Abstract
Non-B DNA structures are a major contributor to the genomic instability associated with repetitive sequences. Immunoglobulin switch Mu (Sμ) region sequence is comprised of guanine-rich repeats and has high potential for forming G4 DNA, in which one strand of DNA folds into an array of guanine quartets. Taking advantage of the genetic tractability of Saccharomyces cerevisiae, we developed a recombination assay to investigate mechanisms involved in maintaining stability of G-rich repetitive sequence. By embedding Sμ sequence within recombination substrates under the control of a tetracycline-regulatable promoter, we demonstrate that the rate and orientation of transcription both affect the stability of Sμ sequence. In particular, the greatest instability was observed under high-transcription conditions when the Sμ sequence was oriented with the C-rich strand as the transcription template. The effect of transcription orientation was enhanced in the absence of the Type IB topoiosmerase Top1, possibly due to enhanced R-loop formation. Loss of Sgs1 helicase and RNase H activity also increased instability, suggesting they may cooperatively function to reduce the formation of non-B DNA structures in highly transcribed regions. Finally, the Sμ sequence was unstable when transcription elongation was perturbed due to a defective THO complex. In a THO-deficient background, there was further exacerbation of orientation-dependent instability associated with the ectopically expressed, single-strand cytosine deaminase AID. The implications of our findings to understanding instability associated with potential G4 DNA forming sequences are discussed.
Keywords: G4 DNA, R-loops, genome instability, transcription, recombination
1. Introduction
Repetitive sequences that can adopt non-B structures in vivo are associated with increased genetic instability [1, 2]. Non-B DNA structures include triplex or H-DNA formed by polypurine tracts (for example, GAA triplet repeats), cruciforms formed by inverted repeats, and left-handed Z-DNA formed by alternating purines and pyrimidines. G-quartets, comprised of four guanines assembled into a planar configuration through Hoogsteen bonding [3], are also an example of a non-B structure forming sequence motifs. When DNA contains more than four tracts of at least three guanines, it can form a higher order structure called G4 DNA, which consists of multiple G-quartets stabilized by stacking on top of each other [4]. Sequences with potential for G4 DNA formation are found throughout the genomes of bacteria, yeast and higher eukaryotes. Notably, telomeres, rDNA loci, immunoglobulin heavy-chain switch regions, and G-rich minisatellites have high potential to form G4 DNA [4, 5]. G4 DNA forming potential also correlates with genomic regions that are relatively unstable, such as proto-oncogenes and sites of frequent translocation breakpoints [6]. In particular, chromosomal translocations involving G-rich immunoglobulin switch regions have been observed in various cancer cell lines [7].
G4 DNA formation requires exposure of the G-rich strand as a single strand, as would occur during transcription, DNA repair or lagging-strand replication. In the case of transcription, the limited single-stranded character of a non-transcribed, G-rich strand can be further enhanced by the relatively stable base pairing between the corresponding deoxyribocytosine (dC)-rich template and riboguanine (rG)-rich transcript. The higher-order DNA structure involving G4 DNA on one strand and an extended RNA:DNA hybrid on the other is referred to as a G-loop [4]. Consistent with a role for RNA:DNA hybrids in G4 DNA formation, disruption of the strong base pairing between rG and dC by substituting ribo-inosine for rG has been shown to reduce G-loop structures in vitro [8]. Although recent reports clearly show that the G-rich sequences of immunoglobulin class switch regions and telomeres can assume G-loop structures in vitro and in bacterial cells [8], the biological significance of such structures is not well understood [5]. Because transcription of the G-rich switch region is required for class-switch recombination (CSR), it has been speculated that co-transcriptionally formed G-loop structures may be relevant to both regulated CSR and to unintended chromosomal breaks and rearrangements [4].
