Abstract
Enterocin is an atypical type II polyketide synthase (PKS) product from the marine actinomycete “Streptomyces maritimus”. The enterocin biosynthesis gene cluster (enc) codes for proteins involved in the assembly and attachment of the rare benzoate primer that initiates polyketide assembly with the addition of seven malonate molecules and culminates in a Favorskii-like rearrangement of the linear poly-β-ketone to give its distinctive non-aromatic, caged core structure. Fundamental to enterocin biosynthesis, which utilizes a single acyl carrier protein (ACP), EncC, for both priming with benzoate and elongating with malonate, involves maintaining the correct balance of acyl-EncC substrates for efficient polyketide assembly. Here we report the characterization of EncL as a type II thioesterase that functions to edit starter unit (mis)priming of EncC. We performed a series of in vivo mutational studies, heterologous expression experiments, in vitro reconstitution studies, and Fourier-transform mass spectrometry-monitored competitive enzyme assays that together support the proposed selective hydrolase activity of EncL toward misprimed acetyl-ACP over benzoyl-ACP to facilitate benzoyl priming of the enterocin PKS complex. While this system resembles the R1128 PKS that also utilizes an editing thioesterase (ZhuC) to purge acetate molecules from its initiation module ACP in favor of alkylacyl groups, the enterocin system is distinct in its usage of a single ACP for both priming and elongating reactions with different substrates.
1. Introduction
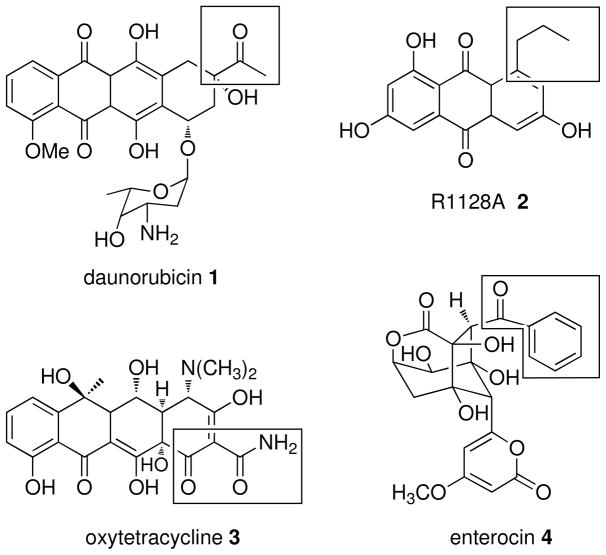
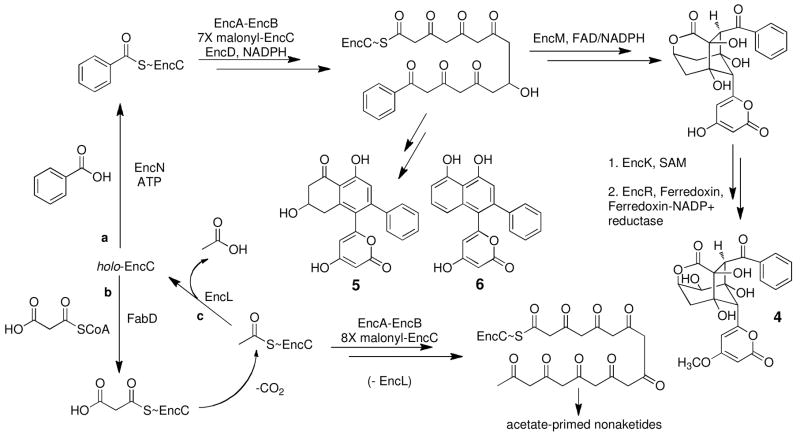
Bacterial type II polyketide synthases (PKSs) initiate the synthesis of polyaromatic antibiotics and other natural products through the iterative condensation of malonyl-CoA molecules.1, 2 Prototypical examples include the streptomycete antibiotics actinorhodin and tetracenomycin. On occasion, type II PKSs initiate polyketide assembly with primer molecules other than acetate derived from the decarboxylation of PKS-bound malonate to achieve structure diversity.3 Alternative primers include aliphatic residues such as propionate and butyrate as in the anticancer agent daunorubicin4 1 and the non- steroidal estrogen receptor antagonist R1128A5, 6 2, respectively, malonamate as in the antibiotic oxytetracycline7 3, and benzoate per the marine antibiotic enterocin8, 9 4 (Figure 1).
Figure 1.
Examples of nonacetate-primed type II PKS products with respective starter units boxed.
