 expression in the liver within 1 h of injection (Figure 4B) as well as resumed feeding after an overnight fast. In contrast, an injection of either Glc or 12AA did not affect hepatic expression of these clock genes. Glc and Glc+12AA induced Gck in the liver but not in the lung (Figure 4B and C). Glc+12AA slightly induced Per1 in the lung, but did not affect Per2, Dec1, and Dec2 (Figure 4C). Levels of Rev-erbα tended to decrease in the liver and lung after an injection of any of the nutrients, but the difference did not reach statistical significance.
Figure 4. Rapid response in clock gene and Gck expression in liver and lung after intraperitoneal injection of nutrients.
(A) Experimental design. BALB/c mice were fasted overnight then intraperitoneally injected with nutrients (Glc: glucose, AA, amino acids; see Materials and methods) or PBS (control) at ZT1. All animals were sampled at ZT2. (B–D) Expression of clock gene and Gck mRNA in liver (B and D) and lung (C) is shown as means ± SEM relative to control group (n = 8, *p<0.05, **p<0.01, ***p<0.001; vs. control; Dunnett's test).
We characterized the amino acid requirement in more detail using three more mixtures generated by subtracting amino acids from the 12AA mixture: 3AA (F, M and T), 6AA (H, I, K, L, R, and V), and 9AA (mixtures 3 plus 6). The time course experiment was repeated in another cohort of BALB/c mice (Figure 4A) that was fasted overnight and then divided into groups that were injected with PBS (control) or Glc+3AA, Glc+6AA, Glc+9AA or Glc+12AA at ZT1. All nutrient mixtures induced Per1 and Gck expression (p<0.01 vs. PBS; Figure 4D). Both Glc+9AA and Glc+12AA, but not Glc+3AA and Glc+6AA induced Per2, and Dec2, and only Glc+12AA induced Dec1 mRNA. Although none of the injected mixtures statistically decreased Rev-erbα, the average expression level gradually declined dependently upon the numbers of injected AA. These results showed that an injection of glucose combined with amino acids elicited an acute response of clock genes in the liver but not in the lung.
Finally, to examine that this acute response induced by an injection of nutrients actually causes a phase delay of the liver clock, we determined the temporal expression of Rev-erbα and Dbp in liver and lung on the next daytime after the nutrients injection (Figure S3A). We used these two clock genes because they sharply peak in the daytime [8], [27], [31], [32], which facilitates detection of the phase shift. Moreover, Dbp would be an appropriate indicator to show the phase of liver clock because it was not acutely affected by the nutritional injection but its expression was expected to be affected in the following cycle as a clock controlled gene. The injection of 12AA+Glc significantly affected Rev-erbα expression around the peak time of the following day in the liver but not in the lung (Figure S3B and C). The expression increased at ZT2 and 5, and decreased at ZT8. Tendency was similar for Dbp (increased at ZT5 and 8, and decreased at ZT11), but without reaching statistical significance. These results seem that an intraperitoneal injection of 12AA+Glc caused phase delay of the liver clock in the following day.
Discussion
Our results showed that a single feeding and a single intraperitoneal injection of nutrients can efficiently induce an acute response of clock genes in the liver but not in the lung, and that a combination of glucose and amino acids is required for this response.
The mice seemed to consume a similar amount or more food during feeding that was delayed by 4 and 8 h to controls, and only the mice with feeding delayed by 13 h seemed to consume less food than the others because the time until sampling was shorter. Despite the decreased food consumption, our results indicated that only 2 h of feeding (feeding delayed by 13 h) is sufficient to delay the mouse liver clock. Moreover, a single intraperitoneal injection of nutrients containing fewer calories than the total daily food intake elicited a response from liver clock genes. Consuming less than the total daily amount of food might entrain the liver clock at least under fasting conditions. This is consistent with a recent study showing that only 30 min of feeding stimuli are sufficient to significantly induce the expression of Per2 and Dec1 within 1 h and alter the transcript levels and circadian phases of other clock genes including Rev-erbα in the liver of rats after a longer interval [12]. The phase delay was larger in the 8- and 13-h, than in the 4-h group. This seemed to fit with phase-response curves as well as the response of the central clock to light [33]. Delayed feeding for 13 h is close to the turnaround point because daytime restricted feeding from ZT5 (17 h-delayed) induces a phase advance of the peripheral clock in the mouse [34]. Consequently, the maximum delay caused by a single feeding might be around 4 h.