In E. coli, formation of G-loop structures requires activated transcription and is inhibited in the presence of the RecQ DNA helicase and RNase H, which degrades the RNA strand of RNA:DNA hybrids [8]. Although individual effects of RecQ and RNase H on G-loop formation were not reported, data are consistent with G4 DNA being directly dissolved by helicase activity and/or its formation inhibited by RNase H. In vivo, the human RecQ family DNA helicase BLM, as well as the yeast homolog Sgs1, is important for general genome stability, especially in maintaining the integrity of G-rich regions such as telomeres and rDNA repeats [9-13]. In in vitro assays, BLM and Sgs1 have been shown to efficiently unwind G4 DNA [14, 15]. The yeast Pif1 helicase has also been shown to unwind G4 DNA, specifically that formed by the human G-rich minisatellite CEB1 [16]. While CEB1 repeats are very unstable in a pif1 mutant background, loss of Sgs1 has no effect on the stability of these particular repeats. Finally, the Dog-1 DNA helicase of C. elegans, which is an ortholog of mammalian FANCJ, stabilizes sequences with high G4-forming potential in this organism [17].
Valuable insight into mechanisms of genomic instability has come through the use of Saccharomyces cerevisiae as a model system to examine specific mammalian sequence motifs. Examples include the CAG/CTG repeat involved in Huntington’s disease and myotonic dystrophy type 1 [18], the G-rich human minisatellite CEB1 [16], the CGG/CCG repeat involved in fragile X syndrome [19], the GAA/TTC repeat involved in Friedreich’s ataxia [20], and the inverted ALU repeats present throughout the human genome [21]. In the current study, we use a recombination assay to analyze whether sequences with high potential to form G4 DNA lead to genomic instability in yeast. Results demonstrate that transcription of the immunoglobulin heavy chain switch Mu (Sμ) sequence is associated with an orientation-dependent increase in recombination in some mutant backgrounds, consistent with the formation of co-transcriptional G-loops.
2. Material and Methods
2.1 Construction of plasmids
pSR886 (lys2::SμF) and pSR887 (lys2::SμR) were constructed by inserting a 750-bp Sau3AI fragment from pT7-Sμ [22] into a unique BglII site within LYS2 sequences of pSR633 (see Figure 1A and 1B). pSR633 contains a 3.5 kb XbaI/BamHI lys2Δ3′ fragment in the URA3-integrating vector pRS306 [23]. pSR953 (lys2::cβ2) was constructed by inserting an 800-bp BglII/BamHI chicken β-globin 2 cDNA (Cβ2) fragment into the BglII site of a plasmid containing the previously described lys2-oligo allele [24]. pSR877 (pRS402-lys2Δ3′) was constructed by inserting a 1.3 kb NotI/SpeI fragment of LYS2 gene into NotI/SpeI-digested pRS402, an ADE2 integrating vector [25].
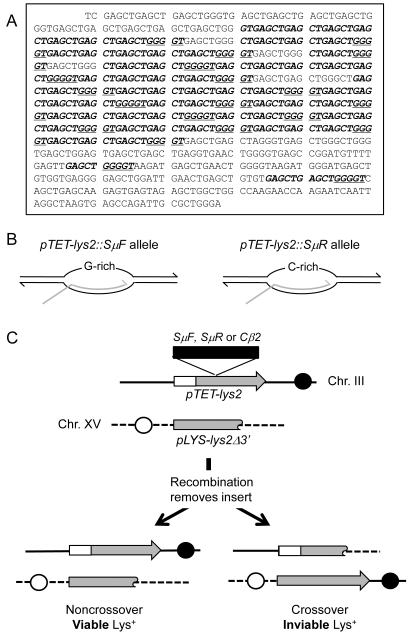
Figure 1.
System for monitoring Sμ-associated recombination.
A. Sequence of the DpnI fragment of murine immunoglobulin Sμ region inserted into the BglII site of LYS2 gene. Uninterrupted (GAGCT)nGGGGT repeat units are indicated in bold italics and 4G runs are underlined.
B. Transcription orientations in the pTET-lys2-SμF and pTET-lys2-SμR constructs. Transcription bubbles are illustrated with the gray line indicating nascent mRNA. The single-stranded, non-transcribed strand is G-rich in the pTET-lys2-SμF construct and C-rich in the pTET-lys2-SμR.