The bacteriostatic agent enterocin 4 from “Streptomyces maritimus” is a distinctive type II PKS product known for its unusual rearranged, non-aromatic core structure.8, 10 Enterocin biosynthesis is encoded by the 20 open reading frame enc gene cluster that harbors the centrally located minimal PKS genes encABC, which code for the EncA-EncB ketosynthase (KS) heterodimer and the acyl carrier protein (ACP) EncC8 (Figure 2a). We previously reported the operation of two independent biosynthetic pathways to the enterocin benzoate starter unit in “S. maritimus” involving an endogenous pathway from L-phenylalanine via cinnamic acid11, 12 and an exogenous pathway from benzoic acid proceeding via a type II nonribosomal peptide synthetase (NRPS)-like priming mechanism catalyzed by the benzoate:ACP ligase EncN.9 As with other type II PKS pathways that incorporate nonacetate starter units, the enc cluster encodes the malonyl-CoA:ACP acyltransferase (MCAT; FabD) homolog EncL that was originally suggested to facilitate the attachment of the dedicated benzoate primer.8 In the case of R1128A 2 biosynthesis, however, the EncL homolog ZhuC was reported to serve as an editing thioesterase (TE) toward misprimed acetyl-ACP in favor of longer chain acyl-ACP primer units.13 Monofunctional type II TEs are common in thiotemplate modular PKSs14, 15 and NRPSs16, 17 where they were first shown to function as repair enzymes to clear stalled intermediates and regenerate functional holo-carrier proteins.18 We thus set out to explore whether EncL serves a similar editing role in the enterocin PKS system, which is distinct from the R1128 PKS system that harbors two ACPs, one for initiation with an alkyl-CoA and the second for iterative extension with malonyl-CoA. The enterocin PKS system, on the other hand, operates with a single ACP, EncC,19 that must function to carry both priming benzoate and extending malonate molecules without being inhibited by acetate that may originate from the decarboxylation of malonate or via the direct transfer from acetyl-CoA. We show here that EncL is a critical repair enzyme that selectively hydrolyzes misprimed acetate molecules from EncC to facilitate benzoate priming in the enterocin system.
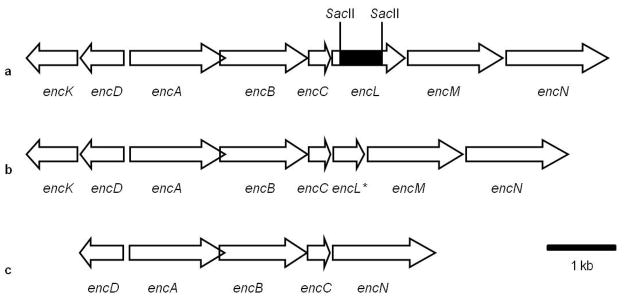
Figure 2.
Partial maps of the enterocin (enc) biosynthesis gene cluster a) in wild-type “S. maritimus” (showing location of 689-bp SacII deletion), b) in the gene knockout mutant “S. maritimus” XL, and c) inserted into heterologous expression plasmid pJK494. Highlighted enc gene functions: encK, methyltransferase; encD, ketoreductase; encA, ketosynthase; encB, chain length factor; encC, acyl carrier protein (ACP); encL, type II thioesterase; encM, favorskiiase flavoprotein; encN, benzoate:ACP ligase.
2. Results
2.1 Probing the in vivo role of EncL
Analysis of enterocin in vivo biosynthesis in wild-type and mutant strains of “S. maritimus” along with expression hosts carrying enc pathway genes previously revealed the faithful production of benzoate-primed pathway products.20, 21 This observation, however, contrasts the in vitro reconstituted pathway in which acetate-primed nonaketides of unknown composition are also produced.19 One of the key differences in the design of these experiments was that EncL was present in all of the prior in vivo experiments yet absent from all the previously reported in vitro experiments. To explore whether the putative acyltransferase EncL functions as an editing type II TE to police benzoate priming, we first explored its in vivo activity. Disruption of encL by double crossover homologous recombination via previously established methodology employing pKC1139-mediated conjugal transfer11 resulted in a 689 bp in-frame deletion of encL to provide the mutant strain “S. maritimus” XL (Figure 2). Fermentation of the ΔencL mutant and wild-type strains and subsequent chromatographic analyses of the organic extracts revealed similar levels of enterocin production, and thereby confirmed that the encL gene product does not function as a dedicated biosynthetic enzyme as originally posited.8
This observation was corroborated by in vivo expression studies in which we synthesized in vivo benzoate-primed polyketides in the absence of encL. The expression plasmid pJK494 harboring the gene set encABCND (Figure 2c) was constructed in the E. coli/Streptomyces shuttle plasmid pSEK4 to give the minimal gene set required for benzoate-primed polyketide production. Heterologous expression of pJK494 in the host S. lividans K4-114 and subsequent growth in benzoate-supplemented media under thiostrepton selection resulted in the production of the benzoate-primed polyketides wailupemycins F 5 and G 6. This result paralleled our earlier observation involving the expression of the encL-containing gene cassette encABCLMND that similarly yielded the wailupemycins.
