Abstract
Nine nicotinic receptor subunits are expressed in the central nervous system indicating that a variety of nicotinic acetylcholine receptors (nAChR) may be assembled. A useful method with which to identify putative nAChR is radioligand binding. In the current study the binding of [125I]α-bungarotoxin, [125I]α-conotoxinMII, 5[125I]-3-((2S)-azetidinylmethoxy)pyridine (A-85380), and [125I]epibatidine has been measured autoradiographically to provide data on many nAChR binding sites. Each binding sites was evaluated semiquantitatively for samples prepared from wild-type and α2, α4, α6, α7, β2, β4, α5 and β3 null mutant mice. Deletion of the α7 subunit completely and selectively eliminated [125I]α-bungarotoxin binding. The binding of [125I]αConotoxinMII was eliminated in most brain regions by deletion of either the α6 or β2 subunit and is reduced by deletion of either the α4 or β3 subunit. The binding of 5[125I]A-85380 was completely eliminated by deletion of the β2 subunit and significantly reduced by deletion of the α4 subunit. Most, but not all, α4-independent sites require expression of the α6 subunit. The effect of gene deletion on total [125I]epibatidine binding was very similar to that on [125I]A-85380 binding. [125I]Epibatidine also labels β4* nAChR, which was readily apparent for incubations conducted in the presence of 100 nM cytisine. The effects of α3 gene deletion could not be evaluated, but persistence of residual sites implies the expression of α3* nAChR. Taken together these results confirm and extend previously published evaluations of the effect of nAChR gene deletion and help to define the nAChR subtypes measurable by ligand binding.
Keywords: nicotinic acetylcholine receptor, null mutant mice, epibatidine, A-85380, α-conotoxin MII, α-bungarotoxin
1. Introduction
1.1. Historical Ligands
Nicotinic acetylcholine receptors (nAChRs) in brain have been studied using ligand binding assays for many years. Early studies of [125I]-α-bungarotoxin (αBgt) [1] and [3H]-nicotine [2] binding to rat brain membrane provided some of the first evidence that nAChRs might be expressed in brain. Subsequently. the demonstration that αBgt and nicotine binding sites had different anatomical distributions [3] and biochemical properties [4] provided the first evidence that more than one putative nAChR subtype is expressed in brain. When the nine nAChR subunit genes expressed in mammalian brain (α2–α7, β2–β4) were cloned and sequenced nearly 20 years ago, the number of potential subtypes expanded dramatically [5–6]. Much of the recent research has attempted to identify the subunit compositions of those nAChR subtypes that are actually expressed (i.e. native receptors) in brain as well as other tissues [7].
1.2. Epibatidine
Although [125I]αBgt and [3H]nicotine (as well as [3H]acetylcholine [8], [3H]cytisine [9] and [3H]methylcarbachol [10]) have been useful in identifying and characterizing putative nAChR binding sites in brain, these ligands do not label a wide array of binding sites. This deficiency was overcome with the discovery and characterization of epibatidine [11]. Epibatidine binds with extraordinarily high affinity to rat brain membranes [11–13]. Saturation binding and differential inhibition studies in rat brain [13–14] and inhibition experiments in mouse brain [15–16] demonstrated that epibatidine binds to multiple nAChR subtypes. Furthermore, epibatidine potently activates α3β2, α3β4, α4β2, α7, and α8 nAChRs expressed in Xenopus laevis oocytes [17]. Epibatidine also binds with very high affinity to heteromeric receptors expressed in Xenopus oocytes [18] and HEK cells [19].
1.3. αConotoxinMII
The ability to measure other nAChR subtypes was expanded by the discovery and initial characterization of αConotoxinMII (αCtxMII) [20] and the demonstration that this ligand labels an unique population of nAChR binding sites in mouse brain that is concentrated in catecholaminergic cells and their terminals and in visual pathways [21–22]. This distribution represents a subset of epibatidine binding sites that can also be visualized as a subset of the sites observed when epibatidine binding is conducted in the presence of a low concentration of cytisine (50 nM) [22].
1.4. A-85380
As part of the nicotinic research program at Abbott Laboratories a potent, high-affinity ligand 3-((2S)-azetidinylmethoxy)pyridine (A-85380) has been developed [23]. Radiolabeled 5-125I-A-85380 labels β2* nAChR selectively [24] and its analogs are extremely useful ligands for positron emission tomography [25–26].
1.5. Knockout Mice
Nicotinic receptor knockout mice have proven to be valuable tools to identify native nAChR expression and function [27–29]. Knockout mice have used to define populations of ligand sites that identify natively expressed nAChR subtypes [22, 30–48]. The results described in this study have extended the information by examining the effect of nAChR subunit gene deletion on the binding of [125I]epibatidine, [125I]A-85380, [125I]αCtxMII and [125I]αBgt in order to provide a comprehensive overview of the expression of native nAChR in mouse brain. Although some compensation is likely to occur following deletion of a nAChR subunit, current evidence indicates that no nAChR subtypes that are not normally present are expressed following deletion of major nAChR subunits such as α3 [49], α4 [30, 46, 50], α6 [22, 51–53], α7 [33, 44], β2 [32, 39, 42, 44–45, 54] and β4 [44, 54]. However, deletion of the auxiliary subunits α5 [51, 55–56] or β3 [51, 55] appears to change the relative ratio of nAChR that mediated synaptosomal dopamine release with differential sensitivity to inhibition by αCtxMII.
2. Materials and Methods
2.1. Materials
The radioligands [125I]epibatidine (specific activity 2200 Ci/mmol), 5[125I]-A-85380 (2200 Ci/mmol) and [125I]α bungatotoxin (αBgt) (250 mCi/mmol) and Kodak MR film were obtained from Perkin-Elmer New England Nuclear, Shelton, CT. αConotoxinMII (αCtxMII) and [125I]αCtx MII (2200 Ci/mmol) were prepared as described previously ([20–21], respectively) were obtained from J. Michael McIntosh, University of Utah, Salt Lake City, UT. Unlabeled I-epibatidine was a gift from Kenneth Kellar, Georgetown University, Washington, DC. Unlabeled 5I-A-85380 was purchased from Tocris Bioscience, Ellisville, MO. HEPES (free acid and Na salt) are products of BDH and were purchased from VWR, Chester, PA. The following chemicals were purchased from Sigma Chemical Co., St. Louis, MO: 2-methylbutane, NaCl, KCl, CaCl2, MgSO4, bovine serum albumin, leupeptin, pepstatin, aprotinin, EDTA, EGTA, and phenylmethylsulfonyl fluoride (PMSF). M-1 Embedding Matrix was purchased from Anatomical Pathology USA, Pittsburgh, PA. Superfrost Plus Microscope Slides were obtained from Fisher Scientific, Fair Lawn, NJ.
2.2. Mice
All procedures involving mice were reviewed and approved by the Animal Care and Utilization Committee of the University of Colorado, Boulder. Mice were bred in the Specific Pathogen Free Colony at the Institute for Behavioral Genetics, University of Colorado, Boulder weaned at 25 days of age and housed with like-sexed littermates. Animals were maintained on a 12 hr light/12 hr dark cycle (lights on 7 AM-7 PM) and allowed free access to food and water. The following nicotinic knockout mice were used in this study: α2 [57]; α4 [50]; α6 [22]; α7 [33]; β2 [32]; β4 [54]; α5 [36] and β3 [35]. Mice differing in nAChR genotype were derived by mating heterozygotes that had been backcrossed to C57BL/6J for at least 10 generations. Tail clippings were obtained from mice about 40 days of age and genotype determined as described previously [51].
2.3. Autoradiography
Mice were sacrificed by cervical dislocation, the brains rapidly removed and quickly frozen by immersion in isopentane (−35°C). Brains were stored at −70°C until sectioning. Brains were sectioned (14 micron thickness) using an IEC or Leica cryostat. Sections were thaw mounted on Fisher Superfrost Plus microscope slides. Slides containing the sections were stored at −70°C until use. For all binding conditions samples were removed from the −70°C freezer and warmed to room temperature under vacuum in a desiccator. After warming the slides were distributed to plastic holders modified to contain 50 slides.
The binding of 6-[125I] epibatidine was conducted as follows [58]. Samples were incubated in binding buffer (NaCl, 140 mM; KCl, 1.5 mM; CaCl2, 2 mM; MgSO4, 1 mM; bovine serum albumin, 1 g/L; HEPES buffer, pH=7.5, 25 mM) containing 500 nM [125I]epibatidine with a final specific activity of 110 Ci/mmol (attained by diluting the commercial [125I]epibatidine (2200 Ci/mmol) with unlabeled 6I-epibatidine) for 2 hr at 22°. Three different incubation conditions were used. The first condition measured total [125I]epibatidine binding and contained no further additions to the buffer. The second condition included 100 nM cytisine. The third condition included 100 nM cytisine and 100 nM α-conotoxin MII. Following the incubation, the slides were washed by immersion in ice-cold protein free binding buffer (2x 30 sec), ice cold 0.1 × protein-free binding buffer (2× 10 sec) and ice-cold 5 mM HEPES, pH 7.5 (2x 5 sec each). Samples were then dried under a gentle stream of air and desiccated overnight before exposure to Kodak MR film for 17 days.
The binding of [125I]A-85380 was conducted as follows [59]. Samples were incubated in binding buffer containing 200 pM [125I]A-85380 with a final specific activity of 110 Ci/mmol (attained by diluting the commercial [125I]A-85380 (2200 Ci/mmol with unlabeled 5I-A-85380) for 2 hr at 22°. Following the incubation, the slides were washed by immersion in ice-cold protein free binding buffer (2x 30 sec), ice cold 0.1 × protein-free binding buffer (2× 10 sec) and ice-cold 5 mM HEPES, pH 7.5 (2×5 sec each). Samples were then dried under a gentle stream of air and desiccated overnight before exposure to Kodak MR film for 17 days.
The binding of [125I]α-CtxMII was conducted as follows [21]. Samples were incubated for 10 min in binding buffer without BSA but including 1 mM PMSF for 10 min. Samples were then incubated with 0.5 nM [125I]αCtxMII in binding buffer that also contained 5 mM EDTA and 5 mM EGTA as well as leupeptin, pepstatin and aprotinin (each 10 μg/ml) for 2 hr at 22°. Following the incubation, samples were incubated twice for 15 min at 22° in binding buffer. Subsequently, the slides were washed by immersion in ice-cold protein free binding buffer (2×30 sec), ice cold 0.1 × protein-free binding buffer (2×10 sec) and ice-cold 5 mM HEPES, pH 7.5 (2×5 sec each). Samples were then dried under a gentle stream of air and desiccated overnight before exposure to Kodak MR film for 5 days.
The binding of [125I]αBgt was conducted as follows [60]. Samples were incubated with 0.25 nM [125I]αBgt in binding buffer for 2 hr at 22°. Following the incubation, samples were incubated twice for 30 min at 22°in binding buffer. Subsequently, the slides were washed by immersion in ice-cold protein free binding buffer (2×30 sec), ice cold 0.1 × protein-free binding buffer (2x 10 sec) and ice-cold 5 mM HEPES, pH 7.5 (2×5 sec each). Samples were then dried under a gentle stream of air and desiccated overnight before exposure to Kodak MR film for 5 days.
2.4. Photography
Films were placed on a Northern Light Precision Illuminator set to full intensity and images were captured using a Nikon D40 camera equipped with a 60 mm microimaging extension lens. Ambient light was kept constant. Subsequently images were transferred to Photoshop for further processing.
2.5 Estimate of Signal Intensity
Visual assessment was used to obtain semiquantitative estimates of signal intensity. Relative binding among regions was determined and graded as follows: ■■■■, highest labeling; ■■■, substantial labeling; ■■, modest labeling; ■, weak labeling; □, no significant labeling. The effect of gene deletion for each brain region relative to that of wild-type mice was graded as follows: ++++, 75–100% of maximum (virtually no effect); +++, 50–75%; ++, 25–50%; +, <25%; and −, no detectable signal remaining.
