Abstract
A novel iterative k-space data-driven technique, namely Parallel Reconstruction Using Null Operations (PRUNO), is presented for parallel imaging reconstruction. In PRUNO, both data calibration and image reconstruction are formulated into linear algebra problems based on a generalized system model. An optimal data calibration strategy is demonstrated by using Singular Value Decomposition (SVD). And an iterative conjugate- gradient approach is proposed to efficiently solve missing k-space samples during reconstruction. With its generalized formulation and precise mathematical model, PRUNO reconstruction yields good accuracy, flexibility, stability. Both computer simulation and in vivo studies have shown that PRUNO produces much better reconstruction quality than autocalibrating partially parallel acquisition (GRAPPA), especially under high accelerating rates. With the aid of PRUO reconstruction, ultra high accelerating parallel imaging can be performed with decent image quality. For example, we have done successful PRUNO reconstruction at a reduction factor of 6 (effective factor of 4.44) with 8 coils and only a few autocalibration signal (ACS) lines.
Keywords: magnetic resonance imaging, parallel imaging, PRUNO, GRAPPA, auto-calibration, high accelerating, ACS, iterative reconstruction
INTRODUCTION
Parallel magnetic resonance imaging (pMRI) techniques have been proposed and widely introduced into clinical applications over the last decade. In pMRI, spatial sensitivity encodings are provided by utilizing an array of multiple receiver surface coils. With this data redundancy, we are able to reconstruct unaliased images from k-space data sampled below the Nyquist rate with fewer k-space phase encodes. This acceleration can be used to save scan time, increase temporal or spatial resolution [1–3], or reduce imaging artifacts [4,5]. Various parallel imaging reconstruction methods have been invented. Among those, the most widely used pMRI techniques are sensitivity encoding (SENSE) [6] and generalized autocalibrating partially parallel acquisition (GRAPPA) [7]. Other Cartesian pMRI reconstruction methods including SMASH [8,9], AUTO-SMASH [10], VD-AUTO-SMASH [11], Generalized SMASH [12], mSENSE [13], PILS [14], and SPACE-RIP [15] have also been demonstrated and implemented into numerous applications. Recently, several non-Cartesian pMRI reconstruction techniques have been investigated as well, such as non-Cartesian SENSE (NC-SENSE) [16] and kSPA [17,18]. All of these reconstruction algorithms can be classified into two categories, based on how the sensitivity encoding is decoded from the multi-channel data, i.e., physically-based reconstruction and data-driven reconstruction [19]. SENSE reconstruction is a typical physically-based algorithm, in which all coils sensitivity maps have to be estimated explicitly because image needs to be unfolded in the image domain based on these maps. On the other hand, GRAPPA is a data-driven method, in which the sensitivity information is merely implicitly derived in the k-space through data calibration, and the missing samples are fitted in k-space based on this information.
As a k-space data-driven method, GRAPPA has become the most successful pMRI technique in recent years, due to its convenience, flexibility and good performance. Compared to SENSE, the cumbersome coil sensitivity measurement is not required in GRAPPA. In addition, neither data calibration nor k-space fitting is computationally demanding. And many experimental results have shown that GRAPPA reconstruction yields relatively good image quality for low accelerating pMRI, for example, with a reduction factor of 2 to 3. However, the performance of GRAPPA reconstruction usually degrades significantly as the reduction rate gets higher [20], unless a very large number of autocalibration signal (ACS) lines are imposed.
In this work, an iterative k-space data-driven pMRI reconstruction algorithm is demonstrated, termed Parallel Reconstruction Using Null Operations (PRUNO). Based on a generalized problem formulation of k-space reconstruction, we show that PRUNO is a more flexible algorithm than GRAPPA. In PRUNO, both data calibration and image reconstruction are formulated into linear algebra problems and an iterative conjugate-gradient (CG) algorithm is proposed to solve the reconstruction equation efficiently. Both simulation and in vivo results show that PRUNO provides significantly improved image quality compared to GRAPPA while requires a reduced number of ACS lines, especially under high reduction factors.
THEROY
In this section, only study the case of 2D Cartesian pMRI is studied for simplicity. The same theory can be easily extended to 3D reconstruction. Let's denote the image size to be Nx × Ny. Without loss of generality, it is assumed that the phase encoding (PE) is along the y direction. Due to the discreteness of data acquisition in MRI, we formulate both acquisition and reconstruction as discrete system problems. Furthermore, two major assumptions are made about the sensitivity encoding:
Assumption 1: All sensitivity maps are band-limited, with a full width of ws in k-space. Due to the smoothness of coil sensitivity maps in nature, ws is usually approximated as a reasonably small number.
Assumption 2: The overall sensitivity encodings yield good orthogonality. That is, the pMRI reconstruction can be treated as an overdetermined problem if Nc > R and the k-space sampling is relatively uniform. Here R is the reduction factor and Nc is the number of coils. (Only the cases of Nc > R are investigated in this work.)
These conditions essentially form the basis behind most k-space data-driven pMRI reconstruction methods, such as GRAPPA [7,21] and kSPA [17,18]. To simplify the following discussion, we treat Nx, Ny and ws as even numbers.
A. Principle of k-Space Based pMRI Reconstruction
For multi-channel acquisition, k-space data acquired from the n-th coil can be approximated as
| [1] |
Here m(kx, ky) stands for the sampled true magnetization to be imaged in the k-space. dn(kx,ky) and sn (kx, ky) represent k-space samples of the acquirable data from the n-th coil and the spectrum of sensitivity map of that coil, respectively. “*” denotes the operator of two-dimensional convolution. Based on our first assumption, sn (kx, ky) is treated as a small kernel. Since a system convolution is essentially a linear operation, the sensitivity encoding can thus be formulated as a system of linear equations,
| [2] |
where d is a vector of concatenated fully-sampled k-space data from all coils (encoded samples) and m is a column vector containing the true magnetization in the k-space (unencoded samples). Hence, S is called sensitivity encoding matrix. The primary goal of a k-space based reconstruction is to compute either m or full presentation of d, from which the final image will be synthesized in image domain. With data undersampling, all samples in d can be classified into two groups according to their existence upon reconstruction, i.e., missing samples and acquired samples. To separate the two groups, masking operators can be applied to Eq. [2]:
| [3] |
Here the subscripts “m” and “a” represent “missing” and “acquired”, respectively. I denotes an identity matrix, which can be decomposed into two diagonal masking matrices, Im and Ia. Each masking only preserves samples from one category by setting entries corresponding to the other category to 0's. We can thus split Eq. [2] into two separate equations: Imd = (ImS)m and Iad = (IaS)m. If rows of all zeroes are eliminated from each equation, they are formulated more compactly as
| [4] |
and
| [5] |
Now dm is the vector including only missing samples and da is the other vector including only acquired ones. Sm and Sa are the corresponding row pruned matrices from ImS and IaS, respectively.