2.2 Probing the in vitro role of EncL
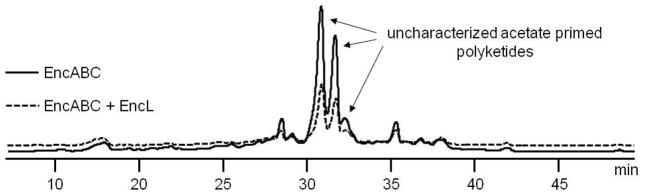
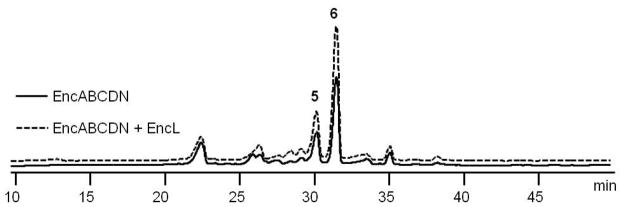
We next explored the in vitro activity of recombinant EncL, which was produced as an octahistidyl-tagged fusion protein in Escherichia coli. A series of in vitro enzymatic assays were performed in which we reconstituted the minimal enterocin PKS in the presence and absence of EncL and evaluated relative polyketide production levels (Table 1). Incubation of EncL with the minimal PKS enzymes EncABC and Streptomyces glaucescens FabD (SgFabD) in the presence of malonyl-CoA resulted in the production of the same set of acetate-primed polyketides, albeit in lower amounts, in comparison to the enzyme assay without EncL.19 Production levels decreased by approximately 40% as determined by analytical HPLC (Figure 3). Upon the addition of the ketoreductase EncD and the benzoate-priming machinery of EncN in the presence of ATP, MgCl2, benzoate and NADPH to the minimal enterocin PKS assay, we measured a distinct increase in the production of the benzoate-primed wailupemycin polyketides 5 and 6 when the assay was run with EncL (Figure 4). Together, these results strongly suggest that EncL suppresses acetate priming on EncC in favor of benzoate priming. In order to explore this hypothesis in greater detail, we next monitored the enzymatic activity of EncL by the Fourier-transform mass spectrometry (FT-MS) analysis of products derived from a series of competitive in vitro assays.
Table 1.
Polyketide products of enzymatic assays
| Enzymesa | Substrates and Cofactorsb | Polyketide Productionc |
|---|---|---|
| EncABC | malonyl-CoA | Acetate-primed enc products |
| EncABCL | malonyl-CoA | Acetate-primed enc products – relative production decreased in presence of EncL |
| EncABCDN | malonyl-CoA, benzoate, ATP/Mg2+, NADPH | Benzoate-primed enc products 5 and 6 |
| EncABCDNL | malonyl-CoA, benzoate, ATP/Mg2+, NADPH | Benzoate-primed enc products 5 and 6 – relative production increased in presence of EncL |
0.2 μM SgFabD (malonyl-CoA:ACP transacylase) included in each assay.
5 mM malonyl-CoA included in each assay.
Polyketide production determined by analytical HPLC. Acetate-primed enc products refer to the uncharacterized nonaketides, and benzoate-primed enc products in this case refer to wailupemycins F 5 and G 6. Enzyme functions EncA–EncB (ketosynthase–chain length factor), holo-EncC (acyl carrier protein (ACP)), EncD (ketoreductase), EncN (benzoate:ACP ligase) and EncL (type II thioesterase).
Figure 3.
HPLC analysis of polyketide products formed from malonyl-CoA and the biosynthesis proteins EncABC (solid line) and with the addition of EncL (dashed line) showing the relative reduction of acetate-primed polyketides in the presence of the EncL TE.
Figure 4.
HPLC analysis of polyketide products formed from benzoyl-CoA, malonyl-CoA and the biosynthesis proteins EncABCN (solid line) and with the addition of EncL (dashed line) showing the relative increase in benzoate-primed 5 and 6 in the presence of the EncL TE.