3. Results
3.1. Autoradiographic Illustration of the Effect of nAChR Gene Deletion on Ligand Binding
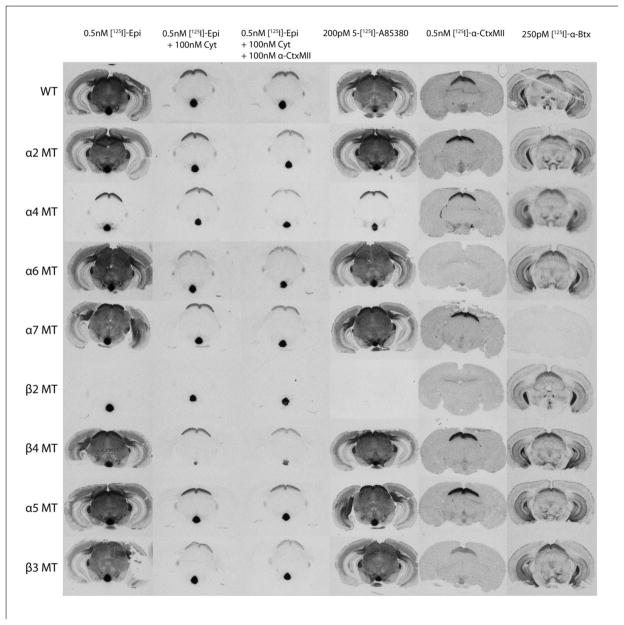
The autoradiograms in Figure 1 illustrate pictorially the effect of deletion of α2, α4, α6, α7, β2, β4, α5 and β3 on the binding of [125I]epibatidine, [125I]A-85380, [125I]αCtxMII and [125I]αBgt at approximately −3.5 mm Bregma. This figure illustrates several features of the data. The binding of each ligand to tissue from wild-type mice is shown in the top row of the figure. The effect of each nAChR gene deletion is shown for each binding condition below that of the wild-type. Similar figures at different anatomical levels (approximate Bregma: −0.1 mm, −0.6 mm, −2.1 mm, −2.5 mm and −5.2 mm) are found in the Supplemental Figures 1–5. These images can also be accessed and downloaded at ibgwww.colorado.edu (under the animal research heading). The descriptions that follow have also been applied to the results summarized in Tables 1–6 in the following sections.
Figure 1.
Autoradiographic images of coronal mouse brain sections at approximately −3.5 mm Bregma. Autoradiograms for each of the ligands [125I]epibatidine (three conditions: total binding, binding in the presence of 100 nM cytisine and binding in the presence of 100 nM cytisine and 100 nM α-conotoxinMII), [125I]A-85380, [125I]α-conotoxinMII and [125I]α-bungarotoxin are shown for wild-type mice and for each of the nAChR null mutants for sections at a level approximately −3.5 mm Bregma.
Table 1.
Semi quantitative visual analysis of coronal mouse brain sections at approximately −0.1 mm Bregma..
| 0.5 nM [125I] - Epibatidine | |||||
|---|---|---|---|---|---|
| Cpu | GP | S Cx | M Cx | OT | |
| ■■■ | ■■■ | ■■■ | ■■■ | ■■■ | |
| WT | + + + + | + + + + | + + + + | + + + + | + + + + |
| α2 | + + + + | + + + + | + + + + | + + + + | + + + + |
| α4 | + | − | − | − | − |
| α6 | + + + + | + + + + | + + + + | + + + + | + + + + |
| α7 | + + + + | + + + + | + + + + | + + + + | + + + + |
| β2 | − | − | − | − | − |
| β4 | + + + + | + + + + | + + + + | + + + + | + + + + |
| α5 | + + + + | + + + + | + + + + | + + + + | + + + + |
| β3 | + + + + | + + + + | + + + + | + + + + | + + + + |
|
| |||||
| 0.5 nM [125I] - Epibatidine + 100 nM Cytisine | |||||
| Cpu | GP | S Cx | M Cx | OT | |
| ■ | ■ | □ | □ | □ | |
| WT | + + + + | + + + + | n/a | n/a | n/a |
| α2 | + + + + | + + + + | n/a | n/a | n/a |
| α4 | − | − | n/a | n/a | n/a |
| α6 | + + + + | + + + + | n/a | n/a | n/a |
| α7 | + + + + | + + + + | n/a | n/a | n/a |
| β2 | − | − | n/a | n/a | n/a |
| β4 | + + + + | + + + + | n/a | n/a | n/a |
| α5 | + + + + | + + + + | n/a | n/a | n/a |
| β3 | + + + + | + + + + | n/a | n/a | n/a |
|
| |||||
| 0.5 nM [125I] - Epibatidine + 100 nM Cytisine + 100 nM α-CtxMII | |||||
| Cpu | GP | S Cx | M Cx | OT | |
| ■ | ■ | □ | □ | □ | |
| WT | + + + + | + + + + | n/a | n/a | n/a |
| α2 | + + + + | + + + + | n/a | n/a | n/a |
| α4 | − | − | n/a | n/a | n/a |
| α6 | + + + + | + + + + | n/a | n/a | n/a |
| α7 | + + + + | + + + + | n/a | n/a | n/a |
| β2 | − | − | n/a | n/a | n/a |
| β4 | + + + + | + + + + | n/a | n/a | n/a |
| α5 | + + + + | + + + + | n/a | n/a | n/a |
| β3 | + + + + | + + + + | n/a | n/a | n/a |
|
| |||||
| 200 pM 5-[125I]A-85380 | |||||
| Cpu | GP | S Cx | M Cx | OT | |
| ■■■ | ■■■ | ■■■ | ■■■ | ■■■ | |
| WT | + + + + | + + + + | + + + + | + + + + | + + + + |
| α2 | + + + + | + + + + | + + + + | + + + + | + + + + |
| α4 | + | + | − | − | − |
| α6 | + + + + | + + + + | + + + + | + + + + | + + + + |
| α7 | + + + + | + + + + | + + + + | + + + + | + + + + |
| β2 | − | − | − | − | − |
| β4 | + + + + | + + + + | + + + + | + + + + | + + + + |
| α5 | + + + + | + + + + | + + + + | + + + + | + + + + |
| β3 | + + + + | + + + + | + + + + | + + + + | + + + + |
|
| |||||
| 0.5 nM [125I] α-ConotoxinMII | |||||
| Cpu | GP | S Cx | M Cx | OT | |
| WT | ■■■ | □ | □ | □ | □ |
| α2 | + + + + | n/a | n/a | n/a | n/a |
| α4 | + + + | n/a | n/a | n/a | n/a |
| α6 | − | n/a | n/a | n/a | n/a |
| α7 | + + + + | n/a | n/a | n/a | n/a |
| β2 | − | n/a | n/a | n/a | n/a |
| β4 | + + + + | n/a | n/a | n/a | n/a |
| α5 | + + + + | n/a | n/a | n/a | n/a |
| β3 | + | n/a | n/a | n/a | n/a |
|
| |||||
| 250 pM [125I] α-Bungarotoxin | |||||
| Cpu | GP | S Cx | M Cx | OT | |
| ■■■ | ■■ | ■■ | ■■ | ■■ | |
| WT | + + + + | + + + + | + + + + | + + + + | + + + + |
| α2 | + + + + | + + + + | + + + + | + + + + | + + + + |
| α4 | + + + + | + + + + | + + + + | + + + + | + + + + |
| α6 | + + + + | + + + + | + + + + | + + + + | + + + + |
| α7 | − | − | − | − | − |
| β2 | + + + + | + + + + | + + + + | + + + + | + + + + |
| β4 | + + + + | + + + + | + + + + | + + + + | + + + + |
| α5 | + + + + | + + + + | + + + + | + + + + | + + + + |
| β3 | + + + + | + + + + | + + + + | + + + + | + + + + |
Autoradiography was performed as described in section 2.3. At bregma = −0.1mm, the following brain regions are visible. Caudate Putamen, CPu; Globus Pallidus, GP; Somatosensory Cortex, S Cx; Motor Cortex, M Cx; Olfactory Tubercules, OT. Regions with no detectable signal above background are designated with □ and relative quantitation was not attempted (n/a).
Table 6.
Semi quantitative visual analysis of coronal mouse brain sections at approximately −5.2 mm Bregma.
| 0.5 nM [125I] - Epibatidine | |||||||
|---|---|---|---|---|---|---|---|
| CIC | DCIC | ECIC | PAG | DT | PN | CEnt | |
| WT | ■■ | ■■■ | ■■ | ■■■ | ■■■■ | ■■■ | ■■■ |
| α2 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α4 | + + + + | + | + | + + | + + + | − | − |
| α6 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α7 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β2 | + | + + + + | − | − | + | − | − |
| β4 | + + | + + | + + | + + + | + + + + | + + + | + + + |
| α5 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β3 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
|
| |||||||
| 0.5 nM [125I] - Epibatidine + 100 nM Cytisine | |||||||
| CIC | DCIC | ECIC | PAG | DT | PN | CEnt | |
| WT | ■ | ■■■ | □ | ■ | □ | □ | □ |
| α2 | + + + + | + + + + | n/a | + + + + | n/a | n/a | n/a |
| α4 | + + | + + + + | n/a | + + + + | n/a | n/a | n/a |
| α6 | − | − | n/a | + + + + | n/a | n/a | n/a |
| α7 | + + + + | + + + + | n/a | + + + + | n/a | n/a | n/a |
| β2 | + + + + | + + + + | n/a | + + + + | n/a | n/a | n/a |
| β4 | − | − | n/a | − | n/a | n/a | n/a |
| α5 | + + + + | + + + + | n/a | + + + + | n/a | n/a | n/a |
| β3 | + + + + | + + + + | n/a | + + + + | n/a | n/a | n/a |
|
| |||||||
| 0.5 nM [125I] - Epibatidine + 100 nM Cytisine + 100 nM α-CtxMII | |||||||
| CIC | DCIC | ECIC | PAG | DT | PN | CEnt | |
| WT | ■ | ■■■ | □ | ■ | □ | □ | □ |
| α2 | + + + + | + + + + | n/a | + + + + | n/a | n/a | n/a |
| α4 | + + | + + + + | n/a | + + + + | n/a | n/a | n/a |
| α6 | − | − | n/a | + + + + | n/a | n/a | n/a |
| α7 | + + + + | + + + + | n/a | + + + + | n/a | n/a | n/a |
| β2 | + + + + | + + + + | n/a | + + + + | n/a | n/a | n/a |
| β4 | − | − | n/a | − | n/a | n/a | n/a |
| α5 | + + + + | + + + + | n/a | + + + + | n/a | n/a | n/a |
| β3 | + + + + | + + + + | n/a | + + + + | n/a | n/a | n/a |
|
| |||||||
| 200 pM 5-[125I]-A85380 | |||||||
| CIC | DCIC | ECIC | PAG | DT | PN | CEnt | |
| WT | ■■ | ■■■ | ■■ | ■■■ | ■■■■ | ■■■ | ■■■ |
| α2 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α4 | + + + + | + | + | + + | + + + | − | − |
| α6 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α7 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β2 | + | + + + + | − | − | + | − | − |
| β4 | + + | + + | + + | + + + | + + + + | + + + | + + + |
| α5 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β3 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
|
| |||||||
| 0.5 nM [125I] α-CtxMII | |||||||
| CIC | DCIC | ECIC | PAG | DT | PN | CEnt | |
| WT | □ | □ | □ | □ | □ | □ | □ |
| α2 | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| α4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| α6 | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| α7 | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| β2 | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| β4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| α5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| β3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
|
| |||||||
| 250 pM [125I] α-Bungarotoxin | |||||||
| CIC | DCIC | ECIC | PAG | DT | PN | CEnt | |
| WT | ■■■■ | ■■■■ | ■■■■ | ■■■■ | ■■■■ | ■■■■ | ■■■■ |
| α2 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α4 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α6 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α7 | − | − | − | − | − | − | − |
| β2 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β4 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α5 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β3 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
Autoradiography was performed as described in section 2.3. At bregma = −5.2mm the following brain regions are visible: CIC - Central nucleus of the Inferior Colliculus; DCIC - Dorsal cortex of the inferior Colliculus; ECIC - External Cortex of the inferior colliculus; DT - Dorsal Tegmental nucleus; PN - Pontine Nucleus; Cent - Caudomedial entorhinal cortex. Regions with no detectable signal above background are designated with □ and relative quantitation was not attempted (n/a).
3.1.1. [125I]αBungarotoxin Binding
The simplest response to nAChR gene deletion is observed for [125I]αBgt binding. Highest binding is observed in superior colliculus and hippocampus with less intense labeling of cortical and midbrain regions. Specific binding is eliminated by deletion of the α7 gene and is virtually unaffected by the other null mutations.
3.1.2. [125I]αConotoxinMII Binding
At this level only the superior colliculus shows intense [125I]αCtxMII binding, although light labeling of the interpeduncular nucleus is also observed. Deletion of α2, α7, β4 or α5 subunits has no detectable effect on these binding sites. In contrast, deletion of the β2 and α6 subunits completely eliminated [125I]αCtxMII binding in superior colliculus. A significant, but not quite complete, reduction was observed in β3 knock-out mice, while a more modest reduction occurred in α4 knock-out mice.