C. Chromosomal configurations of the recombination substrates (see Material and Methods for details). Black and white circles indicate the locations of centromeres relative to the lys2 alleles on Chr. III and Chr. XV, respectively. Recombination initiated by a gap or break within pTET-lys2 allele removes the insert to create a LYS+ pTET-LYS2 allele. Removal of the insert by recombination associated with a crossover event result in unviable acentric and dicentric products.
The plasmid pESC-LEU-hAIDSc [26] was constructed by cloning the human AID gene (hAID) engineered to contain favored yeast codons into BamHI/SalI-digested pESC-LEU (Stratagene) and was a gift from Y. Pavlov (University of Nebraska). To replace the LEU2 maker with a HygR gene, pESC-LEU and pESC-LEU-hAIDSc were digested with EcoRV and BglII and ligated to a 1.3 kb EcoRV/BglII fragment from the plasmid hphMX4 [27].
2.2 Construction of yeast strains
All yeast strains were derived from YPH45 (MATα ura3-52 ade2-101oc trp1Δ1). Construction of a strain with the pTET-LYS2 gene positioned at the HIS4 locus near ARS306 on chromosome III was previously described [28]. The LYS2 wild-type allele placed in opposite orientation relative to ARS306 was replaced with the lys2::SμF, lys2::SμR, or lys2::Cβ2 allele by standard two-step allele replacement (Figure 1C). pSR877 was digested with StuI to target insertion of the lys2Δ3′ allele to the ADE2 locus on chromosome XV; integration of a single copy of pSR877 was confirmed by PCR.
Mutant strains were constructed by one-step gene disruption using PCR-generated deletion cassettes; primers are available upon request. As appropriate, the loxP-flanked URA3Kl orTRP1 marker was deleted following transformation with a Cre recombinase-expressing plasmid [29]. For some strains, the HygR gene originally used to select for deletion of the endogenous LYS2 locus was replaced with the NatR gene using NotI-digested pAG25 [27].
2.3. Recombination rates
Fluctuation analysis was carried out at 30°C using at least two independent isolates of each strain. First, 4-5 colonies were used to inoculate 5 ml of non-selective YEP medium (1% yeast extract, 2% Bacto-peptone, 250 mg/l adenine; 2% agar for plates) supplemented with 2% glycerol plus 2% ethanol (YEPGE). As appropriate, the liquid medium was supplemented with doxycycline hyclate (DOX; obtained from Sigma) to 2000 ng/ml. Following overnight growth, cells were diluted to approximately 250,000 cells/ml in the same liquid medium and six 1-ml aliquots were transferred to separate tubes. Following 3 additional days of growth, appropriately diluted cells were plated on SD-Lys plates to select for recombinants or on YPD to determine the total number of cells in each culture. Colonies were counted after 2 days. Recombination rates were determined using the method of median and 95% confidence intervals were calculated as described previously [30]. 12 to 20 cultures were used for each rate determination.
For the measurement of the hAID-induced recombination rates, two independent isolates of each strain were transformed with pESC-HYG (pSR926) or pESC-HYG-hAIDSc (pSR927). Following selection of transformants on YPD plates supplemented with 0.3 g/l hygromycin B (Invitrogen), 4-5 colonies were used to inoculate a 5 ml overnight culture. The remainder of the fluctuation procedure was as described above, except that YEP liquid media was supplemented with 0.3 g/l hygromycin B, 2 % galactose, 2 % glycerol and 2 % ethanol.
3. Results
3.1. A system for examining effects of switch region sequence on recombination
We chose the immunoglobulin switch region as a model example of a sequence with the potential to form G-loops during transcription [3, 8]. Switch region sequences are required for heavy chain CSR and contain degenerate G-rich repeats of several kb in length [31]. To examine associated instability, we used a 770-bp fragment of the mouse switch mu (Sμ) core sequence containing 20 copies of the repeat (GAGCT)nGGGGT plus additional degenerate repeats (Pubmed Accession #J00442 - Figure 1A). This fragment was inserted in both orientations within a doxycycline (DOX)-repressible pTET-LYS2 gene on chromosome III of a haploid yeast strain. In this system, transcription is activated by tetR-VP16 binding to tetO sequence in absence of DOX and repressed by the tetR’-Ssn6 repressor protein binding to tetO in the presence of DOX [32]. We previously showed that 1000 ng/ml DOX in the growth medium reduced the transcription of the pTET-LYS2 gene by approximately 1000-fold [28]. In the pTET-lys2::SμF “forward” construct, the G-rich sequence is on the top, non-transcribed strand and thus mimics the physiological orientation. In the pTET-lys2::SμR “reverse” construct, the Sμ fragment is in the inverse orientation, resulting in the C-rich sequence being on the top strand (Figure 1B). Because G-loop formation has been shown to occur only when the non-transcribed strand contains the G-rich sequence [8], G-loops have the potential to form when the SμF construct is highly transcribed but not when the SμR construct is highly transcribed.