2.3 FT-MS interrogation of EncL thioesterase activity
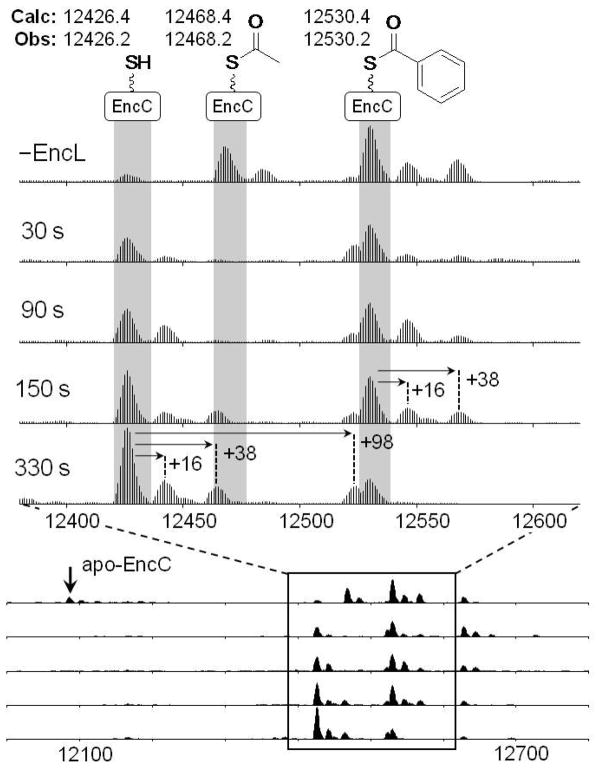
To interrogate whether EncL functions as an anticipated hydrolase and to evaluate its substrate specificity, we next enzymatically synthesized acetyl-EncC and benzoyl-EncC9 and incubated them in equimolar amounts in the presence of EncL. Upon monitoring the real-time reaction by FT-MS, we observed the rapid and selective loss of acetyl-EncC (12468.4 Da) with concomitant production of holo-EncC (12426.4 Da), which is consistent with the removal of the acetyl group (Figure 5). In contrast, benzoyl-EncC (12530.4 Da) remained unchanged during the same time period and thereupon only hydrolyzed at ~10 times slower rate to holo-EncC. These data clearly showed that EncL preferentially hydrolyzes acetyl-EncC over benzoyl-EncC, thereby strongly supporting an editing role in starter unit selection and maintenance whereupon misprimed acetyl-EncC is removed from circulation and holo-EncC is regenerated for benzoylation by EncN and malonation by FabD (Scheme 1).
Figure 5.
Qualitative time course of acetyl- and benzoyl-EncC incubated with EncL at room temperature. Shown are the deconvoluted LTQ-orbitrap mass spectra of the reaction mixture before the addition of EncL (−EncL) and at the indicated times after the addition of EncL. Indicated in Da are the theoretical (Calc) and experimentally observed (Obs) monoisotopic masses of holo-EncC, acetyl-EncC, and benzoyl-EncC. The acetyl-loaded form of EncC was depleted already 30 seconds after the addition of EncL with a corresponding increase of holo-EncC. In contrast, benzoyl-EncC is hydrolyzed relatively slowly. The concentration of each acyl-EncC form was ~20 μM, while the concentration of EncL was ~0.5 μM. Some amount of apo-EncC (calculated mass: 12086.3 Da, observed 12086.1 Da) was clearly visible (vertical arrow) in the control sample without EncL but not in the other samples, probably because of phosphopanteteinylation of apo-EncC by Sfp with residual acyl-CoA. Masses of other strong bands visible in the spectra are consistent with phosphate adducts (+98 Da), potassium adducts (+38), and oxidized forms (+16) of the main species indicated, as shown by the horizontal arrows.
Scheme 1.
Enzymatic biosynthesis of enterocin (4) and the wailupemycins (5 and 6). Assembly of benzoyl-EncC and 5 and 6 from benzoic acid and holo-EncC via route a. Nonaketide assembly from malonyl-CoA and holo-EncC via route b. The editing (hydrolysis) of acyl-EncC to holo-EncC by EncL proposed to occur via route c.