With the two assumptions stated at the beginning of this section, Eq. [5] should be an over-determined linear equation with a very sparse system matrix. If the encoding matrix Sa is known, the image reconstruction will be directly performed by solving m from this equation. However, the knowledge of Sa usually requires a complete estimation of all sensitivity maps. This scenario is a k-space “physically-based” reconstruction, which is essentially similar to Generalized SMASH [12].
To avoid the measurement of sensitivity maps due to its complexity and inaccuracy, we can solve dm first, and then apply a coil-by-coil reconstruction followed by a sum-of-squares (SOS) synthesis in image domain, as in GRAPPA. By using Eq. [4] and [5], this k-space data-driven method can be explicitly formulated as
| [6] |
We use R to denote an overall reconstruction matrix here. And the superscript H is used to label a Hermitian matrix (conjugate transpose). In a more generalized form, Eq. [6] can be rewritten as
| [7] |
And if we re-permute the matrices back according to the original order of all k-space samples, we will end up with
| [8] |
Here N denotes a non-zero system matrix, each row of which can null k-space samples from all coils through multiplication. This equation carries one of the fundamental forms of k-space data-driven reconstruction. Generally speaking, it implies that k-space samples from all receiver channels are essentially linearly correlated through null operations, which results from the intrinsic nature of sensitivity encodings. Once the matrix N is determinable, Eq. [8] can be utilized to solve pMRI reconstruction problem through a procedure that is termed Parallel Reconstruction Using Null Operations (PRUNO).
B. PRUNO
In this section, we will first find a more tractable form of N by directly exploring the linear dependence among local k-space samples. And it will be shown that the resulting null operations may be characterized by a group of 2D convolutions with small kernels. N can thus be feasibly derived through data-calibration. Finally, an iterative conjugate-gradient method will be presented to solve dm from Eq. [8] efficiently.
1) Deriving null operations through convolutions
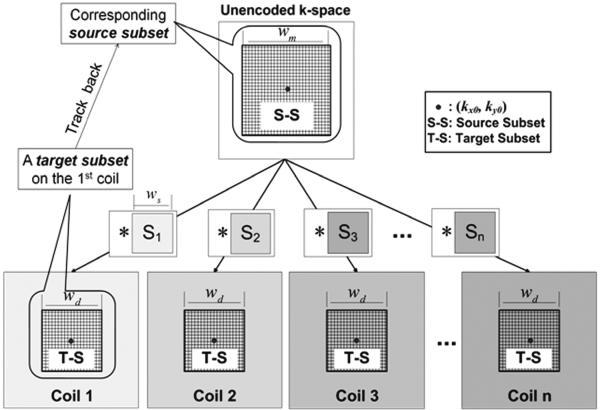
As illustrated in Eq. [1], each encoded sample dn(kx0,ky0) is correlated with m(kx, ky) locally in k-space, i.e., only for , in the unit of Δk, where Δk = 1/ FOV and FOV refers to imaging field of view. Therefore, if we look at a small square-shaped “target subset” from the n-th coil dn(kx, ky), samples of m(kx, ky) that are being encoded into these targets also completely come from a small local “source subset”, as shown in Figure 1. Apparently, this correspondence extends to every coil. If we use wd to denote the width of the target subset, the width of the source subset should be wm = wd + ws − 1.
Fig. 1.
Locality of k-space sensitivity encoding.
Top box: the unencoded k-space data. Bottom boxes: k-space data from receiver coils, which are encoded by spectra of sensitivity maps (small boxes in the middle). Each sensitivity spectrum is approximated to be band-limited with a full width of. The meshed regions in all large boxes label the target subsets (bottom) and their corresponding source subset (top), respectively. When tracking back all of the source samples that correlate to the target ones, the width of the source window is, according to the convolution theorem.
In other words, a local sensitivity encoding relationship can be formulated as
| [9] |
To differentiate from Eq. [2], we use tilde symbols to denote the small local subsets and here. is the corresponding local encoding matrix, which is by . As the size of the target subsets grow, the encoding matrix tends to be a “taller” matrix because the total number of target samples () increases much faster than that of the corresponding source samples () (see Appendix A).
According to the theorem of linear algebra, any “fat” matrix (row < column) has a nonempty null space. Therefore, , the Hermitian of the encoding matrix, would eventually have a non-empty null space if we increase the size of source subsets so that . Furthermore, at least r independent non-zero nulling kernels exist if the following equation is satisfied (see Appendix A)
| [10] |
For each of these nulling kernels, n1, n2, …, nr, . If applying these kernels to both sides of Eq. [9], we will get
| [11] |
Here , r by , turns out to be a local nulling system matrix on . Since sensitivity encoding is essentially shift-invariant in k-space, both and are independent of k-space location. Therefore, we are now able to apply the nulling kernels to all k-space locations so that the full nulling matrix N in Eq. [8], which is r Nx Ny by NcNxNy, can be constructed row by row. On the other hand, if each ni is treated as a weighting kernel, we can conclude that multiplying each ensemble of NxNy rows of N by d is equivalent to a series of 2D filtering (inside each coil) followed by a summation (across all coils) in the 2D k-space. Furthermore, N can be fully characterized by all nulling kernels, that can be feasibly estimated through data calibration.
We call these nulling kernels as PRUNO kernels. And wd is just the kernel size used in PRUNO. Based on the first assumption and empirical approximation, ws is usually estimated to be less than10 for typical imaging FOV and practical coil configurations. For example, if we approximate ws as 8 and use 8 receiver channels, it requires a minimum kernel width of 4 to obtain 8 nulling kernels according to Eq. [10]. This is a feasible number for both data calibration and reconstruction in terms of computational time and the number of required ACS lines, which will be addressed later.
Once N has been determined, the image reconstruction can be proceeded by solving dm from Eq. [8]. The is very similar to the derivation of Eq. [6]. By applying the same masking operators to Eq. [8], we get Nmdm + Nada = 0, or
| [12] |
Here Nm and Na denote the two column pruned matrices, similar to the previous notations of Sm and Sa. In a positive (semi-)definite system form, Eq. [12] turns to be
| [13] |
Essentially, this is another representation of Eq. [6].
2) Data calibration and kernel selection
As stated above, a set of local nulling kernels (columns of ) need to be found in PRUNO through data calibration. This procedure is similar to GRAPPA calibration. For each kernel, a series of linear equations can be established by sliding the kernel window within a certain k-space calibration region. And the calibration is again a linear algebra problem, which is illustrated as
| [14] |
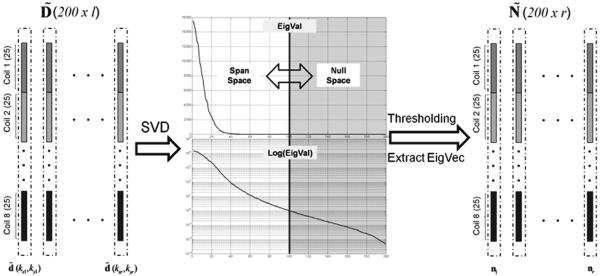
Here l denotes the total number of calibration locations. is a calibration matrix, of which each column is one windowed subset of calibration data, as shown in Figure 2.