3.1.3. [125I]A-85380 Binding
Significantly more sites are labeled by [125I]A-85380 than by either [125I]αBgt or [125I]αCtxMII. The binding of this ligand is completely eliminated by deletion of the β2 subunit. Deletion of the α4 subunit also eliminates many [125I]A-85380 binding sites. However, significant labeling persists in the superficial gray region of the superior colliculus and in the interpeduncular nucleus. A small amount of binding is also seen in the substantia nigra. In contrast to the widespead effects observed following β2 and α4 gene deletion, the effects of deletion of the α6 or β3 gene are restricted to the superficial gray area of the superior colliculus. No significant reductions were noted for α2, α7, β4 and α5 null mutants.
3.1.4. [125I]Epibatidine Binding
The general pattern of [125I]epibatidine binding is similar to that observed with [125I]A-85380 and the effects of β2, α4, α6 and β3 gene deletion are also similar. However, intense labeling in the interpeduncular nucleus persists in the β2 knock-out mice.
It is known that [125I]epibatidine binding is heterogeneous [15–16, 58] with a subset of the sites resistant to inhibition by cytisine. The effect of the null mutations demonstrates the heterogeneity of the cytisine-resistant [125I]epibatidine binding sites. Deletion of β2 eliminates binding in the superior colliculus and deletion of either α6 or β3 significantly reduces this binding. Deletion of α4 has less effect, while no detectable reductions were observed in α2, α7, β4 or α5 null mutants. However, deletion of β4 significantly reduces labeling in the interpeduncular nucleus, while no significant effects were noted for the other null mutants.
A subset of the cytisine-resistant [125I]epibatidine binding sites can be inhibited by αCtxMII [39]. Deletion of the β4 subunit has the most noticeable effect on those residual sites that are resistant to both cytisine and αCtxMII inhibition in interpeduncular nucleus.
3.2. Semiquantitative Analysis of nAChR Gene Deletion on Nicotinic Binding Sites
All nAChR in the CNS must include either the α7, β2 and/or β4 subunit [31, 44, 61]. The heteromeric nAChR include α subunits and may also include the auxiliary α5 and β3 subunits. The results that follow will discuss the effect of the deletion of each nAChR subunit on the various binding sites. The semiquantitative analyses are summarized in Tables 1–6 for analyses of binding at the levels of approximately −0.1 mm, −0.6 mm, −2.1 mm, −2.5 mm, −3.5 and −5.2 mm Bregma, respectively. The symbols for each ligand in the rows for the wild-type mice (□, ■, ■■, ■■■, and ■■■■) illustrate the expression level from undetectable (□) to most intense (■■■■). The symbols for each null mutant (++++, +++, ++, + and −) illustrate the relative effect of that particular gene deletion from no significant change from control (++++) to complete elimination (−).
3.2.1. α7 nAChR Gene Deletion
Deletion of the α7 nAChR gene gives the simplest pattern: Complete elimination of [125I]αBgt binding in every brain region with no significant effect on the binding of [125I]epibatidine, [125I]A-85380, or [125I]αCtxMII.
3.2.2. β2 nAChR Gene Deletion
Deletion of the β2 nAChR gene has the most effect on the ligand binding sites, although it had no noticeable effect on [125I]αBgt binding. Deletion of the β2 nAChR completely eliminates both [125I]A-85380 and high affinity [125I]αCtxMII in every brain region as well as a significant fraction of high affinity [125I]epibatidine binding. Significant [125I]epibatidine binding persists in the medial habenula, fasiculus retroflexus, interpedunular nucleus and inferior colliculus of β2 knock-out mice.
Inclusion of 100 nM cytisine in the binding assays selectively inhibits primarily α4β2* nAChR [14–15, 46] and facilitates identification of less widely expressed nAChR subtypes. With the exception of the binding in the medial habenula, fasiculus retroflexus, and the interpeducular nucleus and a subset of the signal in the optic tracts and inferior colliculus, deletion of the β2 subunit eliminated the cytisine-resistant [125I]epibatidine binding sites.
3.2.3. β4 nAChR Gene Deletion
Deletion of the β4 nAChR gene had no detectable effect on [125I]αBgt, [125I]αCtxMII or [125I]A-85380 binding. In general, deletion of β4 had little effect on total [125I]epibatidine binding with the exception of partial reductions in the signal intensity in the fasiculus retroflexus, interpeduncular nucleus and the inferior colliculus. The effect of β4 gene deletion were more readily observable when 100 nM cytisine was included in the incubation with [125I]epibatidine. Under these experimental conditions, when binding to α4β2* nAChR sites is removed, robust effects of β4 gene deletion could be observed in medial habenula, fasiculus retroflexis, interpeducular nucleus and inferior colliculus, regions that express high levels of cytisine-resistant [125I]epibatidine binding. In addition, detectable reductions in cytisine-resistant [125I]epibatidine binding were also noted in the dorsolateral, ventrolateral and medial geniculate nuclei, olivary pretectal nucleus, superficial gray level of the superior colliculus and the inferior colliculus.
3.2.4. α2 nAChR Gene Deletion
Little effect of the deletion of the α2 nAChR subunit could be noted for any binding sites. However, modest, relatively subtle reductions may have occurred for cytisine-resistant [125I]epibatidine binding sites in several cortical layers, and some regions of the visual tract including the dorsolateral and ventrolateral geniculate nuclei, the olivary pretectal nucleus, the optic tract and the superficial gray area of the superior colliculus.
3.3.5. α4 nAChR Gene Deletion
Deletion of the α4 nAChR gene had no effect on [125I]αBgt binding. All other binding sites were at least partially reduced in α4 knock-out mice.
Significantly less [125I]A-85380 binding was observed in the brain of α4 knock-out mice than wild-type mice. Indeed, deletion of α4 eliminated [125I]A-85380 binding in many brain regions. However, in contrast to the complete elimination of [125I]A-85380 binding following deletion of the β2 subunit, low, but detectable binding, was noted in the caudate putamen, globus pallidus, optic tracts, dorsolateral, ventrolateral and medial geniculate nuclei, substantia nigra pars compacta and ventral tegmental area. In addition, higher levels of [125I]A-85380 binding persisted in medial habenula, olivary pretectal nucleus, substantia nigra pars reticulata and superficial gray region of the superior colliculus in α4 knockouts. Deletion of α4 had little effect on [125I]A-85380 binding in the fasiculus retroflexus and the interpeduncular nucleus.
Deletion of the α4 nAChR subunit generally reduced, but did not eliminate, [125I]αCtxMII binding sites throughout the brain.
Given the complexity of [125I]epibatidine binding sites, the pattern of response to deletion of α4 on these sites was more complex than that for [125I]A-85380 binding. However, those sites that remained in β2 knock-out mice also remained in α4 knock-outs. Furthermore, the additional sites that differed between β2 and α4 knock-outs for [125I]A85380 binding were also noted for total [125I]epibatidine binding. As was the case for the β2 knock-outs, significant signal remained in the medial habenula, fasiculus retroflexus, interpeduncular nucleus and inferior colliculus of α4 knock-out mice. In addition, as was the case for the [125I]A-85380 and [125I]αCtxMII binding, deletion of α4 reduced, but did not eliminate β2 dependent, cytisine-resistant [125I]epibatidine binding sites particularly in the visual system.
3.3.6. α6 nAChR Gene Deletion
Deletion of the α6 nAChR gene had very specific effects on nAChR binding sites. Virtually all [125I]αCtxMII binding was eliminated by α6 gene deletion with the exception of partial reductions in the labeling in the interpedunular nucleus and the optic tracts. While effects of α6 gene deletion for both [125I]A-85380 and total [125I]epibatidine binding were not generally obvious, partial reductions were noted for the dorsolateral and ventrolateral geniculates, the superficial gray layer of the superior colliculus, substantia nigra pars compacta and ventral tegmental areas. Addition of 100 nM cytisine to the [125I]epibatidine binding assay helped to illustrate the effect of α6 gene deletion in the regions listed above as well as in the caudate putamen, the optic tracts and olivary pretectal nucleus.
3.3.7. α5 nAChR Gene Deletion
Deletion of the α5 nAChR gene had little detectable effect on the expression of any of the ligand binding sites.
3.3.8. β3 nAChR Gene Deletion
The patterns for the effect of deletion of the β3 nAChR subunit were very similar to those following deletion of the α6 nAChR subunit: Significant reductions in high affinity [125I]αCtxMII binding, selective reductions [125I]A-85380 binding and selective reductions in [125I]epibatidine binding that were particularly evident in samples including 100 nM cytisine. However, the reduction in signal following deletion of β3 was less complete than that following deletion of α6.
4. Discussion
4.1. Null Mutants and Nicotinic Binding Sites
The examination of the effects of nAChR gene mutation on various nicotinic binding sites has helped to define the nature of the complex nAChR subtypes measured with these ligands.
4.1.1. [125I]α-Bungarotoxin Binding Sites
The binding of [125I]αBgt shows the simplest response to nAChR gene deletion. Deletion of the α7 gene completely eliminates [125I]αBgt binding. No noticeable effect of deletion of any of the other nAChR genes on [125I]αBgt binding was observed. This result is completely consistent with the original cloning of the α7 gene as a αBgt binding protein [62] as well as a functional receptor inhibited by αBgt [63–64]. The original report describing the α7 knockout mouse confirmed that deletion of α7 eliminated [125I]αBgt binding as well as αBgt-sensitive function [33]. Deletion of α7 had no effect on the other binding sites measured here, although a lower-affinity, αBgt-sensitive epibatidine binding site, which measures the same subtype as [125I]αBgt is eliminated by deletion of the α7 gene [39, 44]. Thus, all [125I]αBgt binding sites in mouse brain are α7*-nAChR.
4.1.2. [125I]A-85380 Binding Sites
A-85380 was originally identified as a potent α4β2*-nAChR agonist [23]. It is now recognized as a more general β2* nAChR agonist [24]. Compounds modified by incorporation of a substitution in the 5 position of the pyridine moiety are extremely useful ligands for positron emission tomography [25–26]. A-85380 also serves as the progenitor for the potent agonist, sazitidine, that rapidly desensitizes the α4β2 nAChR [65]. Confirming earlier results [24], deletion of the β2 nAChR subunit completely eliminated [125I]A-85380 binding throughout the mouse brain. The results summarized in this paper also illustrate why A-85380 was originally considered to be an α4β2 nAChR selective ligand: Deletion of the α4 nAChR subunit eliminates most [125I]A-85380 binding sites. However, residual [125I]A-85380 binding sites persist in a few distinct brain areas in the α4 knockout mice. The brain regions remaining in α4 knock-outs include the dopaminergic pathways and visual tracts all of which show partial reductions in signal following deletion of either the α6 or β3 genes. In addition, the medial habenula and interpeduncular nucleus express residual [125I]A-85380 binding sites in α4 knock-out mice which are not obviously reduced by deletion of any of the genes investigated here. These are likely to correspond to α3β2* nAChR that will be discussed in more detail below.
4.1.3. [125I]αConotoxinMII Binding Sites
αCtxMII was originally isolated from the cone snail, Conus magus, and characterized as a inhibitor of α3β2 nAChR expressed in Xenopus oocytes [20]. However, with the exception of the medial habenula, fasiculus retroflexus and interpeduncular nucleus the binding of [125I]αCtxMII is unaffected by deletion of the α3 nAChR subunit [38]. The deletion of the β2 nAChR subunit completely eliminates [125I]αCtxMII binding [39, 52, 55] and that result has been confirmed in the current study. In what was initially a surprise, the binding of [125I]αCtxMII in catecholaminergic and visual pathways was eliminated by deletion of the α6 nAChR subunit [22], an observation that has also been confirmed in the present study. In yet another result that was initially a surprise, deletion of the β3 nAChR subunit also substantially reduced [125I]αCtxMII binding [35] in catecholaminergic and visual pathways. This result has been confirmed with immunochemical experiments [43] and by the results of the current study. In contrast to the effect of α6 nAChR gene deletion, deletion of the β3 nAChR subunit did not completely eliminate [125I]αCtxMII binding or αCtxMII-senstivie function [43, 53], indicating either that a small population of α6β2* nAChR normally exist or that these receptors are assembled in the absence of the β3 subunit. The complexity of [125I]αCtxMII binding sites is further revealed by the demonstration that the α4 nAChR subunit is included in an α4α6β2β3 nAChR [52–53, 66–67] that is also illustrated in the current paper by the partial reduction of [125I]αCtxMII binding in α4 knockout mice. Thus, [125I]αCtxMII binding sites represent a complex mixture of nAChR subtypes. A relatively small proportion of the [125I]αCtxMII binding sites, confined primarily to the medial habenula/interpeduncular nucleus pathway, are α3β2* nAChR. Most of the [125I]αCtxMII binding sites are α6β2* nAChR including α4β6β2β3 nAChR, α6β2β3 nAChR and, perhaps a small native population of α6β2 nAChR, the relative proportions of which vary among dopaminergic terminal regions [67].