3. Discussion
EncL belongs to a family of FabD homologs that are commonly encoded in type II PKS gene clusters associated with nonacetate-primed polyketide products such as 1–4. Two general pathways for nonacetate priming have emerged. The first involves alkylacyl groups such as those in daunorubicin4 1, R1128A5, 6 2, frenolicin,6 and hedamycin.22, 23 These pathways include an additional initiation module specific for starter unit selection that often contain a dedicated ACP distinct from the minimal PKS-associated ACP required for chain elongation. The second group includes oxytetracycline 3 and enterocin 4 that recycle the same ACP for initiation and elongation. Germane to all these pathways is the FabD homolog, which was first shown in the case of 2 with ZhuC to serve as an editing type II TE,13 much like that observed in modular type I PKS and NRPS enzyme systems.14–18 This editing functional role is presumed for other members of the type II PKS-associated family such as DpsD in 1 biosynthesis.24 While DpsD was found to be nonessential for propionate priming, much like the case for EncL in benzoate priming, its absence together with the β-ketoacyl:ACP synthase III DpsC resulted in increased in vivo levels of the acetate-primed desmethylaklanonic acid.25 The production of propionate-primed aklanonic acid is proposed to result from naturally high concentrations of propionyl-CoA in the bacterium and the affinity of the KS heterodimer for propionyl-CoA.25 DpsD is proposed to physically inhibit acetate priming through its strong interaction with the KSα subunit DpsA and may additionally coordinate other subunits of the PKS complex, namely the ketoreductase DpsE and the cyclase DpsY, into close proximity to streamline downstream modifications of the polyketide backbone.26 This scenario may account for the increased in vitro production of benzoate-primed 5 and 6 in the presence of EncL given the absolute requirement of the enterocin ketoreductase EncD in benzoate-primed polyketide biosynthesis.21 In contrast, the deletion of encL in the null mutant had little effect on the production of enterocin and thus it could be presumed that the structural rigidity of the enterocin PKS in the mutant and the wild type strains relative to the in vitro enzyme assay more than compensates for the absence of EncL if indeed its role is also structural. More likely however, DpsD, like ZhuC and EncL, is also a type II TE. The data presented here have allowed us to now fully account for all enc PKS biosynthesis genes in the gene cluster and further our understanding of benzoate priming by the enterocin PKS.
4. Experimental
4.1 Materials
Cosmid pJP15F11,8 which harbors the entire enc biosynthesis gene cluster, was used as the initial source of DNA in the construction of expression plasmids. Restriction enzyme-digested and PCR-amplified DNA fragments were recovered from agarose gel using Qiaquick DNA Purification Kit (Qiagen). Escherichia coli strains XL1-Blue and DH5α served for routine cloning. The temperature sensitive E. coli-streptomycete conjugal transfer vector pKC113927 was used for the construction of the encL deletion mutant. Streptomyces lividans K4-11428 was used as the host for heterologous plasmid (pSEK4-based) expression. pSEK429, 30 was obtained from Professor Chaitan Khosla (Stanford University), and Streptomyces lividans K4-114 was a gift from Kosan Biosciences. E. coli strain BL21(λDE3), Streptomyces lividans TK24 and S. coelicolor YU10531 were used for heterologous protein expression. The expression plasmid pHIS832 was provided by Professor Joseph Noel (Salk Institute for Biological Studies). Enterocin biosynthesis enzymes EncAB, EncC (apo and holo derivatives), EncD, and EncN were prepared as previously described.9, 19 FabD from S. glaucescens was prepared as a polyhistidyl-tagged recombinant protein as previously described,33 and the plasmid encoding hexahistidyl-tagged SgFabD was provided by Professor K. A. Reynolds (Portland State University). Commercially available chemicals were purchased from Sigma as the highest available grade unless stated otherwise.
4.2 General methods
DNA isolation, plasmid preparation, restriction digests, PCR, gel electrophoresis and ligation reactions were conducted according to standard methods.34, 35 The fermentation of wild-type and mutant strains of “S. maritimus” and the streptomycete heterologous host strains, together with their organic extraction and HPLC analysis, followed previously established methods.21
4.3 Construction and characterization of the encL deletion mutant “S. maritimus” XL
This mutant was obtained by gene disruption as follows. A 5.6-kb BamHI fragment containing genes encABCLMN from the cosmid pJP15F11 was subcloned into pGEM.7ZF (Pharmacia) to make pBM30. The 2.0-kb NotI-EcoRV fragment containing encBCL and the 2.8-kb NotI-NdeI containing encBCLM from pBM30 were subcloned into pCRR2.1-TOPOR (Invitrogen) to create pBM31 and pBM32, respectively. After the 689-bp SacII fragment of encL was deleted (deletion shown in Figure 2a) from the 2.0-kb insert in pBM31 by sequential digestion of NotI and SacII, the resultant 1.2-kb NotI-SacII fragment containing encBCL* and its vector were ligated to make pBM33. The 1.3-kb NotI-EcoRV from pBM33 replaced the analogous 2.0-kb fragment in pBM32 to create pBM34. The 2.1-kb NotI-NdeI DNA fragment containing encBCL*M from pBM34 replaced the 2.8-kb fragment in pBM30 to create pBM35. The 4.9-kb BamHI fragment containing genes encABCL*MN from pBM35 was cloned into pKC1139 to create pBM36, which was introduced into “S. maritimus” by conjugal transfer as described for pBM2421 to yield “S. maritimus” XL (Figure 2b).