Fig. 2.
Kernel selection strategies.
An example of SVD kernel selection with an 8-channel coil and 5×5 calibration widows (kernel size). Number of calibration vectors ( ) is around 1000. SVD was performed on the calibration matrix. The intrinsic rank of the vector span space is 200. The magnitudes and logarithms of the eigenvalues are plotted in the top figure and bottom figure, respectively. To approximate the null space of the calibration matrix, a threshold has been applied so that smaller eigenvalues are set to 0. In this example, a cutoff was roughly set in the middle so that the rank of the null space ( ) is 100. Finally, the eigenvectors with 0 eigenvalues form the null space.
To ensure Eq. [12] an overdetermined system, Nm needs to be a tall matrix, i.e., . Although Nc kernels always seem to be sufficient from this perspective, the condition of the final reconstruction system (Eq. [13]) strongly depends on the number of independent equations in Eq. [12], i.e., the number of PRUNO kernels and the orthogonality among them. On the other hand, imposing a larger number of kernels may increase the kernel size and consequently the computational time. We call the procedure of determining the number of kernels and defining the size of kernels as kernel selection.
One simple strategy of kernel selection is inspired by GRAPPA, in which a small number (usually Nc) of template based kernels are used [20]. However, the performance of this method is limited by its inflexibility and instability. An optimal kernel selection strategy is to find all generalized nulling kernels directly from Eq. [14], i.e., from the null space of . This can be easily achieved by performing a Singular Value Decomposition (SVD) on , and extracting the null space from its eigenspace [22]. One example is shown in Figure 2. In this case, a full kernel width of 5 was used on 8-channel simulated phantom data. Around 1000 k-space subsets (vectors) were collected to form the calibration matrix , with a vector length of 200 for each. SVD was then applied on this matrix. The eigenvalue plot indicates that the span of the column vector space is in a much lower rank than 200 if reasonable thresholding is applied to its eigenvalues. Once a cut-off is determined, all nulling kernels are simply eigenvectors corresponding to those eigenvalues that are smaller than the threshold. In this example, around 100 kernels were obtained.
3) Conjugate-Gradient solver
As the nulling matrix has been calculated from data calibration, the remaining objective is to solve Eq. [13]. Considering the huge matrix size, it would usually be computationally infeasible to perform a direct matrix inversion. An alternative indirect solver is thus desired. Using the representation of non-pruned matrices, the system equation can be rewritten as
| [15] |
Now that each forward matrix-vector multiplication in Eq. [15] is either simply a masking operation or essentially composed of 2D convolutions and summations, which are not computationally demanding, a feasible iterative algorithm can be implemented to solve this system. A conjugate-gradient method has been developed and demonstrated in [20], in which matrix-vector multiplications are performed step by step during the reconstruction. This is very similar to NC-SENSE reconstruction [16].
According to the construction of nulling matrix, N can be decomposed into r × Nc blocks,
| [16] |
Here each Nij is a NXNy × NXNy matrix, corresponding to the 2D convolution of the i-th null operation performed on the j-th coil. Eq. [16] illustrates that rNc convolutions are involved in each ensemble of null operations Nx, where the vector x refers to certain k-space data on that the null operations are performed. On the other hand, it is easy to show that in each ensemble of conjugate null operations NH (Nx), rNc convolutions need to be performed as well. Therefore, a total number of 2rNc 2D convolutions are required during each CG loop when applying step-by-step matrix-vector multiplications [20], which constitute the major portion of reconstruction time. In this case, the reconstruction time is nearly proportional to r, which prohibits us from using a large number of kernels in practice, for example, using more than 100 kernels.
Immediately from the decomposition of N (Eq. [16]), we have
| [17] |
This equation shows that one “composite” null operation NHNx is actually equivalent to 2D convolutions, independent of the number of kernels (r). We can thus combine the sequential null operations Nx and their conjugate operations NH (Nx) into single-step convolutions, .i.e., calculate (NHN)x directly. In this case, the representation of a local composite convolution kernel can be directly derived as:
| [18] |
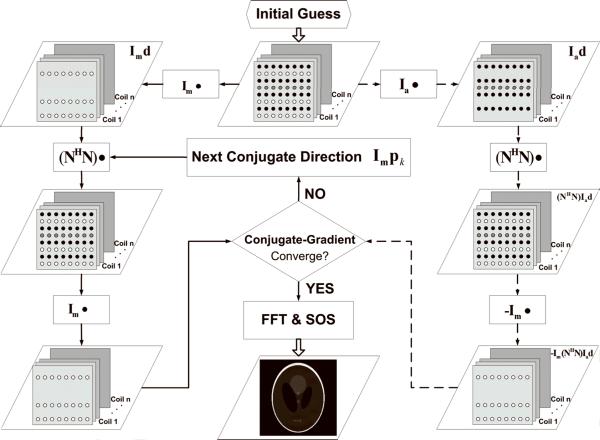
Here n and n* denote a local 2D convolution kernel and its conjugate, respectively. And η is a composite kernel, corresponding to one block in (NHN). Because these convolutions are calculated among very small kernels and only performed once prior to the CG iterations, its computational load is nearly negligible. Although the size of a composite kernel (2wd −1) is larger than that of a nulling kernel, the number of convolutions involved in each CG step will be reduced to if these composite kernels are being used. Therefore, it is favorable to use composite kernels in PRUNO reconstruction because the overall computational time is almost independent of the number of nulling kernels. Such a modified reconstruction algorithm is shown in Figure 3. Details of a CG algorithm can be found in [23] and [16].
Fig. 3.
Conjugate-gradient PRUNO reconstruction.
Black dots: acquired k-space samples; white dots: missing k-space samples; gray dots: ACS samples, which are also regarded as available samples during reconstruction. Each masking matrix (or) is used to include only one particular group of samples for the following operation. At the beginning of reconstruction, both sides of Eq. [15] are evaluated based on the initial guess, and the results are entered into the CG algorithm to calculate the residual error. Afterwards, the forward “composite” null operations will be performed on one conjugate direction ( ) in each CG step (left circle loop in the figure), until any of the stopping criteria has been satisfied. Finally, a sum-of-square image will be synthesized.
4) Summary of PRUNO reconstruction
The procedure of an optimized PRUNO reconstruction is summarized below:
Determine the size of PRUNO kernels (wd).
Construct the calibration matrix from ACS data (Eq. [14]).
Apply SVD on and choose the number of nulling kernels (r) that will be used.
Select the eigenvectors corresponding to the r smallest eigenvalues as PRUNO kernels.
Compute the composite kernels (Eq. [18]).
Provide an initial guess of k-space missing data and perform CG reconstruction (Figure 3).
Generate coil images and synthesize the final image by using SOS.