As a control allele with no G4-forming potential, an 800 bp fragment of chicken β-globin 2 cDNA was inserted into the pTET-LYS2 gene (pTET-lys2::Cβ2 allele). The GC content of this sequence (63%) is similar to that of the Sμ sequence inserted to generate the pTET-lys2::Sμ alleles (62%), but this sequence has no biased distribution of G’s and C’s between the two strands. The Cβ2 sequence contains only four widely separated runs of G’s and six runs of C’s greater than 4 nt, and is not expected to form G4 DNA or G-loops when transcribed. Finally, to provide a donor sequence for the repair of broken or gapped pTET-lys2 reporter alleles by homologous recombination, a lys2 allele with a 3′ deletion and under control of its native promoter (pLYS-lys2Δ3′ allele) was inserted into the ADE2 locus on chromosome XV. It should be noted that, because the pTET-lys2 and pLYS-lys2Δ3′ alleles are in different orientations relative to their respective centromeres, crossover products are inviable (Figure 1C). The systems used here thus measure only rates of noncrossover recombinants.
3.2. Loss of RNase H activity affects SμF- more than SμR-associated recombination
The RNase H class of enzyme degrades the RNA strand of RNA:DNA hybrids, thus promoting the reannealing of the two strands of DNA during transcription. Because formation of a stable RNA:DNA hybrid is a central feature of the G-loop structure, RNase H activity is predicted to limit the formation and/or stability of G-loops. In yeast, three RNase H enzymes have been identified [33]. RNase H1 is a single subunit enzyme encoded by RNH1 gene. RNase H2 is composed of three subunits, with the RNH201 gene (also referred to as RNH35 or RNH2A) encoding the highly conserved catalytic subunit. Finally, RNase H70 was identified based on amino acid homology, but RNase H activity has not been confirmed. Because RNase H1 and RNase H2 appear to have redundant roles in removing RNA:DNA hybrids in yeast [34, 35], we deleted both the RNH1 and RNH201 genes in strains containing the pTET-lys2::SμF, pTET-lys2::SμR or pTET-lys2::Cβ2 construct in order to study the effect of RNase H deficiency on recombination.
Under low-transcription conditions (+DOX) in a wild-type (WT) background, the recombination rate for the pTET-lys2::SμF and pTET-lys2::SμR alleles was indistinguishable, and was approximately 3-fold higher than the recombination rate obtained with the control pTET-lys2::Cβ2 construct (Table 1). RNase H deficiency under low-transcription conditions was associated with a small (~2 fold) elevation in the recombination rates for the pTET-lys2::SμF and pTET-lys2::SμR constructs, and a somewhat larger increase (~10-fold) for the Cβ2 construct. Under high transcription conditions in a WT background, the recombination rates of pTET-lys2::SμF, pTET-lys2::SμR and pTET-lys2::Cβ2 constructs were elevated to similar extents (6.9, 4.6-fold and 3.1, respectively). When transcription was activated in the RNase H-deficient strains, however, the rate of recombination was elevated 87-fold for the pTET-lys::SμF construct but only 16-fold for the pTET-lys:SμR and pTET-lys2::Cβ2 constructs. Under high-transcription conditions in the RNase H-deficient background, the recombination rate associated with the pTET-lys::SμF allele was 5.4- and 3.4-fold greater than that obtained with the pTET-lys:SμR and pTET-lys2::Cβ2 constructs, respectively.
Table 1.