Genomic DNAs from apramycin sensitive colonies were PCR amplified with the primers 5′-GTCGTCGGGCACGCGCGCCAC-3′ (forward) and 5′-GTTAATTAACTGAGGCATCCGTCCGCCC-3′ (reverse). 2.4-kb PCR products were indicative of revertants (wild type) carrying a complete encL gene (in the amplified encLM cassette), whereas 1.7-kb products corresponded to DNA fragments carrying the truncated encL gene (in the amplified encLM cassette) from the mutant.
4.4 Construction of the expression plasmid
pJK494 pJK494 is a derivative of pSEK4 in which encD was cloned into the PacI/HindIII site downfield of the actIII promoter, and encABCN was cloned into the PacI/EcoRI site downfield of the actI promoter. A 2.8-kb fragment harboring encABC was PCR amplified from the cosmid clone pJP15F11 using primers 5′-GACTTAATTAAGCAAGGCGCTCTGGAGC-3′ (forward, PacI underlined) and 5′-CGATCTAGACGGATGCCTCAGTTCCTGC-3′ (reverse, XbaI underlined), and cloned into pCR®-Blunt to create pJK411. The PacI/XbaI fragment harboring encABC was cloned into pNEB193 to yield pJK416. A 1.8-kb XbaI/HindIII fragment from pLX713 harboring encN was cloned into pJK416 downfield of encABC to yield pJK482. In order to remove an unwanted EcoRI located between two XbaI sites between encC and encN, pJK482 was digested with XbaI and then self-ligated to yield pJK487. The ~ 4.7-kb PacI/EcoRI cassette harboring encABCN was cloned into the analogous sites of pSEK4 to yield pJK491. A 2.5-kb PacI/HindIII fragment from pMP6 21 harboring encD was cloned into pJK491 to yield pJK494 (insert as shown in Figure 2c).
4.5 Plasmid expression in the heterologous host S. lividans K4-114 and HPLC analysis
The plasmid pJK494 was introduced into the heterologous host S. lividans K4-114 and grown on solid R2YE (25 ml) at 30°C under thiostrepton (20 μg/ml) selection until sporulation, at which time, spores were spread onto fresh R2YE (supplemented with thiostrepton) for polyketide production. After 24 hours incubation at 30°C, ~1 mg benzoic acid was added, and the culture was allowed to grow for 3 days. The culture was then exhaustively extracted with 5% MeOH/EtOAc, and the extract dried over anhydrous MgSO4 and evaporated to dryness in vacuo. Polyketide production was determined by comparison with pure standards and authentic extracts via analytical HPLC using a Phenomenex C18-RP column (4.5×150 mm, 5μ) employing a linear gradient from 0.15% trifluoroacetic acid to MeOH over 30 min at a flow rate of 1 ml/min with UV detection at 254nm.
4.6 Preparation of recombinant EncL
The gene encL was PCR-amplified from pJP15F11 with primers 5′-CGGAATTCGATGCCCCCACCCCACAC-3′ (forward, BamHI underlined) and 5′-CCCAAGCTTGTCAGAAGTCCGGAGACAG-3′ (reverse, HindIII underlined), cloned into pCRR-Blunt (Invitrogen), sequence verified, digested with BamHI and HindIII, and cloned into the expression plasmid pHIS8. Recombinant EncL was overexpressed as an N-terminal octahistidyl-tagged fusion protein in E. coli BL21(DE3)/pLysS (Invitrogen) and purified by Ni2+-affinity chromatography according to general procedures.11
4.7 Enzymatic synthesis of acetate- and benzoate-primed polyketides
The in vitro reconstitution of enterocin biosynthesis followed previously described protocols19 in which recombinant EncL (5.5 μM) was incubated with different enterocin PKS enzyme sets as summarized in Table 1. Assays were terminated on the addition of EtOAc (1% AcOH) to the reaction vial and extracted as previously reported.19 Extracts were analyzed by reversed phase HPLC using a Phenomenex Luna C18-RP column (4.5×150 mm, 5μ) with a linear gradient of 5% MeCN/water (0.1% trifluoroacetic acid) to 80% MeCN/water (0.1% trifluoroacetic acid) over 40 min at a flow rate of 0.5 ml/min with UV detection at 254 nm.