The expression rhythms of clock- and metabolism-related genes in the liver were attenuated in Clock mutant mice bred under constant light, whereas the circadian expression of about 50 of the 200 genes on this DNA chip was detectable in wild type mice under 12 h LD [27]. This is a useful way to evaluate responses to various stimuli, as it can distinguish them from intrinsic circadian changes in expression. In fact, the microarray data showed that feeding cues inhibit genes related to gluconeogenesis (Pck1, Angptl4, and G6pc), gradually increase the genes associated with fatty acid synthesis (Srebf1, Ldlr, Ppard, and Fasn), and induce acute expression of the clock genes, Per2 and Dec1, among 200 genes related to metabolism in the liver. Insulin secretion caused by glucose uptake in the blood acutely increases Gck and decreases Pck1, genes that are associated with glucose metabolism [35], [36], [37]. Indeed, we found that the feeding cue caused rapid Gck induction and a rapid Pck1 decrease in the liver of Clock mutant mice. Moreover, Gck was also induced by a glucose injection in the livers of wild type mice, which was consistent with the fact that an intraperitoneal injection of glucose induces insulin secretion [38]. However, the expression of clock genes including Per2 and Dec1 was not affected by the glucose injection, suggesting that another pathway induces these clock genes. Such unknown signalling, which is probably modulated by protein modifications such as phosphorylation and acetylation because of the rapidity, might include key molecules that link nutritional signals to circadian rhythms in the liver.
We found here that an intraperitoneal injection of glucose together with amino acids induced rapid changes in clock gene expression in the liver but not in the lungs. We could not exclude the possibility that total osmolarity affects rapid responses to stimuli in the liver. However, the osmolarity of the glucose solution was much higher than that of the amino acid mixture, and it did not induce a rapid response of clock gene expression, suggesting that the response depends on nutritional components rather than on osmolarity. Our results are consistent with the findings that a diet of either glucose or protein alone does not influence the phase of liver clock whereas a diet containing both sugar and protein delays the phase of liver clock [11].
The 12 amino acids that we used are components of Minimal Essential Medium (MEM), that includes all of the essential amino acids and in which mammalian cells are generally cultured. It is possible to record circadian luminescent rhythms from cultured liver samples obtained from Per2::Luc mice for several days using MEM. In fact, injecting 12 amino acids with glucose induces acute clock gene expressions and also affected expression of Rev-erbα even after 24 to 36 h in the liver, suggesting that these amino acids comprise a sufficient nutritional cue for the liver. Moreover, we analyzed the contribution of each of the 12 amino acids classified into 3 groups based on properties such as having a basic or branched chain (HKRILV), hydrophobicity (YW and Cystine), and other (FMT). The combination of 3, 6, 9 and 12 amino acids with glucose induced Per1 expression, and that of 9 and 12 amino acids with glucose also stimulated Per2 and Dec2 expression. The combination of 3 and 6 amino acids with glucose induced expression to some degree although the values did not reach statistical significance. Similarly, all combinations might induce Dec1 and suppress Rev-erbα, but without reaching statistical significance. This implies that any amino acid included in the experiment together with glucose might be sufficient to induce clock gene expression.
Our observations provide an important insight into the rapid response of the liver clock to feeding cues. Namely, digestion or nutrient sensing in the intestine is unnecessary, whereas glucose plus amino acids are essential. Our results suggest that glucose and amino acids digested from proteins circulating in the blood are critical nutrient cues for liver clock. Rapid resetting of the liver clock induced by feeding is associated with the acute induction of Per2 and Dec1 transcription in rats [12]. Our results are consistent with Per2 and Dec1 induction and we also found changes in Per1, Dec2, and Rev-erbα in the liver after short-term feeding and after injecting nutrients. Multiple clock genes are coincidentally induced during rapid resetting of the mammalian liver clock, indicating that this clock is closely associated with energy metabolism. In fact, a deficiency in liver clock function causes low blood glucose levels in fasting phase [39]. On the other hand, nutritional signals are not as critical for the lungs to induce the acute response because the lung clock was not affected by delayed feeding, short-term resumed feeding, and injected nutrients except for Per1 induction by Glc+12AA. Body temperature also works as a universal resetting cue for peripheral clocks [40], and thus the lung clock might be gradually reset by a body temperature rhythm that depends on feeding schedules.