Effect of RNase H, Sgs1 and Top1 on stability of SμF, SμR and Cβ2 substrates
| Recombination rate × 10−8 (CI*) |
||||
|---|---|---|---|---|
| Allele | Genotype | Low transcription | High transcription | High/low |
| lys2::SμF | WT | 2.94 (2.58 – 3.40) |
20.3 (17.2 – 24.4) |
6.9 |
| rnh1 rnh201 | 6.51 (3.88 – 10.4) |
566 (289 – 780) |
87 | |
| sgs1 | 6.61 (4.73 – 12.6) |
38.6 (29.6 – 49.7) |
5.8 | |
| sgs1 rnh1 rnh201 | 57.7 (46.7 - 66.8) |
1500 ( 1190- 2480) |
26 | |
| top1 | 5.11 (4.54 – 5.68) |
128 (113 – 177)) |
25 | |
| lys2::SμR | WT | 2.70 (2.51 – 3.02) |
12.5 (10.4 – 15.6) |
4.6 |
| rnh1 rnh201 | 6.41 (4.92 – 9.76) |
105 (78.8 – 135) |
16 | |
| sgs1 | 3.92 (3.46 – 6.11) |
18.4 (15.4 – 32.7) |
4.7 | |
| sgs1 rnh1 rnh201 | 45.2 (33.0 – 47.0) |
195 (163 – 251) |
4.3 | |
| top1 | 2.68 (2.24 – 3.05) |
12.3 (10.3 – 18.1)) |
4.6 | |
| lys2::Cβ2 | WT | 1.01 (0.89 – 1.35) |
8.55 (8.29 – 12.6) |
8.4 |
| rnh1 rnh201 | 10.3 (8.26 – 16.9) |
165 (134 – 215) |
16 | |
| sgs1 | 5.01 (3.71 – 10.3) |
17.1 (15.7 – 21.7) |
3.4 | |
| sgs1 rnh1 rnh201 | 51.1 (24.4 – 54.9) |
268 (204 – 344) |
5.2 | |
| top1 | 3.19 (2.62 – 3.43) |
14.3 (12.2 – 18.4) |
4.5 | |
CI* - 95 % confidence interval.
3.3. Effect of Sgs1 loss on SμF-, SμR- and Cβ2-associated recombination
Sgs1 is the only member of RecQ DNA helicase family present in yeast, and we examined its effect on stability of the pTET-lys2::SμF, pTET-lys2::SμR and pTET-lys2::Cβ2 reporters under both low- and high-transcription conditions (Table 1). Upon loss of Sgs1 under low-transcription conditions, recombination rates were elevated by 1.5- to 5- fold, consistent with the hyper-recombination phenotype observed in other systems [36]. A further increase in the recombination rate for each construct was observed when transcription was activated in the sgs1 background. Although not statistically significant, we note that the recombination rate for the pTET-lys2::SμF construct was 2-fold higher than the rates obtained with either the pTET-lys2::SμR orpTET-lys2::Cβ2 reporters.
3.4. Increase in SμF-associated instability upon loss of RNase H and Sgs1
Because switch region-associated G-loops in E. coli have been detected in RecQ and RNase H-deficient, but not in WT cells [8], we examined the rates of Sμ-associated recombination in sgs1 rnh1 rnh201 triple mutant yeast strains (Table 1). While the recombination rates for all three constructs were similar under low-transcription conditions, there was a striking difference under high-transcription conditions. The recombination rate for the pTET-lys2::SμF construct was 7.7-fold higher than that associated with the pTET-lys2::SμR reporter, and 5.6-fold higher than obtained with the pTET-lys2::Cβ2 construct. The orientation-dependent effect of the Sμ region on recombination in the RNase H-deficient background thus appears to be enhanced upon additional loss of Sgs1.
3.5. Effect of Top1 topoisomerase on Sμ-associated recombination
Yeast Top1 is a Type IB topoisomerase that can relieve both positive and negative torsional stress in DNA [37]. It covalently attaches to the 3′ end of DNA, creating a single strand nick, which is quickly religated after swiveling of the DNA strands to remove supercoiling. During highly activated transcription, positive and negative supercoiling can build up in front of or behind RNA polymerase complexes, respectively [38]. In absence of Top1, extensive R-loop formation and regions of DNA helix melting have been observed in the highly transcribed rDNA repeat locus [39, 40].