4.1.4. [125I]Epibatidine Binding Sites
By far the most complex collection of nAChR binding sites are those that bind epibatidine. Epibatidine binds with very high affinity to heteromeric nAChR (α2, α3 and α4 assembled with β2 or β4 subunits) expressed in Xenopus oocytes [19] and HEK cells [18]. Epibatidine has also been used to label receptors analyzed by immunoprecipitation [40–41, 43, 47–48, 67–76]. It is an excellent ligand to identify a diverse number of receptor subtypes using this approach. Pharmacological differences among diverse epibatidine binding sites have been used to identify subsets of these sites [14–16]. The combination of the pharmacological approaches with the use of null mutant mice has been used to investigate the diversity of natively expressed receptors analogous to the experiments described here [16, 39, 42, 44, 46, 77–78].
Most [125I]epibatidine binding sites respond to nAChR gene deletion in a manner very similar to, if not quite identical to, that observed for [125I]A-85380: Deletion of either the β2 or the α4 nAChR subunits eliminates most high-affinity [125I]epibatidine binding. Inasmuch as the cytisine-sensitive [125I]epibatidine binding sites are virtually identical to the sites measured by either [3H]nicotine (or [3H]cytisine), the loss of cytisine-sensitive [125I]epibatidine binding by deletion of either the α4 or β2 subunits corresponds to the elimination of [3H]nicotine binding following deletion of these subunits [30, 32].
Inhibition of high affinity [125I]epibatidine binding sites by cytisine is a useful experimental approach to separate subsets of cytisine-sensitive and cytisine-resistant [125I]epibatidine binding sites [14–15, 39, 45–46, 78–80]. Most epibatidine binding sites in brain are sensitive to inhibition by cytisine and represent primarily, if not exclusively, α4β2* nAChR sites [14–15, 46]. Inhibition of the predominant α4β2* nAChR allows detection of effects that may be masked by the large number of cytisine-sensitive sites. The cytisine-resistant epibatidine binding sites represent a diverse population and that is illustrated by the different effects occurring following deletion of the various nAChR genes. Deletion of the α4 reduced the cytisine-resistant [125I]epibatidine binding in several brain regions, particularly those affected by α6 or β3 deletion. Elimination of additional sites following deletion of the β2 subunit demonstrates the existence of β2* nAChR that do not also include the α4 subunit. β2* nAChR that do not include α4 subunits are particularly noticeable in dopaminergic terminal regions and visual tracts. These [125I]epibatidine binding site are substantially reduced by deletion of either the α6 or β3 gene implying the existence of α6β2β3* nAChR in this subset of sites. This assignment parallels that for [125I]αCtxMII binding sites and is consistent with results reported previously for α6 null mutant mice [22]. An effect of β4 gene deletion is also clearly visible for the cytisine-resistant [125I]epibatidine binding sites: A dramatic reduction in signal was also noted in the medial habenula, fasciculus retroflexus, interpeduncular nucleus and inferior colliculus.
Further refinement of the cytisine-resistant [125I]epibatidine binding sites was accomplished by including 100 nM αCtxMII in the incubation. These cytisine-resistant, αCtxMII-sensitive sites detect nAChR that interact with αCtxMII including lower affinity sites that are not measured directly by the binding of [125I]αCtxMII, which are predominantly α6β2β3* nAChR. Indeed, little further inhibition by 100 nM αCtxMII was noted for α6 and β3 knock-out mice in most regions. However, a further reduction in [125I]epibatidine binding was observed in the superior colliculus of α6 and β3 knockout mice with the addition of 100 nM αCtxMII. The existence of additional αCtxMII sensitive sites, likely corresponding to α3β2* nAChR [81], is indicated (see also below).
4.2. Potential Role of Subunits the Deletion of Which Has Little Effect on Ligand Binding
While deletion of most of the nAChR subunits had detectable effects on one or more of the ligand binding sites measured in the current study, deletion of either the α2 or α5 subunit did not have obvious effects.
4.2.1. α3 Subunit
Deletion of the α3 nAChR results in early postnatal death such that null mutants generally do not survive for more than a few weeks [49]. Consequently, α3 knock- out mice were not included in the experiments described here. However, it is evident from the sites that were eliminated by deletion of α2, α4 or α6 nAChR subunits that, by process of elimination, α3* nAChR account for these residual sites. Many of these residual regions including medial habenula, fasiculus retroflexus, interpedunular nucleus and inferior colliculus are found in the cytisine-resistant component of high affinity [125I]epibatidine binding sites, regions that are reduced in β4 knock-out mice. This strongly indicates the presence of α3β4* nAChR in these regions and is consistent with the observations made on the effect of α3 gene deletion in 8 day old α3 knock-out mice [38]. The existence of potential α3β2* nAChR is suggested by the observation that 100 nM αCtxMII decreases [125I]epibatidine binding in the dorso- and ventro-lateral geniculate nuclei and in the superficial gray area of the superior colliculus which is evident particularly in sections from α6 knock-out mice. This observation is reasonable given that αCtxMII is known to interacted with α3β2 nAChR, albeit with lower affinity than with α6β2* nAChR [20, 82]and is consistent with the demonstration that α3* nAChR are detectable in visual pathways [81, 83–84].
4.2.2. α2 Subunit
In situ hybridization studies in either rat [85] or mouse [86] brain indicate that the α2 subunit is sparsely expressed. The experiments describing the binding of [3H]epibatidine to both α2β2 nAChR and α2β4 nAChR expressed in Xenopus oocytes [19] suggests that such sites should be detectable in brain. Indeed, by the use of ligand binding and immunoprecipitation, [125I]epibatidine binding sites including the α2 subunit have been identified in mouse brain [47], however even in regions of highest expression (olfactory bulbs and interpeduncular nucleus) the α2* nAChR are sparsely expressed. There is also evidence that some α2* nAChR are present in visual pathways [81, 83–84]. Given the semi-quantitative nature of these studies relatively small effects of α2 gene deletion on any of the binding sites may have escaped detection. Development of an α2* nAChR selective ligand would facilitate the identification of these receptors.
4.2.3. α5 Subunit
The α5 subunit is an accessory subunit the deletion of which has significant effects on nicotine-mediated responses in vivo [36, 87–88] as well as on nAChR mediated function [48, 51, 89]. Furthermore, immunoprecipitation studies have established that as subset of nAChR measured by epibatidine binding include the α5 subunit [31, 40, 48, 72, 81, 90–91]. Nevertheless, no study, including the current one, has been able to detect a change in the total number epibatidine binding sites [36, 48], despite the significant changes in function, pharmacology and physiology resulting from α5 gene deletion [48, 51, 89]. Among the possible reasons for the failure to detect changes in binding sites include their relative rarity [48, 72, 90] or the possibility that an alternate subunit such as α4 or β2 is now incorporated in nAChR which normally include the α5 subunit thereby preserving measurable binding sites.
4.3. Summary
The results presented here provide a survey of the effects of nAChR gene deletion of nicotinic binding sites measured with [125I]αBgt, [125I]αCtxMII, [125I]A-85380 and [125I]epibatidine under several conditions. Since the experiments were done in one laboratory under standard conditions the effects are easily comparable. The data presented in this paper are completely consistent with the findings of similar experiments with null mutant mice. It should be noted that all comparisons made in the current study are semi-quantitative and therefore provide a general picture of receptor expression and composition. More detailed comparisons will require a quantitative autoradiographic approach with a larger number of replicates than were included here.
Autoradiographic studies have their limitations:
The effects of deletion of genes with relatively low expression (such as the α2 nAChR subunit) are difficult to detect autoradiographically and often require alternative approaches such as immunoprecipitation [47].
Deletion of an auxiliary subunit (such as α5) may have no detectable effect on traditional ligand binding. Consequently its role can escape detection. The lack of a detectable effect on binding could result because the α5* nAChR represent a relatively small fraction of the total heteromeric receptors. However, immunoprecipitation and functional assays have demonstrated an important role for this subunit [48, 89–90], including behavioral consequences [36, 92–93]. The α5 nAChR has been repeatedly shown to have an important role in human tobacco use [92]. Functional assays are essential to define fully the role of nAChR subunits.
Autoradiographic analyses are inadequate to examine effects of subunit deletion, and therefore nAChR composition, when alternate subunits can substitute for the deleted gene. This caveat applies for auxiliary subunits such as α5 as discussed above. It could also apply for alternately assembled nAChR such as an α7* nAChR that includes a β2 subunit [94], which in the absence of β2 could assemble as an α7 nAChR homopentamer.
Autoradiographic analyses also require sufficient signal:noise for detection of binding sites. This requirement can be problematic when ligands with relatively high non-specific binding especially peptide probes such as [125I]αBgt and [125I]αCtxMII. Thus, in ventral tegmental area little [125I]αBgt binding was observed in the current study, consistent with similar observations for mouse [95] or rat [96]. However, functional α7-nAChR have been identified in the ventral tegmental area [94, 97] indicating that the resolution and/or sensitivity achieved with autoradiography is inadequate to identify these binding sites. Indeed, an electron micrograhpic analysis identified α7 nAChR binding sites in the rat ventral tegmental area [98].
Therefore, failure to detect changes in binding site density following gene deletion, particularly for sparsely expressed or auxiliary subunits, does not necessarily establish that nAChR containing this subunits are absent. Additional studies using functional, pharmacological and immunochemical assays, in addition to more quantitative autoradiographic experiments, particularly in concert with null mutant mice, will provide more definitive description of the native nAChR populations to expand on the general picture provided here.
Supplementary Material
Table 2.
Semi quantitative visual analysis of coronal mouse brain sections at approximately −0.6 mm Bregma..
| 0.5 nM [125I] - Epibatidine | ||||||
|---|---|---|---|---|---|---|
| Cpu | GP | Th | MHb | S Cx | M Cx | |
| WT | ■■■ | ■■■ | ■■■ | ■■■■ | ■■■■ | ■■■ |
| α2 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α4 | + | − | − | + + + + | − | − |
| α6 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α7 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β2 | − | − | − | + + + + | − | − |
| β4 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α5 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β3 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
|
| ||||||
| 0.5 nM [125I] - Epibatidine + 100 nM Cytisine | ||||||
| Cpu | GP | Th | MHb | S Cx | M Cx | |
| WT | ■■ | □ | ■■ | ■■■■ | ■ | ■ |
| α2 | + + + | n/a | + + + | + + + + | + + + | + + + |
| α4 | − | n/a | − | + + + + | − | − |
| α6 | − | n/a | + + + + | + + + + | + + + + | + + + + |
| α7 | + + + | n/a | + + + + | + + + + | + + + + | + + + + |
| β2 | − | n/a | − | + + + | − | − |
| β4 | − | n/a | + + + + | + | + + + + | + + + + |
| α5 | + + + | n/a | + + + + | + + + + | + + + + | + + + + |
| β3 | − | n/a | + + + | + + + + | + + + | + + + |
|
| ||||||
| 0.5 nM [125I] - Epibatidine + 100 nM Cytisine + 100 nM α-CtxMII | ||||||
| Cpu | GP | Th | MHb | S Cx | M Cx | |
| WT | ■ | □ | ■■ | ■■■■ | ■ | ■ |
| α2 | + + + | n/a | + + + | + + + + | + + + | + + + |
| α4 | − | n/a | − | + + + + | − | − |
| α6 | − | n/a | + + + + | + + + + | + + + + | + + + + |
| α7 | + + + | n/a | + + + + | + + + + | + + + + | + + + + |
| β2 | − | n/a | − | + + + | − | − |
| β4 | − | n/a | + + + + | + | + + + + | + + + + |
| α5 | + + + | n/a | + + + + | + + + + | + + + + | + + + + |
| β3 | − | n/a | + + + | + + + + | + + + | + + + |
|
| ||||||
| 200 pM 5-[125I]-A85380 | ||||||
| Cpu | GP | Th | MHb | S Cx | M Cx | |
| WT | ■■■ | ■■ | ■■■■ | ■■■■ | ■■■ | ■■■ |
| α2 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α4 | + | − | − | + + | − | − |
| α6 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α7 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β2 | − | − | − | − | − | − |
| β4 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α5 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β3 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
|
| ||||||
| 0.5 nM [125I]α-ConotoxinMII | ||||||
| Cpu | GP | Th | MHb | S Cx | M Cx | |
| WT | ■■■■ | □ | □ | □ | □ | □ |
| α2 | + + + + | n/a | n/a | n/a | n/a | n/a |
| α4 | + | n/a | n/a | n/a | n/a | n/a |
| α6 | − | n/a | n/a | n/a | n/a | n/a |
| α7 | + + + + | n/a | n/a | n/a | n/a | n/a |
| β2 | − | n/a | n/a | n/a | n/a | n/a |
| β4 | + + + + | n/a | n/a | n/a | n/a | n/a |
| α5 | + + + + | n/a | n/a | n/a | n/a | n/a |
| β3 | + | n/a | n/a | n/a | n/a | n/a |
|
| ||||||
| 250 pM [125I] α-Bungarotoxin | ||||||
| Cpu | GP | Th | MHb | S Cx | M Cx | |
| WT | ■■ | ■■ | ■■ | ■■ | ■■ | ■■■ |
| α2 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α4 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α6 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α7 | − | − | − | − | − | − |
| β2 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β4 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α5 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β3 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
Autoradiography was performed as described in section 2.3. At bregma = −0.6 mm the following brain regions are visible: Caudate Putamen, CPu; Globus Pallidus, GP; Thalamus, Th; Medial Habenular, MHb; Somatosensory Cortex, S Cx; Motor Cortex, M Cx. Regions with no detectable signal above background are designated with □ and relative quantitation was not attempted (n/a).