MATERIALS AND METHODS
For comparison, PRUNO and GRAPPA reconstructions have been applied to Cartesian pMRI for both computer simulated phantom and in vivo studies. A wide range of reduction factors were tested with 8-channel coils. Both data simulation and image reconstruction were implemented by using MATLAB 7.7 (The Mathworks, Inc, Natick, MA, USA) on a Linux PC with a 3.20 GHz Intel Pentium 4 CPU and 4GB RAM.
Phantom Simulations
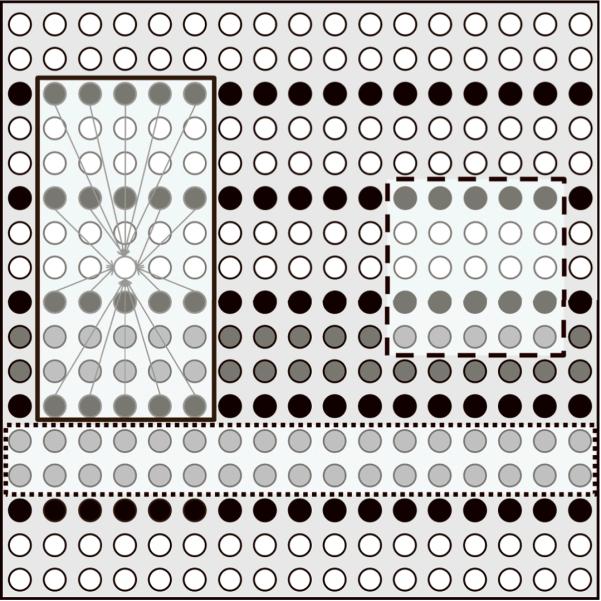
A 256×256 Shepp-Logan image was used for the simulation. Pre-measured sensitivity maps from an eight-channel receiving coil were used for sensitivity encodings [6,24–26]. Full k-space data of each coil were generated by applying fast Fourier transform to the encoded coil image, which was produced by multiplying the original image by the corresponding sensitivity map. Complex white Gaussian noises were added to the phantom image during the simulation, which corresponds to a signal to noise ratio of 25. Uniform data undersampling was then performed while preserving a small number of ACS lines near k-space center. The reduction factors were varied from 2 to 7 in these experiments. Two calibration data blocks (Nb=2), or 2 (R-1) ACS lines, were used in the cases of 2 to 4-fold. And three blocks were used if R is greater than 4. A calibration block refers to one unsampled slab in k-space, as shown in Figure 4. The exact number of ACS lines and the effective reduction factor of each study are listed in Table 1. The same k-space data were used for both GRAPPA and PRUNO calibration and reconstruction, with all ACS lines preserved during reconstruction.
Fig. 4.
GRAPPA kernel and PRUNO kernel.
A reduction factor of 3 is used in this example and k-space data from one coil is displayed. The notations of different dots are the same as Fig. 3. Red window (solid line): one of the 4×5 GRAPPA kernels (there are always (R-1) different kernels, each of which has different fitting coefficients) and the width is defined based on the acquired/fitted data (black dots) only. Green window (dashed line): a PRUNO kernel, which is always a continuous window and will be applied at all k-space locations. Blue window (dotted line): one calibration window block, which is composed of (R-1) ACS lines. Two successive calibration blocks exist in this example.
Table 1.
Configurations of PRUNO reconstructions.
| R = 2 | R = 3 | R = 4 | R = 5 | R = 6 | R = 7 | |
|---|---|---|---|---|---|---|
| # of Calibration Blocks (Nb) | 2 | 2 | 2 | 3 | 3 | 3 |
| # of ACS Lines | 2 | 4 | 6 | 12 | 15 | 18 |
| Effective Reduction Rate | 1.97 | 2.87 | 3.66 | 4.05 | 4.44 | 4.69 |
| Simulation | ||||||
| GRAPPA | ||||||
| Kernel Size | 4×5 | 4×5 | 2×5 | 4×5 | 4×5 | 4×5 |
| PRUNO | ||||||
| Kernel Width (wd) | 5 | 5 | 5 | 7 | 7 | 7 |
| # of Kernels (r) | 100 | 100 | 100 | 200 | 200 | 200 |
| Recon Time (min) | 0.5 | 1 | 3.5 | 8 | 12.5 | 12.5 |
| In vivo | ||||||
| GRAPPA | ||||||
| Kernel Size | 2×3 | 2×5 | 2×5 | 4×5 | 4×5 | |
| PRUNO | ||||||
| Kernel Width (wd) | 5 | 5 | 7 | 7 | 7 | / |
| # of Kernels (r) | 50 | 50 | 150 | 150 | 150 | / |
| Recon Time (min) | 1.5 | 2 | 4 | 9 | 15 | / |
In Vivo Experiments
In vivo brain images of a healthy volunteer were acquired on a 3T whole-body scanner (GE MR750; GE Healthcare, Waukesha, WI, USA) with an array of 8 receiver coils. A 2D FSE T2-FLAIR sequence was used to obtain axial images. In this experiment, the following scan parameters were used: FOV=24cm, TE/TR =50ms/2s, rBW(receiver bandwidth)=15.63kHz, slice-thickness=5mm, and matrix size = 256×256. Full Cartesian k-space samples were acquired from scanner without undersampling. The data were uniformly undersampled along the phase encoding direction offline, by using reduction factors ranging from 2 to 6. In each experiment, the same strategy was used to keep necessary ACS lines, as in the phantom simulation.
Image Reconstruction
Numerous parameter configurations were first tested for both reconstruction algorithms, including kernel width of GRAPPA, kernel selection of PRUNO (size and number), initial guess and stopping criteria of the CG algorithm. The stopping criterion is defined as a condition that either the relative residual RMS error is lower than an error bound or a predefined maximum loop number has been reached. In both GRAPPA and PRUNO, the ACS blocks are always preserved during the reconstruction. The effective reduction factors (Reff) of all experiments are calculated, as shown in Table 1.
For GRAPPA, we have evaluated various kernel sizes, including 2×3, 2×5, 4×3, and 4×5 for each experiment. The kernel size mentioned here is defined on acquired samples only, as shown in Figure 4. In each case, the best GRAPPA image was picked according to its minimum RMS reconstruction error. A similar scenario applies to the PRUNO kernel selection as well. The optimal PRUNO kernel selection strategies varied among different studies. Detailed optimal GRAPPA and PRUNO kernel configurations for each study are listed in Table 1. The LSQR algorithm was used for each data calibration. No other regularization was applied during the following image reconstruction so that we could compare the intrinsic reconstruction power of GRAPPA and PRUNO.
In each experiment, a GRAPPA reconstruction was performed first. The output k-space data of GRAPPA was used as a good initial guess to the following PRUNO reconstruction because it usually offers fast algorithm convergence. For all phantom experiments, the CG error bound was set to 0.01% and the maximum iteration number was 200. While for all in vivo experiments, the error bound was relaxed to be 0.1% and the maximum step size was 300.