In order to determine the effect of accumulated torsional stress on the stability of G-run containing sequence, we examined the rates of Sμ-associated recombination in Top1-deficient strains (Table 1). Under low transcription conditions, there were slight increases (less than 2-fold) in the recombination rates for the pTET-lys2::SμF and pTET-lys2::Cβ2 constructs. When transcription was activated in the top1 background, the recombination rates for the pTET-lys2::SμR and pTET-lys2::Cβ2 constructs were elevated by ~4-fold. A more striking increase (25-fold) was observed for the recombination rate of the pTET-lys2::SμF construct in a top1 background under high-transcription conditions. The resulting recombination rate for the pTET-lys2::SμF allele was 10- and 9- fold higher than the recombination rates for pTET-lys2::SμR or pTET-lys2::Cβ2 constructs, respectively.
3.6. Effect of Pif1 helicase on Sμ-associated recombination
Given the role of Pif1 in maintaining stability of the human CEB1 minisatellite [16], we explored whether Pif1 is likewise important for limiting recombination associated with the pTET-lys2::SμF, pTET-lys2::SμR or pTET-lys2::Cβ2 reporters (Table 2). Under low-transcription conditions, loss of Pif1 elevated pTET-lys2::Cβ2 - associated recombination 45-fold, but increased pTET-lys2::SμF- and pTET-lys2::SμR - associated recombination only 8.5-fold. High levels of transcription in the pif1 background stimulated recombination involving the pTET-lys2::SμF and pTET-lys2::SμR constructs approximately 4.5-fold, and had a slightly smaller effect (2.5-fold) on recombination associated with the pTET-lys2::Cβ2 reporter. The stabilizing effect of Pif1 on recombination in our system thus does not appear to be related to the potential of GC-rich sequences to form co-transcriptional G4 DNA.
Table 2.
Effects of Mft1 and Pif1 on stability of SμF, SμR and Cβ2 substrates
| Recombination rate × 10−8 (CI*) |
|||
|---|---|---|---|
| Allele | Genotype | Low transcription | High transcription |
| lys2::SμF | WT | 2.94 (2.58 – 3.40) |
20.3 (17.2 – 24.4) |
| pif1 | 24.9 (17.8 - 27.9) |
107 (87 - 1640) |
|
| mft1 | 124 (115 – 134) |
181 (163 – 213) |
|
| mft1 sgs1 | 24.2 (13.2 – 36.0) |
320 (242 – 422) |
|
| lys2::SμR | WT | 2.70 (2.51 – 3.02) |
12.5 (10.4 – 15.6) |
| pif1 | 23.3 (16.3 - 24.4) |
111 (80 - 218) |
|
| mft1 | 116 (86.1 – 124) |
358 (309 – 410) |
|
| mft1 sgs1 | 37.5 (34.4 – 61.9) |
1070 (965 – 1590) |
|
| lys2::Cβ2 | WT | 1.01 (0.89 – 1.35) |
8.55 (8.29 – 12.6) |
| pif1 | 45.5 (33.0 - 75.1) |
114 (88 - 157) |
|
| mft1 | 16.3 (13.0 – 17.8) |
67.3 (48.7 – 136) |
|
| mft1 sgs1 | 10.0 (3.57 – 11.6) |
124 (21.8 – 456) |
|
CI* - 95 % confidence interval.
3.7. Effect of Mft1 loss on Sμ-associated recombination
THO is a multi-functional protein complex involved in mRNA processing and mRNP export, as well as in efficient elongation by the RNAP II holoenzyme (reviewed in [41]). Disabling the THO complex promotes R-loop formation and generally leads to a strong hyperrecombination phenotype [35, 42]. Mft1 is a component of the THO complex and deletion of the MFT1 gene results in THO-defective, hyper-recombination phenotype [41]. In an mft1 background under low-transcription conditions, we observed a large, ~40-fold increase in recombination rates for the pTET-lys2::Sμ constructs, but only a 16-fold increase for the pTET-lys2::Cβ2 allele (Table 2). Although high-transcription further elevated recombination in the mft1 background, the effect was greater with the pTET-lys2::SμR than with the pTET-lys2::SμF construct (3.1- and 1.5-fold increases, respectively). It should be noted that this is the reverse of the orientation-dependent effect observed in the rnh1 rnh201 double mutant.