Table 3.
Semi quantitative visual analysis of coronal mouse brain sections at approximately −2.1 mm Bregma..
| 0.5 nM [125I] - Epibatidine | ||||||||
|---|---|---|---|---|---|---|---|---|
| Th | opt | DLG | MHb | fr | HP | Au1 | RSG | |
| WT | ■■■■ | ■■■■ | ■■■■ | ■■■■ | ■■■■ | ■■■ | ■■■ | ■■■ |
| α2 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α4 | − | + + | + + | + + + + | + + + + | − | − | − |
| α6 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α7 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β2 | − | − | − | + + + + | + + + + | − | − | − |
| β4 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α5 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β3 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
|
| ||||||||
| 0.5 nM [125I] - Epibatidine + 100 nM Cytisine | ||||||||
| Th | opt | DLG | MHb | fr | HP | Au1 | RSG | |
| WT | ■ | ■■ | ■■■ | ■■■■ | ■■■■ | □ | □ | □ |
| α2 | + + + + | + + + | + + + + | + + + + | + + + + | n/a | n/a | n/a |
| α4 | + + + + | − | + + + | + + | + + | n/a | n/a | n/a |
| α6 | + + + + | + + + | + + + | + + + + | + + + + | n/a | n/a | n/a |
| α7 | + + + + | + + + | + + + + | + + + + | + + + + | n/a | n/a | n/a |
| β2 | + + + + | − | − | + + + | + + + | n/a | n/a | n/a |
| β4 | + + + + | − | + + + | + | + | n/a | n/a | n/a |
| α5 | + + + + | + + + | + + + | + + + + | + + + + | n/a | n/a | n/a |
| β3 | + + + + | − | + + + | + + + + | + + + + | n/a | n/a | n/a |
| 0.5 nM [125I] - Epibatidine + 100 nM Cytisine + 100 nM α-CtxMII | ||||||||
| Th | opt | DLG | MHb | fr | HP | Au1 | RSG | |
| WT | ■ | ■■ | ■■■ | ■■■■ | ■■■■ | □ | □ | □ |
| α2 | + + + + | + + + | + + + + | + + + + | + + + + | n/a | n/a | n/a |
| α4 | + + + + | − | + + + | + + | + + | n/a | n/a | n/a |
| α6 | + + + + | + + + | + + + | + + + + | + + + + | n/a | n/a | n/a |
| α7 | + + + + | + + + | + + + + | + + + + | + + + + | n/a | n/a | n/a |
| β2 | + + + + | − | − | + + + | + + + | n/a | n/a | n/a |
| β4 | + + + + | − | + + + | + | + | n/a | n/a | n/a |
| α5 | + + + + | + + + | + + + | + + + + | + + + + | n/a | n/a | n/a |
| β3 | + + + + | − | + + + | + + + + | + + + + | n/a | n/a | n/a |
|
| ||||||||
| 200 pM 5-[125I]-A85380 | ||||||||
| Th | opt | DLG | MHb | fr | HP | Au1 | RSG | |
| WT | ■■■■ | ■■■■ | ■■■■ | ■■■■ | ■■■■ | ■■■ | ■■■ | ■■■ |
| α2 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α4 | − | + | + | + + + | + + | − | − | − |
| α6 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α7 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β2 | − | − | − | − | − | − | − | − |
| β4 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α5 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β3 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
|
| ||||||||
| 0.5 nM [125I] α-ConotoxinMII | ||||||||
| Th | opt | DLG | MHb | fr | HP | Au1 | RSG | |
| WT | □ | ■■■■ | ■■ | ■■ | □ | □ | □ | □ |
| α2 | n/a | + + + + | + + + + | + + + + | n/a | n/a | n/a | n/a |
| α4 | n/a | + + | + | + + | n/a | n/a | n/a | n/a |
| α6 | n/a | + + | − | − | n/a | n/a | n/a | n/a |
| α7 | n/a | + + + + | + + + + | + + + + | n/a | n/a | n/a | n/a |
| β2 | n/a | + + | − | + | n/a | n/a | n/a | n/a |
| β4 | n/a | + + + + | + + + + | + + + | n/a | n/a | n/a | n/a |
| α5 | n/a | + + | + + + + | + + + + | n/a | n/a | n/a | n/a |
| β3 | n/a | + + | + | + + | n/a | n/a | n/a | n/a |
|
| ||||||||
| 250 pM [125I] α-Bungarotoxin | ||||||||
| Th | opt | DLG | MHb | fr | HP | Au1 | RSG | |
| WT | ■■■■ | ■■■ | ■■■■ | ■■■ | ■■■■ | ■■■■ | ■■ | ■■ |
| α2 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α4 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α6 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α7 | − | − | − | − | − | − | − | − |
| β2 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β4 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α5 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β3 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
Autoradiography was performed as described in section 2.3. At bregma = −2.1 mm the following brain regions are visible: Thalamus, Th; Optic Tract, opt; Dorsal Lateral Geniculates, DLG; Medial Habenular, MHb; Fasciculus Retroflexus, fr; Hippocampus, HP; Primary Auditory Cortex, Au1; Retrosplenial Granular Cortex, RSG. Regions with no detectable signal above background are designated with □ and relative quantitation was not attempted (n/a).
Table 4.
Semi quantitative visual analysis of coronal mouse brain sections. at approximately −2.5 mm Bregma.
| 0.5 nM [125I] - Epibatidine | ||||||||
|---|---|---|---|---|---|---|---|---|
| DS | VLG | DLG | OPT | fr | HP | V1 | RSG | |
| WT | ■■■■ | ■■■■ | ■■■■ | ■■■■ | ■■■■ | ■■■■ | ■■■■ | ■■■■ |
| α2 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α4 | − | + + | + + | + + + + | + + + + | − | − | − |
| α6 | + + + + | + | + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α7 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β2 | − | − | − | − | + + + + | − | − | − |
| β4 | + + + + | + + + + | + + + + | + + + + | + | + + + + | + + + + | + + + + |
| α5 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β3 | + + + + | + | + | + + + + | + + + | + + + + | + + + + | + + + + |
|
| ||||||||
| 0.5 nM [125I] - Epibatidine + 100 nM Cytisine | ||||||||
| DS | VLG | DLG | OPT | fr | HP | V1 | RSG | |
| WT | □ | ■■■■ | ■■■■ | ■■■■ | ■■■■ | □ | □ | □ |
| α2 | n/a | + + + | + + + | + + + | + + + + | n/a | n/a | n/a |
| α4 | n/a | + + | + + | + + | + + + | n/a | n/a | n/a |
| α6 | n/a | + + | + + | + | + + + + | n/a | n/a | n/a |
| α7 | n/a | + + + | + + + | + + + | + + + + | n/a | n/a | n/a |
| β2 | n/a | − | − | − | + + + + | n/a | n/a | n/a |
| β4 | n/a | + + | + + | + + | + | n/a | n/a | n/a |
| α5 | n/a | + + + + | + + + + | + + + + | + + + | n/a | n/a | n/a |
| β3 | n/a | + + | + + | + | + + + | n/a | n/a | n/a |
|
| ||||||||
| 0.5 nM [125I] - Epibatidine + 100 nM Cytisine + 100 nM α-CtxMII | ||||||||
| DS | VLG | DLG | OPT | fr | HP | V1 | RSG | |
| WT | □ | ■■■ | ■■■ | ■■■ | ■■■■ | □ | □ | □ |
| α2 | n/a | + + + + | + + + + | + + + | + + + + | n/a | n/a | n/a |
| α4 | n/a | + + + | + + + | + + | + + + + | n/a | n/a | n/a |
| α6 | n/a | + + + | + + + | + | + + + + | n/a | n/a | n/a |
| α7 | n/a | + + + + | + + + + | + + + | + + + + | n/a | n/a | n/a |
| β2 | n/a | − | − | − | + + + + | n/a | n/a | n/a |
| β4 | n/a | + + + | + + + | + + | + + + | n/a | n/a | n/a |
| α5 | n/a | + + + + | + + + + | + + + + | + + + + | n/a | n/a | n/a |
| β3 | n/a | + + | + + | + | + + + + | n/a | n/a | n/a |
|
| ||||||||
| 200 pM 5-[125I]-A85380 | ||||||||
| DS | VLG | DLG | OPT | fr | HP | V1 | RSG | |
| WT | ■■■■ | ■■■■ | ■■■■ | ■■■■ | ■■■■ | ■■■■ | ■■■■ | ■■■■ |
| α2 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α4 | − | + + | + + | + + | + + + + | − | − | − |
| α6 | + + + + | + + | + + + | + + | − | + + + + | + + + + | + + + + |
| α7 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β2 | − | − | − | − | − | − | − | − |
| β4 | + + + + | + + + + | + + + + | + + | + + + + | + + + + | + + + + | + + + + |
| α5 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β3 | + + + + | + + | + + + | + + | − | + + + + | + + + + | + + + + |
|
| ||||||||
| 0.5 nM [125I] α-ConotoxinMII | ||||||||
| DS | VLG | DLG | OPT | fr | HP | V1 | RSG | |
| WT | □ | ■■■■ | ■■■■ | ■■■■ | □ | □ | □ | □ |
| α2 | n/a | + + + + | + + + + | + + + + | n/a | n/a | n/a | n/a |
| α4 | n/a | + + | + + | + + | n/a | n/a | n/a | n/a |
| α6 | n/a | − | − | − | n/a | n/a | n/a | n/a |
| α7 | n/a | + + + + | + + + + | + + + + | n/a | n/a | n/a | n/a |
| β2 | n/a | − | − | − | n/a | n/a | n/a | n/a |
| β4 | n/a | + + + + | + + + + | + + + + | n/a | n/a | n/a | n/a |
| α5 | n/a | + + + + | + + + + | + + + + | n/a | n/a | n/a | n/a |
| β3 | n/a | + | + | + | n/a | n/a | n/a | n/a |
|
| ||||||||
| 250 pM [125I] α-Bungarotoxin | ||||||||
| DS | VLG | DLG | OPT | fr | HP | V1 | RSG | |
| WT | ■■■■ | ■■■■ | ■■■■ | ■■■■ | ■■■■ | ■■■■ | ■■■■ | ■■■■ |
| α2 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α4 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α6 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α7 | − | − | − | − | − | − | − | − |
| β2 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β4 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α5 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β3 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
Autoradiography was performed as described in section 2.3. At bregma = −2.5 mm the following brain regions are visible: Dorsal Subiculum, DS; Ventral Lateral Geniculates, VLG; Thalamus, Dorsal Lateral Geniculates, DLG; Olivary Pretectal Nucleus, OPT; Fasciculus Retroflexus, fr; Hippocampus, HP; Primary Visual Cortex, V1; Retrosplenial Granular Cortex, RSG Regions with no detectable signal above background are designated with □ and relative quantitation was not attempted (n/a).
Table 5.