Peripheral clocks dominate not only local physiology but also energy metabolism and hormonal secretion at the whole body level [39], [41]. Because nutritional cues are critical for peripheral clock entrainment, biological rhythms disrupted by irregular feeding might lead to various physical and mental disorders [1], [7]. Understanding the mechanisms underlying the entrainment of peripheral clocks with food is linked to the prevention of many lifestyle-related diseases and to improving the quality of life.
Supporting Information
Representative data of temporal food consumption immediately before sampling in experiment shown Figure 1 . Bars show amount of food consumed during 15 min. Y axis shows 0.5 g. Total onsumption is shown at right of graph. Another independent assay showed total consumption of 2.3 g (Control), 2.7 g (Delayed 4 h), 2.5 g (Delayed 8 h), and 1.5 g (Delayed 13 h).
(TIF)
Dbp and Bmal1 expression in liver 1 h after resumed feeding. Expression of mRNA for Dbp in liver under the condition in shown Figure 3. Means ± SEM (n = 8). There was no significant difference in both genes.
(TIF)
Intraperitoneal injection of nutrients delays phase of liver, but not lung clock. (A) Experimental design. BALB/c mice fasted overnight and then were intraperitoneally injected with nutrients (12AA+Glc) or PBS (control) at ZT1. All animals were sampled at next day ZT2, 5, 8, and 11. (B and C) Temporal expression of Rev-erbα and Dbp mRNA in liver (B) and lung (C) is shown as means ± SEM (n = 3, *p<0.05, **p<0.01 Student's t-test).
(TIF)
Gene expression data in DNA microarray analysis.
(PDF)
Acknowledgments
We thank Yokogawa Electric Co. for the loan of the DNA microarray reader and technical advice.
Footnotes
Competing Interests: The authors, including those employed by Mitsubishi Rayon Company Ltd, have declared that no competing interests exist.
Funding: This study was supported by KAKENHI [21700778; Grant-in-Aid for Young Scientists (B)] to HO. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hastings M, Reddy A, Maywood E. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 2.Reppert S, Weaver D. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 3.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 4.Dardente H, Cermakian N. Molecular circadian rhythms in central and peripheral clocks in mammals. Chronobiol Int. 2007;24:195–213. doi: 10.1080/07420520701283693. [DOI] [PubMed] [Google Scholar]
- 5.Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, et al. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 6.Mendoza J. Circadian clocks: setting time by food. J Neuroendocrinol. 2007;19:127–137. doi: 10.1111/j.1365-2826.2006.01510.x. [DOI] [PubMed] [Google Scholar]
- 7.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stokkan K, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 10.Wakamatsu H, Yoshinobu Y, Aida R, Moriya T, Akiyama M, et al. Restricted-feeding-induced anticipatory activity rhythm is associated with a phase-shift of the expression of mPer1 and mPer2 mRNA in the cerebral cortex and hippocampus but not in the suprachiasmatic nucleus of mice. Eur J Neurosci. 2001;13:1190–1196. doi: 10.1046/j.0953-816x.2001.01483.x. [DOI] [PubMed] [Google Scholar]
- 11.Hirao A, Tahara Y, Kimura I, Shibata S. A balanced diet is necessary for proper entrainment signals of the mouse liver clock. PLoS One. 2009;4:e6909. doi: 10.1371/journal.pone.0006909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu T, Ni Y, Kato H, Fu Z. Feeding-induced rapid resetting of the hepatic circadian clock is associated with acute induction of Per2 and Dec1 transcription in rats. Chronobiol Int. 2010;27:1–18. doi: 10.3109/07420520903398625. [DOI] [PubMed] [Google Scholar]