We also examined the effect of transcription on recombination in an mft1 sgs1 double mutant background. Interestingly, Sgs1 loss had opposing effects under low-versus high-transcription conditions in the mft1 background. Recombination rates were uniformly reduced in the mft1 sgs1 double relative to the mft1 single mutant under low-transcription conditions, but uniformly elevated under high-transcription conditions. We currently have no explanation for this difference. While transcription in the mft1 sgs1 background stimulated recombination with all three substrates, the effect was again slightly greater for the pTET-lys2::SμR than for the pTET-lys2::SμF or pTET-lys2::Cβ2 construct (29- versus 12-fold, respectively).
3.8. Effect of AID expression on Sμ-associated recombination rates
Activation-induced deaminase (AID) is a single-strand specific deaminase that converts cytosine to uracil [43] and is required for the somatic hypermutation (SHM) of immunoglobulin variable region genes as well as the heavy chain class switch recombination (CSR) [44]. Transcription is required for both SHM and CSR, and is thought to facilitate access of AID to single-stranded DNA. AID has previously been shown to stimulate recombination and mutagenesis in a transcription-dependent manner in THO-defective yeast strains [45]. It was also recently shown that a defective THO complex together with AID expression enhances chromosomal translocations mediated by non-homologous end joining (NHEJ), presumably by inducing double-strand breaks in the murine Sμ sequence [46]. In order to examine the effect of AID expression on homologous recombination, we ectopically expressed the human AID (hAID) in strains containing the pTET-lys2::Sμ constructs. While there was no stimulation of recombination in a WT background with the pTET-lys2::SμF construct, there was a 7-fold stimulation of recombination with the pTET-lys2::SμR construct (Table 3). Deletion of the uracil DNA glycosylase gene (UNG1) eliminated the hAID-stimulated recombination observed with the pTET-lys2::SμR construct, confirming that the elevated recombination occurs through the processing of uracil intermediates.
Table 3.
hAID and stability of SμF and SμR under high-transcription conditions
| Recombination rate × 10−8 (CI*) |
|||
|---|---|---|---|
| Genotype | Plasmid | lys2::SμF | lys2::SμR |
| WT | Empty vector | 19.2 (17.4 – 27.8) |
14.8 (12.8 – 17.0) |
| Vector + hAID | 20.9 (19.3 – 22.5) |
102 (73.2 – 114) |
|
| ung1 | Empty vector | 21.8 (20.1 – 23.8) |
11.7 (8.58 – 13.9) |
| Vector + hAID | 20.1 (19.2 – 22.3) |
16.7 (10.6 – 21.6) |
|
| mft1 | Empty vector | 89.7 (51.4 – 134) |
359 (265 – 594) |
| Vector + hAID | 3020 (1190 – 5400) |
26000 (19100 – 29200) |
|
| top1 | Empty vector | 138 (99.7 – 174) |
18.0 (15.8 – 19.9) |
| Vector + hAID | 171 (123 – 293) |
282 (253 – 294) |
|
CI* - 95 % confidence interval.
In an mft1 mutant background where extensive R-loop formation was previously reported [35], expression of hAID was accompanied by a large stimulation of recombination for both constructs (Table 3). As in the WT background, however, hAID expression was associated with an orientation-dependent bias in the stimulation of recombination in the mft1 background. With hAID expression, the recombination rate of pTET-lys2::SμR construct was ~ 9-fold higher than that of the pTET-lys2-SμF construct. In a top1 background, hAID expression was associated with similar orientation bias (Table 3). While there was no stimulation of recombination of the pTET-lys2-SμF construct, the rate of recombination for the pTET-lys2::SμR construct was stimulated by ~ 16-fold with hAID expression.