Semi quantitative visual analysis of coronal mouse brain sections at approximately −3.5 mm Bregma.
| 0.5 nM [125I] - Epibatidine | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IPN | MG | SuG | DpG | SNpr | SNpc | VTA | V1 | HP | DS | |
| WT | ■■■■ | ■■■ | ■■■■ | ■ | ■ | ■■■ | ■■■ | ■■■ | ■ | ■■■ |
| α2 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α4 | + + + + | + | + + + | − | − | + | + | − | − | − |
| α6 | + + + + | + + | + + | + + + + | + + + + | + + | + + | + + + + | + + + + | + + + + |
| α7 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + | + + + | + + + + | + + + + | + + + + |
| β2 | + + + + | − | − | − | − | − | − | − | − | − |
| β4 | + + | + + + | + + + + | + + + + | + + + + | + + + | + + + | + + + + | + + + + | + + + + |
| α5 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β3 | + + + + | + + | + + | + + + + | + + + + | + + | + + | + + + + | + + + + | + + + + |
|
| ||||||||||
| 0.5 nM [125I] - Epibatidine + 100 nM Cytisine | ||||||||||
| IPN | MG | SuG | DpG | SNpr | SNpc | VTA | V1 | HP | DS | |
| WT | ■■■■ | ■ | ■■■ | □ | □ | □ | □ | □ | □ | □ |
| α2 | + + + + | + + + + | + + + + | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| α4 | + + + + | + + + + | + + + | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| α6 | + + + + | + + + + | + + | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| α7 | + + + + | + + + + | + + + + | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| β2 | + + + + | − | − | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| β4 | + | − | + + + | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| α5 | + + + + | + + + + | + + + + | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| β3 | + + + + | + + + + | + + | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
|
| ||||||||||
| 0.5 nM [125I] - Epibatidine + 100 nM Cytisine + 100 nM α-CtxMII | ||||||||||
| IPN | MG | SuG | DpG | SNpr | SNpc | VTA | V1 | HP | DS | |
| WT | ■■■■ | ■ | ■■■ | □ | □ | □ | □ | □ | □ | □ |
| α2 | + + + + | + + + + | + + + | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| α4 | + + + + | + + | + + | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| α6 | + + + + | + + + + | + + + | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| α7 | + + + + | − | + + + | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| β2 | + + + + | − | − | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| β4 | + | − | + + | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| α5 | + + + + | + + + + | + + + + | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| β3 | + + + + | + + + + | + + | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
|
| ||||||||||
| 200 pM 5-[125I]-A85380 | ||||||||||
| IPN | MG | SuG | DpG | SNpr | SNpc | VTA | V1 | HP | DS | |
| WT | ■■■■ | ■■■ | ■■■■ | ■■■ | ■■ | ■■■ | ■■■ | ■■ | ■■ | ■■■ |
| α2 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α4 | + + + + | + | + + | − | + + + | + | + | − | − | − |
| α6 | + + + + | + + + + | ‘+ | + + + + | + + + + | + + | + + | + + + + | + + + + | + + + + |
| α7 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β2 | − | − | − | − | − | − | − | − | − | − |
| β4 | + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| α5 | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + | + + + + |
| β3 | + + + + | + + + + | ‘+ | + + + + | + + + + | + + | + + | + + + + | + + + + | + + + + |
|
| ||||||||||
| 0.5 nM [125I] α-ConotoxinMII | ||||||||||
| IPN | MG | SuG | DpG | SNpr | SNpc | VTA | V1 | HP | DS | |
| WT | ■■ | □ | ■■■■ | □ | □ | □ | □ | □ | □ | □ |
| α2 | + + + + | n/a | + + + + | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| α4 | + + | n/a | + + + | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| α6 | + + + | n/a | − | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| α7 | + + + | n/a | + + + + | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| β2 | − | n/a | − | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| β4 | + + + | n/a | + + + + | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| α5 | + + + | n/a | + + + + | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| β3 | + + + | n/a | + | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
|
| ||||||||||
| 250 pM [125I] α-Bungarotoxin | ||||||||||
| IPN | MG | SuG | DpG | SNpr | SNpc | VTA | V1 | HP | DS | |
| WT | ■■ | ■■■■ | ■■■■ | ■■ | ■■ | □ | □ | ■ | ■ | ■ |
| α2 | + + + + | + + + + | + + + + | + + + + | + + + + | n/a | n/a | + + + + | + + + + | + + + + |
| α4 | + + + + | + + + + | + + + + | + + + + | + + + + | n/a | n/a | + + + + | + + + + | + + + + |
| α6 | + + + + | + + + + | + + + + | + + + + | + + + + | n/a | n/a | + + + + | + + + + | + + + + |
| α7 | − | − | − | − | − | n/a | n/a | − | − | − |
| β2 | + + + + | + + + + | + + + + | + + + + | + + + + | n/a | n/a | + + + + | + + + + | + + + + |
| β4 | + + + + | + + + + | + + + + | + + + + | + + + + | n/a | n/a | + + + + | + + + + | + + + + |
| α5 | + + + + | + + + + | + + + + | + + + + | + + + + | n/a | n/a | + + + + | + + + + | + + + + |
| β3 | + + + + | + + + + | + + + + | + + + + | + + + + | n/a | n/a | + + + + | + + + + | + + + + |
Autoradiography was performed as described in section 2.3. At bregma = −3.52mm the following brain regions are visible: Interpeduncular Nucleus, IPN; Medial Geniculates, MG; Superior Colliculus Superficial Grey layer, SuG; Superior Colliculus deep grey layer, DpG; Substantia Nigra pars reticulata, SNpr; Substantia Nigra pars compacta, SNpc; Ventral Tegmental Area, VTA; Hippocampus, HP; Primary Visual Cortex, V1; Dorsal Subiculum, DS. Regions with no detectable signal above background are designated with □ and relative quantitation was not attempted (n/a).
Acknowledgments
This work was supported by the following grants from the National Institutes on Drug Abuse of the National Institutes of Health of the United States of America: R01 DA003194, R01 DA012242 and P30 DA015663.
The authors thank Dr. Sharon R. Grady and Dr. Heidi C. O’Neill for critical reading of the manuscript and helpful comments. The authors also thank the original providers of the null mutant mice: Dr. Jim Boulter, UCLA—α2; Dr. Richard Paylor, Baylor University School of Medicine—α5, α7 and β4; Dr. John Drago, University of Melbourne—α4; Dr. Marina Picciotto—Yale University—β2; Dr. Stephan Heinemann, Salk Institute—β3; Dr. Uwe Maskos, Pasteur Institute—α6
Abbreviations
- nAChR
nicotinic cholinergic receptor
- αBgt
α-bungarotoxin
- A-85380
3-((2S)-azetidinylmethoxy)pyridine
- αCtxMII
α-conotoxinMII
Footnotes
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Patrick J, Stallcup B. alpha-Bungarotoxin binding and cholinergic receptor function on a rat sympathetic nerve line. J Biol Chem. 1977;252:8629–33. [PubMed] [Google Scholar]
- 2.Romano C, Goldstein A. Stereospecific nicotine receptors on rat brain membranes. Science. 1980;210:647–50. doi: 10.1126/science.7433991. [DOI] [PubMed] [Google Scholar]
- 3.Clarke PB, Schwartz RD, Paul SM, Pert CB, Pert A. Nicotinic binding in rat brain: autoradiographic comparison of [3H]acetylcholine, [3H]nicotine, and [125I]-alpha-bungarotoxin. J Neurosci. 1985;5:1307–15. doi: 10.1523/JNEUROSCI.05-05-01307.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marks MJ, Collins AC. Characterization of nicotine binding in mouse brain and comparison with the binding of alpha-bungarotoxin and quinuclidinyl benzilate. Mol Pharmacol. 1982;22:554–64. [PubMed] [Google Scholar]
- 5.Sargent PB. The diversity of neuronal nicotinic acetylcholine receptors. Annu Rev Neurosci. 1993;16:403–43. doi: 10.1146/annurev.ne.16.030193.002155. [DOI] [PubMed] [Google Scholar]
- 6.Lindstrom J, Schoepfer R, Conroy W, Whiting P, Das M, Saedi M, et al. The nicotinic acetylcholine receptor gene family: structure of nicotinic receptors from muscle and neurons and neuronal alpha-bungarotoxin-binding proteins. Adv Exp Med Biol. 1991;287:255–78. doi: 10.1007/978-1-4684-5907-4_22. [DOI] [PubMed] [Google Scholar]
- 7.Gotti C, Clementi F, Fornari A, Gaimarri A, Guiducci S, Manfredi I, et al. Structural and functional diversity of native brain neuronal nicotinic receptors. Biochem Pharmacol. 2009;78:703–11. doi: 10.1016/j.bcp.2009.05.024. [DOI] [PubMed] [Google Scholar]
- 8.Schwartz RD, McGee R, Jr, Kellar KJ. Nicotinic cholinergic receptors labeled by [3H]acetylcholine in rat brain. Mol Pharmacol. 1982;22:56–62. [PubMed] [Google Scholar]
- 9.Pabreza LA, Dhawan S, Kellar KJ. [3H]cytisine binding to nicotinic cholinergic receptors in brain. Mol Pharmacol. 1991;39:9–12. [PubMed] [Google Scholar]
- 10.Abood LG, Grassi S. [3H]methylcarbamylcholine, a new radioligand for studying brain nicotinic receptors. Biochem Pharmacol. 1986;35:4199–202. doi: 10.1016/0006-2952(86)90695-7. [DOI] [PubMed] [Google Scholar]
- 11.Badio B, Daly JW. Epibatidine, a potent analgetic and nicotinic agonist. Mol Pharmacol. 1994;45:563–9. [PubMed] [Google Scholar]
- 12.Davila-Garcia MI, Musachio JL, Perry DC, Xiao Y, Horti A, London ED, et al. [125I]IPH, an epibatidine analog, binds with high affinity to neuronal nicotinic cholinergic receptors. J Pharmacol Exp Ther. 1997;282:445–51. [PubMed] [Google Scholar]
- 13.Houghtling RA, Davila-Garcia MI, Kellar KJ. Characterization of (+/−)(−)[3H]epibatidine binding to nicotinic cholinergic receptors in rat and human brain. Mol Pharmacol. 1995;48:280–7. [PubMed] [Google Scholar]
- 14.Perry DC, Xiao Y, Nguyen HN, Musachio JL, Davila-Garcia MI, Kellar KJ. Measuring nicotinic receptors with characteristics of alpha4beta2, alpha3beta2 and alpha3beta4 subtypes in rat tissues by autoradiography. J Neurochem. 2002;82:468–81. doi: 10.1046/j.1471-4159.2002.00951.x. [DOI] [PubMed] [Google Scholar]
- 15.Marks MJ, Smith KW, Collins AC. Differential agonist inhibition identifies multiple epibatidine binding sites in mouse brain. J Pharmacol Exp Ther. 1998;285:377–86. [PubMed] [Google Scholar]
- 16.Zoli M, Lena C, Picciotto MR, Changeux JP. Identification of four classes of brain nicotinic receptors using beta2 mutant mice. J Neurosci. 1998;18:4461–72. doi: 10.1523/JNEUROSCI.18-12-04461.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerzanich V, Peng X, Wang F, Wells G, Anand R, Fletcher S, et al. Comparative pharmacology of epibatidine: a potent agonist for neuronal nicotinic acetylcholine receptors. Mol Pharmacol. 1995;48:774–82. [PubMed] [Google Scholar]
- 18.Xiao Y, Baydyuk M, Wang HP, Davis HE, Kellar KJ. Pharmacology of the agonist binding sites of rat neuronal nicotinic receptor subtypes expressed in HEK 293 cells. Bioorg Med Chem Lett. 2004;14:1845–8. doi: 10.1016/j.bmcl.2003.09.105. [DOI] [PubMed] [Google Scholar]
- 19.Parker MJ, Beck A, Luetje CW. Neuronal nicotinic receptor beta2 and beta4 subunits confer large differences in agonist binding affinity. Mol Pharmacol. 1998;54:1132–9. [PubMed] [Google Scholar]
- 20.Cartier GE, Yoshikami D, Gray WR, Luo S, Olivera BM, McIntosh JM. A new alpha-conotoxin which targets alpha3beta2 nicotinic acetylcholine receptors. J Biol Chem. 1996;271:7522–8. doi: 10.1074/jbc.271.13.7522. [DOI] [PubMed] [Google Scholar]
- 21.Whiteaker P, McIntosh JM, Luo S, Collins AC, Marks MJ. 125I-alpha-conotoxin MII identifies a novel nicotinic acetylcholine receptor population in mouse brain. Mol Pharmacol. 2000;57:913–25. [PubMed] [Google Scholar]