4.8 Preparation of acetyl- and benzoyl-EncC
To reduce phosphate adducts in the mass spectra of EncC, a 0.2 mM stock solution of apo-EncC was diluted to 50 μM in 50 mM Tris-HCl pH 7.5 and dialyzed with 7000-MW cutoff Slide-A-Lyzer Mini Dialysis Units (Pierce) against 1 L of 50 mM Tris-HCl pH 7.5 with stirring at room temperature for 5 h. To prepare the acetyl- and benzoyl-loaded forms of EncC, 42 μM dialyzed apo-EncC was incubated at room temperature for 1 h with 520 μM acetyl-CoA or benzoyl-CoA, 5.2 mM tris(2-carboxyethyl)-phosphine (TCEP) (Fluka BioChemika), 5.2 mM MgCl2 and 2 μM Sfp in a total volume of 96 μL. Upon mixing the reagents, the reaction pH was measured with colorpHast® pH-indicator strips (EMD Chemicals) and adjusted to 7.0 – 7.5 by addition of 1 M Tris-HCl pH 8.8. The reaction was terminated after 1 h by retaining unreacted acyl-CoA on Micro Bio-Spin® 6 size-exclusion columns (Bio-Rad) pre-equilibrated with 50 mM Tris-HCl pH 7.5. The protein-containing flow-through was temporarily kept on ice before incubation with EncL.
4.9 Incubation of acetyl- and benzoyl-EncC with EncL
A 150 μL mixture of 21 μM acetyl-EncC and 21 μM benzoyl-EncC was obtained by mixing equal volumes of the acyl-EncC forms prepared as above. A 20 μL control sample of this mixture was set aside and mixed with 5 μL 10% formic acid on ice. To start the reaction, 2 μL of 32 μM EncL was added to the remaining 130 μL of the acyl- EncC solution. At 30 s intervals, five 20 μL aliquots of the reaction mixture were quenched with 5 μL 10% formic acid on ice. The remaining reaction mixture was quenched 5.5 min after the start. The six time-point samples were cooled to −80°C, lyophilized, and stored at −80°C.
4.10 MS sample preparation and analysis
Each lyophilized sample from the acyl-EncC/EncL reaction was dissolved in 20 μL of equilibration solution (1% AcOH in water) and desalted on a C4 ZipTip® pipette tip (Millipore) by sequentially injecting the following solutions into the back side of the ZipTip with a standard 200 μL pipette tip: 50 μL wetting solution (100% MeCN), 50 μL equilibration solution, 20 μL redissolved reaction sample, 100 μL wash solution (1% AcOH in water), and 20 μL elution solution (85% MeCN, 2% AcOH, in water). The desalted samples were ionized using a Nanomate BiVersa chip-based electrosprayer (Advion Biosystems, Ithaca, NY) and analyzed on an LTQ-orbitrap mass spectrometer (Thermo-Electron Corporation, San Jose, CA). To obtain monoisotopic masses, the mass spectra were deconvoluted using Xtract software (Thermo).
Acknowledgments
This research was supported by NIH grants AI47818 to B.S.M. and GM085283 to P.C.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. Nat Prod Rep. 2007;24:162. doi: 10.1039/b507395m. [DOI] [PubMed] [Google Scholar]
- 2.Das A, Khosla C. Acc Chem Res. 2009;42:631. doi: 10.1021/ar8002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore BS, Hertweck C. Nat Prod Rep. 2002;19:70. doi: 10.1039/b003939j. [DOI] [PubMed] [Google Scholar]
- 4.Bao W, Sheldon PJ, Hutchinson CR. Biochemistry. 1999;38:9752. doi: 10.1021/bi990751h. [DOI] [PubMed] [Google Scholar]
- 5.Meadows ES, Khosla C. Biochemistry. 2001;40:14855. doi: 10.1021/bi0113723. [DOI] [PubMed] [Google Scholar]
- 6.Tang Y, Lee TS, Kobayashi S, Khosla C. Biochemistry. 2003;42:6588. doi: 10.1021/bi0341962. [DOI] [PubMed] [Google Scholar]
- 7.Pickens LB, Tang Y. J Biol Chem. 2010;285:27509. doi: 10.1074/jbc.R110.130419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piel J, Hertweck C, Shipley P, Hunt DS, Newman MS, Moore BS. Chem Biol. 2000;7:943. doi: 10.1016/s1074-5521(00)00044-2. [DOI] [PubMed] [Google Scholar]