RESULTS
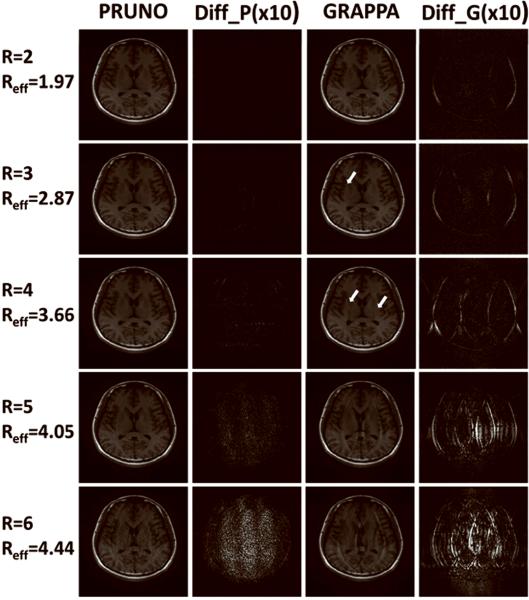
Figure 5 and Figure 6 compare PRUNO and GRAPPA reconstructions of phantom simulations and in vivo experiments, respectively. Each reconstructed image was also compared with a reference image, which was reconstructed under full k-space sampling, and the image difference (×10) was taken for evaluating the quality of this reconstruction. The actual reconstruction time of each study is listed in Table 1.
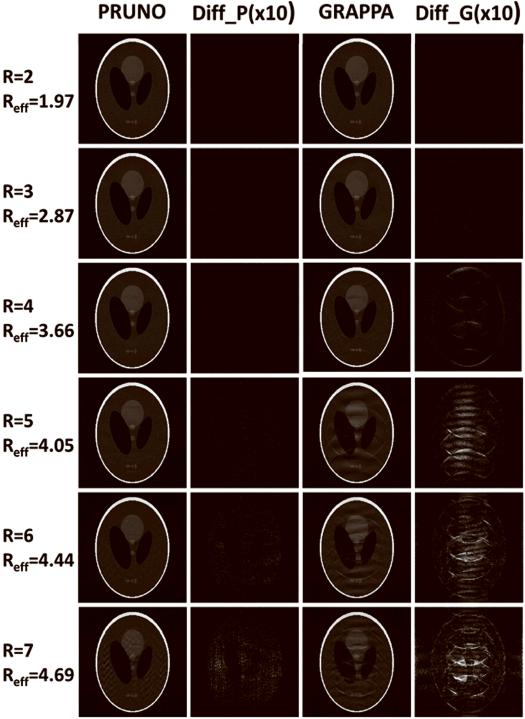
Fig. 5.
PRUNO and GRAPPA reconstructions of the phantom data.
Reduction factor ranging from 2 to 7 were used for both PRUNO and GRAPPA reconstructions. A reference sum-of-square image was also reconstructed with full k-space data, so that an exaggerated (10×) difference image was generated for each pMRI reconstruction and displayed for comparison. ACS data were always included for each reconstruction. GRAPPA reconstruction always yields larger residual errors, which can be seen from the difference images. Intolerable aliasing can be seen for GRAPPA starting from R=4; while PRUNO yields good image reconstruction up to R=5.
Fig. 6.
PRUNO and GRAPPA reconstructions of the in vivo data.
Reduction factor of 2 to 6 were used. Difference images were computed and compared at 10× as well. ACS data were included for both PRUNO and GRAPPA reconstruction. For GRAPPA, noticeable aliasing start showing up at R=3 and 4 (white arrows); and larger artifacts corrupt the reconstructed images completely as the reduction factor further increases. For PRUNO, images are consistently reconstructed with good quality as long as R<6 ( ). Besides, the reconstruction errors (aliasing) appear relatively uniformly distributed across PRUNO images, which tend to produce smaller visual artifacts.
Figure 5 shows that the reconstruction error of PRUNO is consistently smaller than that of GRAPPA, which indicates that a solution that is closer to the actual missing data has been derived in the CG algorithm by utilizing the equations established from null operations. GRAPPA performs very similar to PRUNO at R = 2, with good image quality. As the reduction factor increases, the reconstruction quality degrades for both algorithms. However, the performance of GRAPPA reconstruction drops much more rapidly than that of PRUNO. It can be seen that there are obvious unfolded aliasing artifacts in GRAPPA images as R exceeds 3. On the contrary, PRUNO is capable of producing much better images with the same data availability. Nearly no visible artifacts are observed on PRUNO images for high accelerating cases, up to R = 5. Even if the reduction factor is pushed into 6, the PRUNO reconstruction still yields reasonably good image quality. However, as R grows even further, the image aliasing can hardly be eliminated because the PRUNO reconstruction tends to be a severely ill-posed problem. Furthermore, it can be seen that the reconstruction errors and noises are distributed more uniformly in PRUNO.
Very similar phenomena are observed in the in vivo studies as well, as demonstrated in Figure 6. GRAPPA yields comparable performance to PRUNO at R = 2. Small visible aliasing artifacts are present in the GRAPPA image at R = 3. At reduction rates higher than 3, the image quality of GRAPPA reconstruction is not acceptable given the number ACS lines we have. On the other hand, PRUNO works robustly as long as the reduction factor is no greater than 5. At R = 6, some high-frequency reconstruction errors can be seen in PRUNO due to the poor system condition in Eq. [12].
DISCUSSION
We have shown that PRUNO is an iterative k-space data-driven parallel imaging reconstruction algorithm that is more robust than GRAPPA, especially for ultra-high accelerating MRI. With its generalized formulation and precise mathematical model, PRUNO offers excellent flexibility and accuracy. Our experimental results have shown that PRUNO performs fairly stable against various influencing factors such as reduction factor and low SNR. In addition, the PRUNO algorithm usually requires a smaller number of ACS lines to achieve acceptable reconstruction quality, which can help further reduce scan time.
Kernel Selection
The width and content of all kernels have an important impact to the system condition according to Eq. [12]. A good ensemble of null operations is expected to capture sufficient impendent linear equations between missing samples and acquired samples. In general, larger kernels are always favorable because more samples can be correlated in each null operation. However, using larger kernels demands more ACS lines and longer reconstruction time. On the other hand, PRUNO kernel width cannot be too small either, as addressed in Eq. [10]. Our empirical choice of kernel width is 5 to 7 for typical imaging protocols, which has been tested by using our 8-channel coils in site. At small reduction rates such as 2 or 3, a width of 5 should be used to lower the ACS requirement, without clearly degrading the performance of the algorithm. However, larger kernels are usually necessary as R gets larger, which ensures that missing samples and acquired samples are coupled with each other in a long distance (along y).