- 22.Champtiaux N, Han ZY, Bessis A, Rossi FM, Zoli M, Marubio L, et al. Distribution and pharmacology of alpha 6-containing nicotinic acetylcholine receptors analyzed with mutant mice. J Neurosci. 2002;22:1208–17. doi: 10.1523/JNEUROSCI.22-04-01208.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sullivan JP, Donnelly-Roberts D, Briggs CA, Anderson DJ, Gopalakrishnan M, Piattoni-Kaplan M, et al. A-85380 [3-(2(S)-azetidinylmethoxy) pyridine]: in vitro pharmacological properties of a novel, high affinity alpha 4 beta 2 nicotinic acetylcholine receptor ligand. Neuropharmacology. 1996;35:725–34. doi: 10.1016/0028-3908(96)84644-2. [DOI] [PubMed] [Google Scholar]
- 24.Mukhin AG, Gundisch D, Horti AG, Koren AO, Tamagnan G, Kimes AS, et al. 5-Iodo-A-85380, an alpha4beta2 subtype-selective ligand for nicotinic acetylcholine receptors. Mol Pharmacol. 2000;57:642–9. doi: 10.1124/mol.57.3.642. [DOI] [PubMed] [Google Scholar]
- 25.Rueter LE, Donnelly-Roberts DL, Curzon P, Briggs CA, Anderson DJ, Bitner RS. A-85380: a pharmacological probe for the preclinical and clinical investigation of the alphabeta neuronal nicotinic acetylcholine receptor. CNS Drug Rev. 2006;12:100–12. doi: 10.1111/j.1527-3458.2006.00100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sihver W, Nordberg A, Langstrom B, Mukhin AG, Koren AO, Kimes AS, et al. Development of ligands for in vivo imaging of cerebral nicotinic receptors. Behav Brain Res. 2000;113:143–57. doi: 10.1016/s0166-4328(00)00209-6. [DOI] [PubMed] [Google Scholar]
- 27.Picciotto MR. Nicotine as a modulator of behavior: beyond the inverted U. Trends Pharmacol Sci. 2003;24:493–9. doi: 10.1016/S0165-6147(03)00230-X. [DOI] [PubMed] [Google Scholar]
- 28.Cordero-Erausquin M, Marubio LM, Klink R, Changeux JP. Nicotinic receptor function: new perspectives from knockout mice. Trends Pharmacol Sci. 2000;21:211–7. doi: 10.1016/s0165-6147(00)01489-9. [DOI] [PubMed] [Google Scholar]
- 29.Champtiaux N, Changeux JP. Knockout and knockin mice to investigate the role of nicotinic receptors in the central nervous system. Prog Brain Res. 2004;145:235–51. doi: 10.1016/s0079-6123(03)45016-4. [DOI] [PubMed] [Google Scholar]
- 30.Marubio LM, del Mar Arroyo-Jimenez M, Cordero-Erausquin M, Lena C, Le Novere N, de Kerchove d’Exaerde A, et al. Reduced antinociception in mice lacking neuronal nicotinic receptor subunits. Nature. 1999;398:805–10. doi: 10.1038/19756. [DOI] [PubMed] [Google Scholar]
- 31.Millar NS, Gotti C. Diversity of vertebrate nicotinic acetylcholine receptors. Neuropharmacology. 2009;56:237–46. doi: 10.1016/j.neuropharm.2008.07.041. [DOI] [PubMed] [Google Scholar]
- 32.Picciotto MR, Zoli M, Lena C, Bessis A, Lallemand Y, Le Novere N, et al. Abnormal avoidance learning in mice lacking functional high-affinity nicotine receptor in the brain. Nature. 1995;374:65–7. doi: 10.1038/374065a0. [DOI] [PubMed] [Google Scholar]
- 33.Orr-Urtreger A, Goldner FM, Saeki M, Lorenzo I, Goldberg L, De Biasi M, et al. Mice deficient in the alpha7 neuronal nicotinic acetylcholine receptor lack alpha-bungarotoxin binding sites and hippocampal fast nicotinic currents. J Neurosci. 1997;17:9165–71. doi: 10.1523/JNEUROSCI.17-23-09165.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Champtiaux N, Changeux JP. Knock-out and knock-in mice to investigate the role of nicotinic receptors in the central nervous system. Curr Drug Targets CNS Neurol Disord. 2002;1:319–30. doi: 10.2174/1568007023339247. [DOI] [PubMed] [Google Scholar]
- 35.Cui C, Booker TK, Allen RS, Grady SR, Whiteaker P, Marks MJ, et al. The beta3 nicotinic receptor subunit: a component of alpha-conotoxin MII-binding nicotinic acetylcholine receptors that modulate dopamine release and related behaviors. J Neurosci. 2003;23:11045–53. doi: 10.1523/JNEUROSCI.23-35-11045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salas R, Orr-Urtreger A, Broide RS, Beaudet A, Paylor R, De Biasi M. The nicotinic acetylcholine receptor subunit alpha 5 mediates short-term effects of nicotine in vivo. Mol Pharmacol. 2003;63:1059–66. doi: 10.1124/mol.63.5.1059. [DOI] [PubMed] [Google Scholar]
- 37.Salas R, Pieri F, De Biasi M. Decreased signs of nicotine withdrawal in mice null for the beta4 nicotinic acetylcholine receptor subunit. J Neurosci. 2004;24:10035–9. doi: 10.1523/JNEUROSCI.1939-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whiteaker P, Peterson CG, Xu W, McIntosh JM, Paylor R, Beaudet AL, et al. Involvement of the alpha3 subunit in central nicotinic binding populations. J Neurosci. 2002;22:2522–9. doi: 10.1523/JNEUROSCI.22-07-02522.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marks MJ, Laverty DS, Whiteaker P, Salminen O, Grady SR, McIntosh JM, et al. John Daly’s compound, epibatidine, facilitates identification of nicotinic receptor subtypes. J Mol Neurosci. 2010;40:96–104. doi: 10.1007/s12031-009-9264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grady SR, Moretti M, Zoli M, Marks MJ, Zanardi A, Pucci L, et al. Rodent habenulo-interpeduncular pathway expresses a large variety of uncommon nAChR subtypes, but only the alpha3beta4* and alpha3beta3beta4* subtypes mediate acetylcholine release. J Neurosci. 2009;29:2272–82. doi: 10.1523/JNEUROSCI.5121-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gotti C, Moretti M, Meinerz NM, Clementi F, Gaimarri A, Collins AC, et al. Partial deletion of the nicotinic cholinergic receptor alpha 4 or beta 2 subunit genes changes the acetylcholine sensitivity of receptor-mediated 86Rb+ efflux in cortex and thalamus and alters relative expression of alpha 4 and beta 2 subunits. Mol Pharmacol. 2008;73:1796–807. doi: 10.1124/mol.108.045203. [DOI] [PubMed] [Google Scholar]
- 42.Marks MJ, Whiteaker P, Grady SR, Picciotto MR, McIntosh JM, Collins AC. Characterization of [(125) I]epibatidine binding and nicotinic agonist-mediated (86) Rb(+) efflux in interpeduncular nucleus and inferior colliculus of beta2 null mutant mice. J Neurochem. 2002;81:1102–15. doi: 10.1046/j.1471-4159.2002.00910.x. [DOI] [PubMed] [Google Scholar]
- 43.Gotti C, Moretti M, Clementi F, Riganti L, McIntosh JM, Collins AC, et al. Expression of nigrostriatal alpha 6-containing nicotinic acetylcholine receptors is selectively reduced, but not eliminated, by beta 3 subunit gene deletion. Mol Pharmacol. 2005;67:2007–15. doi: 10.1124/mol.105.011940. [DOI] [PubMed] [Google Scholar]
- 44.Marks MJ, Whiteaker P, Collins AC. Deletion of the alpha7, beta2, or beta4 nicotinic receptor subunit genes identifies highly expressed subtypes with relatively low affinity for [3H]epibatidine. Mol Pharmacol. 2006;70:947–59. doi: 10.1124/mol.106.025338. [DOI] [PubMed] [Google Scholar]
- 45.Marks MJ, Stitzel JA, Grady SR, Picciotto MR, Changeux JP, Collins AC. Nicotinic-agonist stimulated (86)Rb(+) efflux and [(3)H]epibatidine binding of mice differing in beta2 genotype. Neuropharmacology. 2000;39:2632–45. doi: 10.1016/s0028-3908(00)00115-5. [DOI] [PubMed] [Google Scholar]
- 46.Marks MJ, Meinerz NM, Drago J, Collins AC. Gene targeting demonstrates that alpha4 nicotinic acetylcholine receptor subunits contribute to expression of diverse [3H]epibatidine binding sites and components of biphasic 86Rb+ efflux with high and low sensitivity to stimulation by acetylcholine. Neuropharmacology. 2007;53:390–405. doi: 10.1016/j.neuropharm.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whiteaker P, Wilking JA, Brown RW, Brennan RJ, Collins AC, Lindstrom JM, et al. Pharmacological and immunochemical characterization of alpha2* nicotinic acetylcholine receptors (nAChRs) in mouse brain. Acta Pharmacol Sin. 2009;30:795–804. doi: 10.1038/aps.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown RW, Collins AC, Lindstrom JM, Whiteaker P. Nicotinic alpha5 subunit deletion locally reduces high-affinity agonist activation without altering nicotinic receptor numbers. J Neurochem. 2007;103:204–15. doi: 10.1111/j.1471-4159.2007.04700.x. [DOI] [PubMed] [Google Scholar]
- 49.Xu W, Gelber S, Orr-Urtreger A, Armstrong D, Lewis RA, Ou CN, et al. Megacystis, mydriasis, and ion channel defect in mice lacking the alpha3 neuronal nicotinic acetylcholine receptor. Proc Natl Acad Sci U S A. 1999;96:5746–51. doi: 10.1073/pnas.96.10.5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ross SA, Wong JY, Clifford JJ, Kinsella A, Massalas JS, Horne MK, et al. Phenotypic characterization of an alpha 4 neuronal nicotinic acetylcholine receptor subunit knock-out mouse. J Neurosci. 2000;20:6431–41. doi: 10.1523/JNEUROSCI.20-17-06431.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salminen O, Murphy KL, McIntosh JM, Drago J, Marks MJ, Collins AC, et al. Subunit composition and pharmacology of two classes of striatal presynaptic nicotinic acetylcholine receptors mediating dopamine release in mice. Mol Pharmacol. 2004;65:1526–35. doi: 10.1124/mol.65.6.1526. [DOI] [PubMed] [Google Scholar]
- 52.Salminen O, Whiteaker P, Grady SR, Collins AC, McIntosh JM, Marks MJ. The subunit composition and pharmacology of alpha-Conotoxin MII-binding nicotinic acetylcholine receptors studied by a novel membrane-binding assay. Neuropharmacology. 2005;48:696–705. doi: 10.1016/j.neuropharm.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 53.Salminen O, Drapeau JA, McIntosh JM, Collins AC, Marks MJ, Grady SR. Pharmacology of alpha-conotoxin MII-sensitive subtypes of nicotinic acetylcholine receptors isolated by breeding of null mutant mice. Mol Pharmacol. 2007;71:1563–71. doi: 10.1124/mol.106.031492. [DOI] [PubMed] [Google Scholar]
- 54.Xu W, Orr-Urtreger A, Nigro F, Gelber S, Sutcliffe CB, Armstrong D, et al. Multiorgan autonomic dysfunction in mice lacking the beta2 and the beta4 subunits of neuronal nicotinic acetylcholine receptors. J Neurosci. 1999;19:9298–305. doi: 10.1523/JNEUROSCI.19-21-09298.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grady SR, Salminen O, Laverty DC, Whiteaker P, McIntosh JM, Collins AC, et al. The subtypes of nicotinic acetylcholine receptors on dopaminergic terminals of mouse striatum. Biochem Pharmacol. 2007;74:1235–46. doi: 10.1016/j.bcp.2007.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grady SR, Salminen O, McIntosh JM, Marks MJ, Collins AC. Mouse striatal dopamine nerve terminals express alpha4alpha5beta2 and two stoichiometric forms of alpha4beta2*-nicotinic acetylcholine receptors. J Mol Neurosci. 2010;40:91–5. doi: 10.1007/s12031-009-9263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakauchi S, Brennan RJ, Boulter J, Sumikawa K. Nicotine gates long-term potentiation in the hippocampal CA1 region via the activation of alpha2* nicotinic ACh receptors. Eur J Neurosci. 2007;25:2666–81. doi: 10.1111/j.1460-9568.2007.05513.x. [DOI] [PubMed] [Google Scholar]