- 9.Izumikawa M, Cheng Q, Moore BS. J Am Chem Soc. 2006;128:1428. doi: 10.1021/ja0559707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiang L, Kalaitzis JA, Moore BS. Proc Natl Acad Sci USA. 2004;101:15609. doi: 10.1073/pnas.0405508101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang L, Moore BS. J Biol Chem. 2002;277:32505. doi: 10.1074/jbc.M204171200. [DOI] [PubMed] [Google Scholar]
- 12.Xiang L, Moore BS. J Bacteriol. 2003;185:399. doi: 10.1128/JB.185.2.399-404.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang Y, Koppisch AT, Khosla C. Biochemistry. 2004;43:9546. doi: 10.1021/bi049157k. [DOI] [PubMed] [Google Scholar]
- 14.Heathcote ML, Staunton J, Leadlay PF. Chem Biol. 2001;8:207. doi: 10.1016/s1074-5521(01)00002-3. [DOI] [PubMed] [Google Scholar]
- 15.Kim BS, Cropp TA, Beck BJ, Sherman DH, Reynolds KA. J Biol Chem. 2002;277:48028. doi: 10.1074/jbc.M207770200. [DOI] [PubMed] [Google Scholar]
- 16.Linne U, Schwarzer D, Schroeder GN, Marahiel MA. Eur J Biochem. 2004;271:1536. doi: 10.1111/j.1432-1033.2004.04063.x. [DOI] [PubMed] [Google Scholar]
- 17.Schwarzer D, Mootz HD, Linne U, Marahiel MA. Proc Natl Acad Sci USA. 2002;99:14083. doi: 10.1073/pnas.212382199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koglin A, Löhr F, Bernhard F, Rogov VV, Frueh DP, Strieter ER, Mofid MR, Güntert P, Wagner G, Walsh CT, Marahiel MA, Dötsch V. Nature. 2008;454:907. doi: 10.1038/nature07161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng Q, Xiang L, Izumikawa M, Meluzzi D, Moore BS. Nat Chem Biol. 2007;3:557. doi: 10.1038/nchembio.2007.22. [DOI] [PubMed] [Google Scholar]
- 20.Piel J, Hoang K, Moore BS. J Am Chem Soc. 2000;122:5415. [Google Scholar]
- 21.Hertweck C, Xiang L, Kalaitzis JA, Cheng Q, Palzer M, Moore BS. Chem Biol. 2004;11:461. doi: 10.1016/j.chembiol.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 22.Bililign T, Hyun CG, Williams JS, Czisny AM, Thorson JS. Chem Biol. 2004;11:959. doi: 10.1016/j.chembiol.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 23.Das A, Khosla C. Chem Biol. 2009;16:1197. doi: 10.1016/j.chembiol.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otten SL, Stutzman-Engwall KL, Hutchinson CR. J Bacteriol. 1990;172:3427. doi: 10.1128/jb.172.6.3427-3434.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajgarhia VB, Priestly ND, Strohl WR. Metabol Engin. 2001;3:49. doi: 10.1006/mben.2000.0173. [DOI] [PubMed] [Google Scholar]
- 26.Castaldo G, Zucko J, Heidelberger S, Vujaklija D, Hranueli D, Cullum J, Wattana-Amorn P, Crump MP, Crosby J, Long PF. Chem Biol. 2008;15:1156. doi: 10.1016/j.chembiol.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 27.Bierman J, Logan R, O’Brien K, Seno ET, Rao RN, Schoner BE. Gene. 1992;116:43. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 28.Ziermann R, Betlach MC. BioTechniques. 1999;26:106. doi: 10.2144/99261st05. [DOI] [PubMed] [Google Scholar]
- 29.McDaniel R, Ebert-Khosla S, Fu H, Hopwood DA, Khosla C. Proc Natl Acad Sci USA. 1994;91:11542. doi: 10.1073/pnas.91.24.11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDaniel R, Ebert-Khosla S, Hopwood DA, Khosla C. Science. 1993;262:1546. doi: 10.1126/science.8248802. [DOI] [PubMed] [Google Scholar]
- 31.Yu TW, Hopwood DA. Microbiology. 1995;141:2779. doi: 10.1099/13500872-141-11-2779. [DOI] [PubMed] [Google Scholar]
- 32.Jez JM, Ferrer JL, Bowman ME, Dixon RA, Noel JP. Biochemistry. 2000;39:890. doi: 10.1021/bi991489f. [DOI] [PubMed] [Google Scholar]
- 33.Han L, Lobo S, Reynolds KA. J Bacteriol. 1998;180:4481. doi: 10.1128/jb.180.17.4481-4486.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hopwood DA, Bibb MJ, Chater KF, Kieser T, Bruton CJ, Kieser HM, Lydiate DJ, Smith CP, Ward JM, Schrempf H. Genetic Manipulations of Streptomyces: A Laboratory Manual. Norwich: The John Innes Foundation; 1985. [Google Scholar]
- 35.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, a Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]