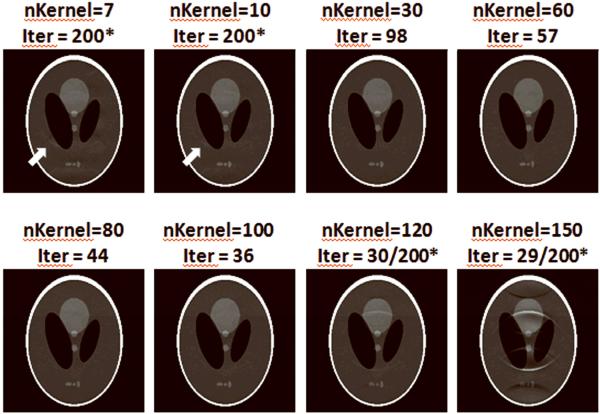
There is another tradeoff between the number of kernels (or system condition) and the accuracy of null operations resulting from these kernels. Less PRUNO kernels ensures a more strict null space, thus produces smaller residual errors. On the contrary, a larger number of nulling kernels form more equations in Eq. [12] that improves the condition of the linear system and speeds up the convergence of CG. Choosing a moderate number of kernels can thus optimize the performance of PRUNO reconstruction. In practice, however, the image quality of the final reconstruction is not quite sensitive to the number of kernels if it is chosen in a reasonable range. This can be seen from Figure 7. In this R = 4 example, we may choose the number of kernels from a really wide range without significantly affecting the reconstruction quality. But more kernels are usually preferred to speed up the reconstruction speed. To have a better convergence rate, the number of PRUNO kernels was empirically determined in our experiments, based on the eigenvalues of the calibration data. A threshold that roughly corresponds to 0.1% of the maximum eigenvalue was used in each case to separate null space and span space. Alternatively, the necessary number of kernels may also be estimated if we have a reasonable estimation of the maximum bandwidth of sensitivity maps (ws) [17], as discussed in Appendix A, such that
| [19] |
In phantom experiments, ws is approximated to be 6. In this case, Eq. [19] returns r ≤ 100 if the kernel width is 5, and it returns r ≤ 248 if the kernel width is 7. For in vivo experiments, a slightly larger ws is assumed to improve the accuracy of data calibration. If ws = 8 is used, Eq. [19] returns r ≤ 56 when wd=5, and r ≤ 196 when wd=7. All these numbers are close to our actual selections, as shown in Table 1.
Fig. 7.
Relationship between number of PRUNO kernels and image quality.
The same R=4 data set as Figure 5 was used in this case. Number of used PRUNO kernels and the total number of required iterations are labeled on top of each image. The asterisk marks the cases in with the maximum number of iterations were reached. Reconstructed images are in similar quality with moderate number of kernels (30~100). But the convergence speed increases as the number of kernels increases. If too few kernels are used (7 or 10), the algorithm may not converge well enough as the system condition is poor. In these cases, small residual aliasing can be observed (white arrows). On the other hand, using too many kernels (120 or 150) leads to inaccurate modeling. For the iteration counts in these last two cases, the first number labels the number of steps required to reach the error bound of CG. However, images shown here are generated after 200 CG steps after lowering the error bounds to verify whether it is due to insufficient looping.
Reconstruction Time
In these experiments, a PRUNO reconstruction usually takes ten to hundreds of CG steps to converge with our stopping criteria. The computational time of a single CG loop varies from 1 to 3 seconds in the actual implementations, depending on the image size, number of coils, and the width of PRUNO kernels. Therefore, a typical PRUNO reconstruction takes several minutes. The actual converging speed of the CG algorithm is determined by several factors, which have been mostly addressed already, including the reduction factor, the exact sampling pattern, the number and quality of PRUNO kernels, the initial guess of missing data, and the image noise level.
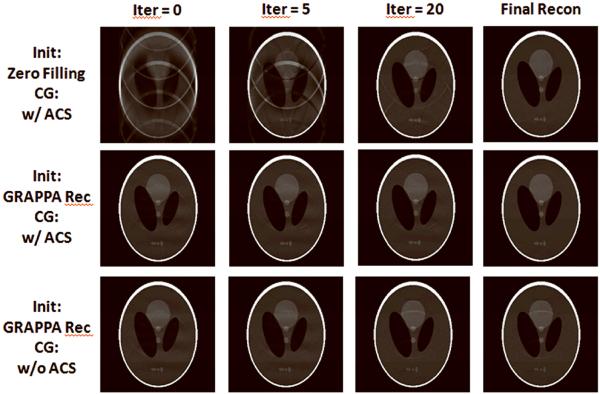
There are a few options in generating the initial guess data. The simplest strategy is to set values of all missing samples to 0's, which doesn't require any extra effort. Another straightforward strategy is to perform a certain interpolation at the missing k-space locations. More accurate estimates can be made as well, such as the approach used in our demonstrated experiments. That is, a GRAPPA reconstruction is performed first and the fitted k-space data are used as the initial guess of the PRUNO reconstruction. According to our evaluations, the choice of the initial guess does not necessarily have a significant impact onto the quality of the final image in most cases. However, a good starting guess does help improving the converging speed of the algorithm. This can be clearly observed from the top two rows of Figure 8. It would thus be worthy of generating the initial guess data through a rough GRAPPA reconstruction, as the computational time of GRAPPA is much shorter compared with that of PRUNO.
Fig. 8.
Effect of initial guess and ACS signal inclusion on PRUNO reconstruction.
The same R=4 data set as Figure 5 was used here as well. First row: zero-filling was used as the initial guess of CG. 62 iterations were taken during the reconstruction. Second row: GRAPPA reconstruction was used as the initial guess of CG. 36 iterations were taken in this case. Third row: GRAPPA reconstruction was also used as the initial but the ACS blocks were removed during CG. It can be seen that the reconstruction doesn't converge to a good solution even after 200 CG steps.
Revisit to GRAPPA
As another k-space coil-by-coil reconstruction algorithm, GRAPPA reconstruction also yields a formulation of Eq. [6]. In GRAPPA, the reconstruction matrix R itself is assumed to be sparse and in a shift-invariant pattern, so that it can be directly approximated from data calibration. In this case, each missing k-space sample is fitted from multiple neighboring acquired samples, which is equivalent to applying a template based PRUNO kernel. Therefore, GRAPPA requires that there exists a nulling kernel for each particular template. For small reduction rates, this fitting model may be a good approximation since the target sample and the fitting samples are close to each other, so that there would be strong linear correlation between them. However, as the reduction rate goes higher, the distance between target sample and fitting samples increases as well. Consequently, correlation between these samples becomes weaker and the template based nulling kernel may not be applicable any longer. To obtain an accurate fitting kernel, many more fitting samples must be involved (larger kernels) which requires a larger number of ACS lines. In addition, the distances between fitting samples and the target differ significantly among each other under high acceleration rates. Because the k-space data exhibit exponential decay in distance, the data calibration in GRAPPA would tend to be a highly ill-posed problem. These factors make GRAPPA an invalid reconstruction algorithm for ultra-high accelerating pMRI.
On the contrary, PRUNO always uses a local convolution kernel regardless of the reduction factor. With a proper kernel size, each nulling kernel in PRUNO yields better fitting accuracy due to the greater number of involved samples and better locality. With iterative reconstruction, each missing sample will eventually be fitted by using all available k-space data. This explains why PRUNO always performs better than GRAPPA, especially under high reduction rates.