- 58.Whiteaker P, Jimenez M, McIntosh JM, Collins AC, Marks MJ. Identification of a novel nicotinic binding site in mouse brain using [(125)I]-epibatidine. Br J Pharmacol. 2000;131:729–39. doi: 10.1038/sj.bjp.0703616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mineur YS, Brunzell DH, Grady SR, Lindstrom JM, McIntosh JM, Marks MJ, et al. Localized low-level re-expression of high-affinity mesolimbic nicotinic acetylcholine receptors restores nicotine-induced locomotion but not place conditioning. Genes Brain Behav. 2009;8:257–66. doi: 10.1111/j.1601-183X.2008.00468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pauly JR, Marks MJ, Gross SD, Collins AC. An autoradiographic analysis of cholinergic receptors in mouse brain after chronic nicotine treatment. J Pharmacol Exp Ther. 1991;258:1127–36. [PubMed] [Google Scholar]
- 61.Millar NS. Assembly and subunit diversity of nicotinic acetylcholine receptors. Biochem Soc Trans. 2003;31:869–74. doi: 10.1042/bst0310869. [DOI] [PubMed] [Google Scholar]
- 62.Schoepfer R, Conroy WG, Whiting P, Gore M, Lindstrom J. Brain alpha-bungarotoxin binding protein cDNAs and MAbs reveal subtypes of this branch of the ligand-gated ion channel gene superfamily. Neuron. 1990;5:35–48. doi: 10.1016/0896-6273(90)90031-a. [DOI] [PubMed] [Google Scholar]
- 63.Couturier S, Bertrand D, Matter JM, Hernandez MC, Bertrand S, Millar N, et al. A neuronal nicotinic acetylcholine receptor subunit (alpha 7) is developmentally regulated and forms a homo-oligomeric channel blocked by alpha-BTX. Neuron. 1990;5:847–56. doi: 10.1016/0896-6273(90)90344-f. [DOI] [PubMed] [Google Scholar]
- 64.Seguela P, Wadiche J, Dineley-Miller K, Dani JA, Patrick JW. Molecular cloning, functional properties, and distribution of rat brain alpha 7: a nicotinic cation channel highly permeable to calcium. J Neurosci. 1993;13:596–604. doi: 10.1523/JNEUROSCI.13-02-00596.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiao Y, Fan H, Musachio JL, Wei ZL, Chellappan SK, Kozikowski AP, et al. Sazetidine-A, a novel ligand that desensitizes alpha4beta2 nicotinic acetylcholine receptors without activating them. Mol Pharmacol. 2006;70:1454–60. doi: 10.1124/mol.106.027318. [DOI] [PubMed] [Google Scholar]
- 66.Champtiaux N, Gotti C, Cordero-Erausquin M, David DJ, Przybylski C, Lena C, et al. Subunit composition of functional nicotinic receptors in dopaminergic neurons investigated with knock-out mice. J Neurosci. 2003;23:7820–9. doi: 10.1523/JNEUROSCI.23-21-07820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gotti C, Guiducci S, Tedesco V, Corbioli S, Zanetti L, Moretti M, et al. Nicotinic acetylcholine receptors in the mesolimbic pathway: primary role of ventral tegmental area alpha6beta2* receptors in mediating systemic nicotine effects on dopamine release, locomotion, and reinforcement. J Neurosci. 2010;30:5311–25. doi: 10.1523/JNEUROSCI.5095-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yeh JJ, Yasuda RP, Davila-Garcia MI, Xiao Y, Ebert S, Gupta T, et al. Neuronal nicotinic acetylcholine receptor alpha3 subunit protein in rat brain and sympathetic ganglion measured using a subunit-specific antibody: regional and ontogenic expression. J Neurochem. 2001;77:336–46. doi: 10.1046/j.1471-4159.2001.t01-1-00259.x. [DOI] [PubMed] [Google Scholar]
- 69.Hernandez SC, Vicini S, Xiao Y, Davila-Garcia MI, Yasuda RP, Wolfe BB, et al. The nicotinic receptor in the rat pineal gland is an alpha3beta4 subtype. Mol Pharmacol. 2004;66:978–87. doi: 10.1124/mol.104.002345. [DOI] [PubMed] [Google Scholar]
- 70.Turner JR, Kellar KJ. Nicotinic cholinergic receptors in the rat cerebellum: multiple heteromeric subtypes. J Neurosci. 2005;25:9258–65. doi: 10.1523/JNEUROSCI.2112-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marritt AM, Cox BC, Yasuda RP, McIntosh JM, Xiao Y, Wolfe BB, et al. Nicotinic cholinergic receptors in the rat retina: simple and mixed heteromeric subtypes. Mol Pharmacol. 2005;68:1656–68. doi: 10.1124/mol.105.012369. [DOI] [PubMed] [Google Scholar]
- 72.Mao D, Perry DC, Yasuda RP, Wolfe BB, Kellar KJ. The alpha4beta2alpha5 nicotinic cholinergic receptor in rat brain is resistant to up-regulation by nicotine in vivo. J Neurochem. 2008;104:446–56. doi: 10.1111/j.1471-4159.2007.05011.x. [DOI] [PubMed] [Google Scholar]
- 73.Martin-Ruiz CM, Court JA, Molnar E, Lee M, Gotti C, Mamalaki A, et al. Alpha4 but not alpha3 and alpha7 nicotinic acetylcholine receptor subunits are lost from the temporal cortex in Alzheimer’s disease. J Neurochem. 1999;73:1635–40. doi: 10.1046/j.1471-4159.1999.0731635.x. [DOI] [PubMed] [Google Scholar]
- 74.Balestra B, Vailati S, Moretti M, Hanke W, Clementi F, Gotti C. Chick optic lobe contains a developmentally regulated alpha2alpha5beta2 nicotinic receptor subtype. Mol Pharmacol. 2000;58:300–11. doi: 10.1124/mol.58.2.300. [DOI] [PubMed] [Google Scholar]
- 75.Zoli M, Moretti M, Zanardi A, McIntosh JM, Clementi F, Gotti C. Identification of the nicotinic receptor subtypes expressed on dopaminergic terminals in the rat striatum. J Neurosci. 2002;22:8785–9. doi: 10.1523/JNEUROSCI.22-20-08785.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moretti M, Mugnaini M, Tessari M, Zoli M, Gaimarri A, Manfredi I, et al. A comparative study of the effects of the intravenous self-administration or subcutaneous minipump infusion of nicotine on the expression of brain neuronal nicotinic receptor subtypes. Mol Pharmacol. 2010;78:287–96. doi: 10.1124/mol.110.064071. [DOI] [PubMed] [Google Scholar]
- 77.Marks MJ, Whiteaker P, Calcaterra J, Stitzel JA, Bullock AE, Grady SR, et al. Two pharmacologically distinct components of nicotinic receptor-mediated rubidium efflux in mouse brain require the beta2 subunit. J Pharmacol Exp Ther. 1999;289:1090–103. [PubMed] [Google Scholar]
- 78.Whiteaker P, Marks MJ, Grady SR, Lu Y, Picciotto MR, Changeux JP, et al. Pharmacological and null mutation approaches reveal nicotinic receptor diversity. Eur J Pharmacol. 2000;393:123–35. doi: 10.1016/s0014-2999(00)00052-2. [DOI] [PubMed] [Google Scholar]
- 79.Nguyen HN, Rasmussen BA, Perry DC. Subtype-selective up-regulation by chronic nicotine of high-affinity nicotinic receptors in rat brain demonstrated by receptor autoradiography. J Pharmacol Exp Ther. 2003;307:1090–7. doi: 10.1124/jpet.103.056408. [DOI] [PubMed] [Google Scholar]
- 80.Salas R, Pieri F, Fung B, Dani JA, De Biasi M. Altered anxiety-related responses in mutant mice lacking the beta4 subunit of the nicotinic receptor. J Neurosci. 2003;23:6255–63. doi: 10.1523/JNEUROSCI.23-15-06255.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gotti C, Moretti M, Zanardi A, Gaimarri A, Champtiaux N, Changeux JP, et al. Heterogeneity and selective targeting of neuronal nicotinic acetylcholine receptor (nAChR) subtypes expressed on retinal afferents of the superior colliculus and lateral geniculate nucleus: identification of a new native nAChR subtype alpha3beta2(alpha5 or beta3) enriched in retinocollicular afferents. Mol Pharmacol. 2005;68:1162–71. doi: 10.1124/mol.105.015925. [DOI] [PubMed] [Google Scholar]
- 82.McIntosh JM, Olivera BM, Cruz LJ. Conus peptides as probes for ion channels. Methods Enzymol. 1999;294:605–24. doi: 10.1016/s0076-6879(99)94034-x. [DOI] [PubMed] [Google Scholar]
- 83.Moretti M, Vailati S, Zoli M, Lippi G, Riganti L, Longhi R, et al. Nicotinic acetylcholine receptor subtypes expression during rat retina development and their regulation by visual experience. Mol Pharmacol. 2004;66:85–96. doi: 10.1124/mol.66.1.85. [DOI] [PubMed] [Google Scholar]
- 84.Cox BC, Marritt AM, Perry DC, Kellar KJ. Transport of multiple nicotinic acetylcholine receptors in the rat optic nerve: high densities of receptors containing alpha6 and beta3 subunits. J Neurochem. 2008;105:1924–38. doi: 10.1111/j.1471-4159.2008.05282.x. [DOI] [PubMed] [Google Scholar]
- 85.Wada E, Wada K, Boulter J, Deneris E, Heinemann S, Patrick J, et al. Distribution of alpha 2, alpha 3, alpha 4, and beta 2 neuronal nicotinic receptor subunit mRNAs in the central nervous system: a hybridization histochemical study in the rat. J Comp Neurol. 1989;284:314–35. doi: 10.1002/cne.902840212. [DOI] [PubMed] [Google Scholar]
- 86.Marks MJ, Pauly JR, Gross SD, Deneris ES, Hermans-Borgmeyer I, Heinemann SF, et al. Nicotine binding and nicotinic receptor subunit RNA after chronic nicotine treatment. J Neurosci. 1992;12:2765–84. doi: 10.1523/JNEUROSCI.12-07-02765.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Salas R, Sturm R, Boulter J, De Biasi M. Nicotinic receptors in the habenulo-interpeduncular system are necessary for nicotine withdrawal in mice. J Neurosci. 2009;29:3014–8. doi: 10.1523/JNEUROSCI.4934-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jackson KJ, Marks MJ, Vann RE, Chen X, Gamage TF, Warner JA, et al. Role of alpha5 nicotinic acetylcholine receptors in pharmacological and behavioral effects of nicotine in mice. J Pharmacol Exp Ther. 2010;334:137–46. doi: 10.1124/jpet.110.165738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bailey CD, De Biasi M, Fletcher PJ, Lambe EK. The nicotinic acetylcholine receptor alpha5 subunit plays a key role in attention circuitry and accuracy. J Neurosci. 2010;30:9241–52. doi: 10.1523/JNEUROSCI.2258-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Heath CJ, King SL, Gotti C, Marks MJ, Picciotto MR. Cortico-thalamic connectivity is vulnerable to nicotine exposure during early postnatal development through alpha4/beta2/alpha5 nicotinic acetylcholine receptors. Neuropsychopharmacology. 2010;35:2324–38. doi: 10.1038/npp.2010.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gotti C, Moretti M, Gaimarri A, Zanardi A, Clementi F, Zoli M. Heterogeneity and complexity of native brain nicotinic receptors. Biochem Pharmacol. 2007;74:1102–11. doi: 10.1016/j.bcp.2007.05.023. [DOI] [PubMed] [Google Scholar]
- 92.Gangitano D, Salas R, Teng Y, Perez E, De Biasi M. Progesterone modulation of alpha5 nAChR subunits influences anxiety-related behavior during estrus cycle. Genes Brain Behav. 2009;8:398–406. doi: 10.1111/j.1601-183X.2009.00476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fowler CD, Lu Q, Johnson PM, Marks MJ, Kenny PJ. Habenular alpha5 nicotinic receptor subunit signalling controls nicotine intake. Nature. 2011;471:597–601. doi: 10.1038/nature09797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu Q, Huang Y, Xue F, Simard A, DeChon J, Li G, et al. A novel nicotinic acetylcholine receptor subtype in basal forebrain cholinergic neurons with high sensitivity to amyloid peptides. J Neurosci. 2009;29:918–29. doi: 10.1523/JNEUROSCI.3952-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pauly JR, Stitzel JA, Marks MJ, Collins AC. An autoradiographic analysis of cholinergic receptors in mouse brain. Brain Res Bull. 1989;22:453–9. doi: 10.1016/0361-9230(89)90072-5. [DOI] [PubMed] [Google Scholar]
- 96.Clarke PB, Pert CB, Pert A. Autoradiographic distribution of nicotine receptors in rat brain. Brain Res. 1984;323:390–5. doi: 10.1016/0006-8993(84)90320-2. [DOI] [PubMed] [Google Scholar]
- 97.Mansvelder HD, McGehee DS. Long-term potentiation of excitatory inputs to brain reward areas by nicotine. Neuron. 2000;27:349–57. doi: 10.1016/s0896-6273(00)00042-8. [DOI] [PubMed] [Google Scholar]
- 98.Jones IW, Wonnacott S. Precise localization of alpha7 nicotinic acetylcholine receptors on glutamatergic axon terminals in the rat ventral tegmental area. J Neurosci. 2004;24:11244–52. doi: 10.1523/JNEUROSCI.3009-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.