- 13.Hirota T, Okano T, Kokame K, Shirotani-Ikejima H, Miyata T, et al. Glucose down-regulates Per1 and Per2 mRNA levels and induces circadian gene expression in cultured Rat-1 fibroblasts. J Biol Chem. 2002;277:44244–44251. doi: 10.1074/jbc.M206233200. [DOI] [PubMed] [Google Scholar]
- 14.Stephan FK, Davidson AJ. Glucose, but not fat, phase shifts the feeding-entrained circadian clock. Physiol Behav. 1998;65:277–288. doi: 10.1016/s0031-9384(98)00166-8. [DOI] [PubMed] [Google Scholar]
- 15.Saito M, Nishimura K, Kato H. Modifications of circadian cortisol rhythm by cyclic and continuous total enteral nutrition. J Nutr Sci Vitaminol (Tokyo) 1989;35:639–647. doi: 10.3177/jnsv.35.639. [DOI] [PubMed] [Google Scholar]
- 16.Saito M, Kato H, Suda M, Yugari Y. Parenteral feeding abolishes the circadian adrenocortical rhythm in rats. Experientia. 1981;37:754–755. doi: 10.1007/BF01967962. [DOI] [PubMed] [Google Scholar]
- 17.Kato H, Saito M, Shimazu T. Attenuated blood corticosterone rhythm in rats with jejunal resection. Life Sci. 1984;34:331–335. doi: 10.1016/0024-3205(84)90620-9. [DOI] [PubMed] [Google Scholar]
- 18.Miki H, Yano M, Iwanaga H, Tsujinaka T, Nakayama M, et al. Total parenteral nutrition entrains the central and peripheral circadian clocks. Neuroreport. 2003;14:1457–1461. doi: 10.1097/00001756-200308060-00010. [DOI] [PubMed] [Google Scholar]
- 19.Sei H, Oishi K, Morita Y, Ishida N. Mouse model for morningness/eveningness. Neuroreport. 2001;12:1461–1464. doi: 10.1097/00001756-200105250-00033. [DOI] [PubMed] [Google Scholar]
- 20.Yoo S, Yamazaki S, Lowrey P, Shimomura K, Ko C, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamazaki S, Takahashi JS. Real-time luminescence reporting of circadian gene expression in mammals. Methods Enzymol. 2005;393:288–301. doi: 10.1016/S0076-6879(05)93012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oike H, Kobori M, Suzuki T, Ishida N. Caffeine lengthens circadian rhythms in mice. Biochem Biophys Res Commun. 2011;410:654–658. doi: 10.1016/j.bbrc.2011.06.049. [DOI] [PubMed] [Google Scholar]
- 23.Pezuk P, Mohawk JA, Yoshikawa T, Sellix MT, Menaker M. Circadian organization is governed by extra-SCN pacemakers. J Biol Rhythms. 2010;25:432–441. doi: 10.1177/0748730410385204. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura TJ, Sellix MT, Kudo T, Nakao N, Yoshimura T, et al. Influence of the estrous cycle on clock gene expression in reproductive tissues: effects of fluctuating ovarian steroid hormone levels. Steroids. 2010;75:203–212. doi: 10.1016/j.steroids.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abe M, Herzog ED, Yamazaki S, Straume M, Tei H, et al. Circadian rhythms in isolated brain regions. J Neurosci. 2002;22:350–356. doi: 10.1523/JNEUROSCI.22-01-00350.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobori M, Nakayama H, Fukushima K, Ohnishi-Kameyama M, Ono H, et al. Bitter gourd suppresses lipopolysaccharide-induced inflammatory responses. J Agric Food Chem. 2008;56:4004–4011. doi: 10.1021/jf800052y. [DOI] [PubMed] [Google Scholar]
- 27.Oike H, Nagai K, Fukushima T, Ishida N, Kobori M. High-salt diet advances molecular circadian rhythms in mouse peripheral tissues. Biochem Biophys Res Commun. 2010;402:7–13. doi: 10.1016/j.bbrc.2010.09.072. [DOI] [PubMed] [Google Scholar]