However, unlike in GRAPPA, ACS blocks should be always added into the PRUNO reconstruction. The existence of a continuous ACS band in CG not only helps increase the SNR of the reconstruction, but also significantly improves the condition of the entire linear system. Otherwise, the original under sampling pattern may easily lead Eq. [12] into a seriously ill-posed system. In other words, the ACS samples are essentially strong and important constraint conditions when solving this linear system by using a least square approach. This effect can be clearly seen if we compare the second and third rows of Figure 8.
Future Directions
PRUNO doesn't require any regularization during either data calibration or image reconstruction. However, a proper regularization during CG loops might further improve the performance of the algorithm, by either improving the image quality or achieving faster converging rate. For instance, the noise-like high-frequency reconstruction errors observed in the R = 6 case of in vivo study (Figure 6) are expected to be suppressed if the CG algorithm is combined with an image domain smoothness constraint, such as total variance (TV) regularization [27]. Hench, an even higher reduction factor might be achieved with regularized PRUNO reconstruction. This is beyond the scope of this work and will be investigated in our future research.
Only 1D-PRUNO has been discussed in this work. However, there is not any restriction about the undersampling pattern in the formulation of PRUNO. It hence suggests that PRUNO reconstruction could also deal with any 2D undersampling problem. Although issues such as system condition of 2D-PRUNO and optimal 2D sampling scheme still need to be investigated, we believe PRUNO will offer many opportunities in ultra-fast 3D parallel imaging as well.
CONCLUSION
In this work, an iterative parallel imaging reconstruction algorithm termed PRUNO has been demonstrated, which is based on a generalized formulation of k-space data-driven pMRI reconstruction. Both simulation and in vivo studies have shown that PRUNO offers better image quality than GRAPPA with very light ACS requirement, especially under high accelerating rates. Data calibration and image reconstruction in PRUNO are both tractable linear algebra problems and hence easy to implement. With its robustness and flexibility, PRUNO is expected to be applied to various clinical applications to offer improved pMRI reconstruction.
ACKNOWLEDGEMENTS
The authors thank Dr. Jing Chen for proofreading the manuscript.
Grant Sponsors: National Institute of Health, Grant number R00EB007182, R01NS04760705 and NCRR P41 RR09784; Center of Advanced MR Technology of Stanford, Lucas Foundation.
APPENDIX
k-Space Locality and Existence of Nulling Kernels
If we use Ld and Lm to denote the vector length of and in Eq. [9] respectively, we will have
and
Apparently, if multiple receiver coils are used, i.e., Nc > 1, Ld increases more rapidly than Lm as wd gets larger. In another word, we may make Ld > Lm if wd is sufficiently large. In this case, (Lm × Ld) is a flat matrix, having a non-empty null space. If we denote r0 as the rank of its null space, according to the rank-nullity theorem, we will have
Therefore, existence of r independent non-zero nulling kernels in Eq. [11] requires that
| [A.1] |
This is exactly Eq. [19]. We can also solve wd from this equation to get Eq. [10].
REFERENCES
- 1.Tsao J, Hansen MS, Kozerke S. Accelerated parallel imaging by transform coding data compression with k-t SENSE. Conf Proc IEEE Eng Med Biol Soc. 2006;1:372. doi: 10.1109/IEMBS.2006.259751. [DOI] [PubMed] [Google Scholar]
- 2.Bauer JS, Banerjee S, Henning TD, Krug R, Majumdar S, Link TM. Fast high-spatial-resolution MRI of the ankle with parallel imaging using GRAPPA at 3 T. AJR Am J Roentgenol. 2007;189(1):240–245. doi: 10.2214/AJR.07.2066. [DOI] [PubMed] [Google Scholar]
- 3.Quick HH, Vogt FM, Maderwald S, Herborn CU, Bosk S, Gohde S, Debatin JF, Ladd ME. High spatial resolution whole-body MR angiography featuring parallel imaging: initial experience. Rofo. 2004;176(2):163–169. doi: 10.1055/s-2004-817623. [DOI] [PubMed] [Google Scholar]
- 4.Fautz HP, Honal M, Saueressig U, Schafer O, Kannengiesser SA. Artifact reduction in moving-table acquisitions using parallel imaging and multiple averages. Magn Reson Med. 2007;57(1):226–232. doi: 10.1002/mrm.21117. [DOI] [PubMed] [Google Scholar]
- 5.Winkelmann R, Bornert P, Dossel O. Ghost artifact removal using a parallel imaging approach. Magn Reson Med. 2005;54(4):1002–1009. doi: 10.1002/mrm.20640. [DOI] [PubMed] [Google Scholar]
- 6.Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med. 1999;42(5):952–962. [PubMed] [Google Scholar]
- 7.Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA) Magn Reson Med. 2002;47(6):1202–1210. doi: 10.1002/mrm.10171. [DOI] [PubMed] [Google Scholar]
- 8.McKenzie CA, Ohliger MA, Yeh EN, Price MD, Sodickson DK. Coil-by-coil image reconstruction with SMASH. Magn Reson Med. 2001;46(3):619–623. doi: 10.1002/mrm.1236. [DOI] [PubMed] [Google Scholar]
- 9.Sodickson DK, Griswold MA, Jakob PM. SMASH imaging. Magn Reson Imaging Clin N Am. 1999;7(2):237–254. vii–viii. [PubMed] [Google Scholar]
- 10.Jakob PM, Griswold MA, Edelman RR, Sodickson DK. AUTO-SMASH: a self-calibrating technique for SMASH imaging. SiMultaneous Acquisition of Spatial Harmonics. MAGMA. 1998;7(1):42–54. doi: 10.1007/BF02592256. [DOI] [PubMed] [Google Scholar]
- 11.Heidemann RM, Griswold MA, Haase A, Jakob PM. VD-AUTO-SMASH imaging. Magn Reson Med. 2001;45(6):1066–1074. doi: 10.1002/mrm.1141. [DOI] [PubMed] [Google Scholar]
- 12.Bydder M, Larkman DJ, Hajnal JV. Generalized SMASH imaging. Magn Reson Med. 2002;47(1):160–170. doi: 10.1002/mrm.10044. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Klugel K. Parrel acquisition techniques with modified SENSE reconstruction (mSENSE). 1st Workshop on Parallel MRI: Basics and Clinical Applications; Wurzberg, Germany. 2001. p. 92. [Google Scholar]