- 28.Hohjoh H, Fukushima T. Expression profile analysis of microRNA (miRNA) in mouse central nervous system using a new miRNA detection system that examines hybridization signals at every step of washing. Gene. 2007;391:39–44. doi: 10.1016/j.gene.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 29.Oishi K, Miyazaki K, Ishida N. Functional CLOCK is not involved in the entrainment of peripheral clocks to the restricted feeding: entrainable expression of mPer2 and BMAL1 mRNAs in the heart of Clock mutant mice on Jcl:ICR background. Biochem Biophys Res Commun. 2002;298:198–202. doi: 10.1016/s0006-291x(02)02427-0. [DOI] [PubMed] [Google Scholar]
- 30.Pitts S, Perone E, Silver R. Food-entrained circadian rhythms are sustained in arrhythmic Clk/Clk mutant mice. Am J Physiol Regul Integr Comp Physiol. 2003;285:R57–67. doi: 10.1152/ajpregu.00023.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 32.Yang X, Downes M, Yu RT, Bookout AL, He W, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 33.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 34.Hara R, Wan K, Wakamatsu H, Aida R, Moriya T, et al. Restricted feeding entrains liver clock without participation of the suprachiasmatic nucleus. Genes Cells. 2001;6:269–278. doi: 10.1046/j.1365-2443.2001.00419.x. [DOI] [PubMed] [Google Scholar]
- 35.Hanson RW, Reshef L. Regulation of phosphoenolpyruvate carboxykinase (GTP) gene expression. Annu Rev Biochem. 1997;66:581–611. doi: 10.1146/annurev.biochem.66.1.581. [DOI] [PubMed] [Google Scholar]
- 36.Granner D, Andreone T, Sasaki K, Beale E. Inhibition of transcription of the phosphoenolpyruvate carboxykinase gene by insulin. Nature. 1983;305:549–551. doi: 10.1038/305549a0. [DOI] [PubMed] [Google Scholar]
- 37.Iynedjian PB, Pilot PR, Nouspikel T, Milburn JL, Quaade C, et al. Differential expression and regulation of the glucokinase gene in liver and islets of Langerhans. Proc Natl Acad Sci U S A. 1989;86:7838–7842. doi: 10.1073/pnas.86.20.7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andrikopoulos S, Blair AR, Deluca N, Fam BC, Proietto J. Evaluating the glucose tolerance test in mice. Am J Physiol Endocrinol Metab. 2008;295:E1323–1332. doi: 10.1152/ajpendo.90617.2008. [DOI] [PubMed] [Google Scholar]
- 39.Lamia K, Storch K, Weitz C. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buhr ED, Yoo SH, Takahashi JS. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330:379–385. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Son GH, Chung S, Choe HK, Kim HD, Baik SM, et al. Adrenal peripheral clock controls the autonomous circadian rhythm of glucocorticoid by causing rhythmic steroid production. Proc Natl Acad Sci U S A. 2008;105:20970–20975. doi: 10.1073/pnas.0806962106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative data of temporal food consumption immediately before sampling in experiment shown Figure 1 . Bars show amount of food consumed during 15 min. Y axis shows 0.5 g. Total onsumption is shown at right of graph. Another independent assay showed total consumption of 2.3 g (Control), 2.7 g (Delayed 4 h), 2.5 g (Delayed 8 h), and 1.5 g (Delayed 13 h).
(TIF)
Dbp and Bmal1 expression in liver 1 h after resumed feeding. Expression of mRNA for Dbp in liver under the condition in shown Figure 3. Means ± SEM (n = 8). There was no significant difference in both genes.
(TIF)
Intraperitoneal injection of nutrients delays phase of liver, but not lung clock. (A) Experimental design. BALB/c mice fasted overnight and then were intraperitoneally injected with nutrients (12AA+Glc) or PBS (control) at ZT1. All animals were sampled at next day ZT2, 5, 8, and 11. (B and C) Temporal expression of Rev-erbα and Dbp mRNA in liver (B) and lung (C) is shown as means ± SEM (n = 3, *p<0.05, **p<0.01 Student's t-test).
(TIF)
Gene expression data in DNA microarray analysis.
(PDF)