- 14.Griswold MA, Jakob PM, Nittka M, Goldfarb JW, Haase A. Partially parallel imaging with localized sensitivities (PILS) Magn Reson Med. 2000;44(4):602–609. doi: 10.1002/1522-2594(200010)44:4<602::aid-mrm14>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 15.Kyriakos WE, Panych LP, Kacher DF, Westin CF, Bao SM, Mulkern RV, Jolesz FA. Sensitivity profiles from an array of coils for encoding and reconstruction in parallel (SPACE RIP) Magn Reson Med. 2000;44(2):301–308. doi: 10.1002/1522-2594(200008)44:2<301::aid-mrm18>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 16.Pruessmann KP, Weiger M, Bornert P, Boesiger P. Advances in sensitivity encoding with arbitrary k-space trajectories. Magn Reson Med. 2001;46(4):638–651. doi: 10.1002/mrm.1241. [DOI] [PubMed] [Google Scholar]
- 17.Liu C, Bammer R, Moseley ME. Parallel imaging reconstruction for arbitrary trajectories using k-space sparse matrices (kSPA) Magn Reson Med. 2007;58(6):1171–1181. doi: 10.1002/mrm.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu C, Zhang J, Moseley ME. Auto-Calibrated Parallel Imaging Reconstruction Using K-Space Sparse Matrices (KSPA). ISMRM 16th Annual Meeting; Toronto, Canada. 2008. p. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brau AC, Beatty PJ, Skare S, Bammer R. Comparison of reconstruction accuracy and efficiency among autocalibrating data-driven parallel imaging methods. Magn Reson Med. 2008;59(2):382–395. doi: 10.1002/mrm.21481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Liu C, Moseley ME. Parallel Reconstruction Using Null Operations (PRUNO). ISMRM 16th Annual Meeting; Toronto, Canada. 2008. p. 9. [Google Scholar]
- 21.Griswold MA, Blaimer M, Breuer F, Heidemann RM, Mueller M, Jakob PM. Parallel magnetic resonance imaging using the GRAPPA operator formalism. Magn Reson Med. 2005;54(6):1553–1556. doi: 10.1002/mrm.20722. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Liu C, Moseley ME. Generalized PRUNO Kernel Selection by Using Singular Value Decomposition (SVD). ISMRM 18th Annual Meeting; Stockholm, Sweden. 2010. p. 4907. [Google Scholar]
- 23.Magnus RH, Eduard S. Methods of Conjugate Gradients for Solving Linear Systems. Journal of Research of the National Bureau of Standards. 1952;49(6):28. [Google Scholar]
- 24.Lin FH, Chen YJ, Belliveau JW, Wald LL. A wavelet-based approximation of surface coil sensitivity profiles for correction of image intensity inhomogeneity and parallel imaging reconstruction. Hum Brain Mapp. 2003;19(2):96–111. doi: 10.1002/hbm.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bydder M, Larkman DJ, Hajnal JV. Combination of signals from array coils using image-based estimation of coil sensitivity profiles. Magn Reson Med. 2002;47(3):539–548. doi: 10.1002/mrm.10092. [DOI] [PubMed] [Google Scholar]
- 26.Griswold MA, Breuer F, Blaimer M, Kannengiesser S, Heidemann RM, Mueller M, Nittka M, Jellus V, Kiefer B, Jakob PM. Autocalibrated coil sensitivity estimation for parallel imaging. NMR Biomed. 2006;19(3):316–324. doi: 10.1002/nbm.1048. [DOI] [PubMed] [Google Scholar]
- 27.Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58(6):1182–1195. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- 28.Blaimer M, Breuer F, Mueller M, Heidemann RM, Griswold MA, Jakob PM. SMASH, SENSE, PILS, GRAPPA: how to choose the optimal method. Top Magn Reson Imaging. 2004;15(4):223–236. doi: 10.1097/01.rmr.0000136558.09801.dd. [DOI] [PubMed] [Google Scholar]
- 29.Blaimer M, Breuer FA, Mueller M, Seiberlich N, Ebel D, Heidemann RM, Griswold MA, Jakob PM. 2D-GRAPPA-operator for faster 3D parallel MRI. Magn Reson Med. 2006;56(6):1359–1364. doi: 10.1002/mrm.21071. [DOI] [PubMed] [Google Scholar]
- 30.Breuer FA, Kannengiesser SA, Blaimer M, Seiberlich N, Jakob PM, Griswold MA. General formulation for quantitative G-factor calculation in GRAPPA reconstructions. Magn Reson Med. 2009;62(3):739–746. doi: 10.1002/mrm.22066. [DOI] [PubMed] [Google Scholar]
- 31.Heidemann RM, Griswold MA, Seiberlich N, Kruger G, Kanneng SA, Kiefer B, Wiggins G, Wald LL, Jakob PM. Direct parallel image reconstructions for spiral trajectories using GRAPPA. Magn Reson Med. 2006;56(2):317–326. doi: 10.1002/mrm.20951. [DOI] [PubMed] [Google Scholar]
- 32.Heidemann RM, Griswold MA, Seiberlich N, Nittka M, Kannengiesser SA, Kiefer B, Jakob PM. Fast method for 1D non-cartesian parallel imaging using GRAPPA. Magn Reson Med. 2007;57(6):1037–1046. doi: 10.1002/mrm.21227. [DOI] [PubMed] [Google Scholar]
- 33.Huo D, Wilson D. Using the Perceptual Difference Model (PDM) to Optimize GRAPPA Reconstruction. Conf Proc IEEE Eng Med Biol Soc. 2005;7:7409–7412. doi: 10.1109/IEMBS.2005.1616224. [DOI] [PubMed] [Google Scholar]
- 34.Huo D, Wilson DL. Robust GRAPPA reconstruction and its evaluation with the perceptual difference model. J Magn Reson Imaging. 2008;27(6):1412–1420. doi: 10.1002/jmri.21352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kreitner KF, Romaneehsen B, Krummenauer F, Oberholzer K, Muller LP, Duber C. Fast magnetic resonance imaging of the knee using a parallel acquisition technique (mSENSE): a prospective performance evaluation. Eur Radiol. 2006;16(8):1659–1666. doi: 10.1007/s00330-006-0288-0. [DOI] [PubMed] [Google Scholar]
- 36.Lin FH, Kwong KK, Belliveau JW, Wald LL. Parallel imaging reconstruction using automatic regularization. Magn Reson Med. 2004;51(3):559–567. doi: 10.1002/mrm.10718. [DOI] [PubMed] [Google Scholar]
- 37.Lustig M, Pauly JM. Iterative GRAPPA: a General Solution for the GRAPPA Reconstruction from Arbitrary k-Space Sampling. Berlin, Germany: 2007. p. 333. [Google Scholar]
- 38.Zhao T, Hu X. Iterative GRAPPA (iGRAPPA) for improved parallel imaging reconstruction. Magn Reson Med. 2008;59(4):903–907. doi: 10.1002/mrm.21370. [DOI] [PubMed] [Google Scholar]
- 39.Sodickson DK. Tailored SMASH image reconstructions for robust in vivo parallel MR imaging. Magn Reson Med. 2000;44(2):243–251. doi: 10.1002/1522-2594(200008)44:2<243::aid-mrm11>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 40.Sodickson DK, McKenzie CA, Ohliger MA, Yeh EN, Price MD. Recent advances in image reconstruction, coil sensitivity calibration, and coil array design for SMASH and generalized parallel MRI. MAGMA. 2002;13(3):158–163. doi: 10.1007/BF02678591. [DOI] [PubMed] [Google Scholar]