4. Discussion
Homologous recombination is a high-fidelity repair process that is initiated by single-strand gaps or double-strand breaks in duplex DNA, and is a sensitive indicator of DNA damage. In the current study, a yeast recombination assay has been used to examine genetic instability associated with the repetitive mouse immunoglobulin Sμ sequence, as well as with a control, nonrepetitive sequence (Cβ2) of comparable size and GC content. Each test sequence was engineered into a full-length, chomosomal LYS2 gene, which served as the recipient of genetic information provided by a truncated lys2 allele on a different chromosome. According to standard models of recombination, the recipient allele is the site of the recombination-initiating lesion. By placing the Sμ sequence in both directions within a pTET-regulated LYS2 gene, we have also been able to discern orientation-specific effects of this sequence on mitotic recombination rates. The Sμ sequence is in its physiological orientation in the pTET-lys2::SμF allele, with the G-rich strand being the non-transcribed strand; the G-rich strand is the transcribed strand in the pTET-lys2::SμR allele.
In the absence of highly activated transcription in a WT background, both Sμ inserts were associated with a higher recombination rate than was the control Cβ2 sequence. Given the similar sizes and GC contents of these sequences, and the further enhancement of Sμ-Cβ2 difference in an mft1 background (see below), we suggest that this likely reflects the repetitive nature of the Sμ sequence. When the transcription was turned on to a high level, the recombination rate associated with each construct increased a comparable amount (5-10 fold). Under high-transcription conditions in RNase H-defective background, however, there were many more recombinants generated with the pTET-lys2::SμF allele than with the pTET-lys2::SμR or pTET-lys2::Cβ2 allele. We suggest that the difference reflects more efficient formation and/or enhanced stability of co-transcriptional RNA:DNA hybrids (R-loops) with the pTET-lys2::SμF allele. Increased R-loops could be due to the greater stability of RNA:DNA hybrids when the RNA strand is the purine-rich strand [47], enhanced nucleation of R-loop formation by clusters of guanines on the nascent transcript [48], and/or a stabilizing feature of the single strand of DNA excluded from the R-loop. With regard to the latter possibility, it should be noted that additional elimination of the Sgs1 helicase further enhanced the orientation-dependent effect of Sμ sequences on recombination. An intriguing possibility is that lack of RNase H in yeast provides greater opportunity for G4 DNA to form on the nontranscribed strand of the pTET-lys2::SμF allele, and once formed, Sgs1 can unwind at least some of the G4 DNA. We suggest a situation analogous to that reported previously in E. coli, where a physiologically oriented murine Sμ sequence forms co-transcriptional G-loops in cells deficient in RNase H and RecQ [8]. Finally, the recent demonstration that the Sμ sequence efficiently blocks RNA polymerase when the G-rich sequence is present on the nontranscribed strand [49] suggests another possible contributor to the orientation-dependent instability of the Sμ sequence in our system.
The absence of Top1 results in a build up of helical torsion in highly transcribed areas and, in E. coli, results in extensive R-loop formation that can be relieved by overexpression of RNase H [50]. In the highly transcribed rDNA locus in yeast, Top1 deficiency results in extensive formation of R-loops and “topo bubbles” which are regions of strand separation due to the accumulation of negative supercoils [39, 40]. Similar to the effect of RNase H deficiency, an orientation-specific increase in instability was observed with the highly transcribed pTET-lys2::Sμ alleles in a top1 background, with the pTET-lys2:: SμF allele generating 10-fold more recombinants than the pTET-lys2::SμR. This difference can be explained by enhanced G4 DNA formation due to more extensive RNA-DNA hybrids on the transcribed strand and increased single-stranded character on the G-containing non-transcribed strand of the pTET-lys2::SμF construct.
Another way to promote RNA:DNA hybrid formation is through disruption of the THO complex [35], which is important for processing nascent transcripts [41]. Unexpectedly, mft1 mutants with a defective THO complex exhibited a recombination phenotype in our system that was distinct from that observed in the absence of RNase H activity or Top1. First, under low-transcription conditions in an rnh1 rnh201 background, the Sμ and Cβ2 constructs were associated with similar recombination rates. By contrast, the Cβ2-containing construct produced 7-fold fewer recombinants than either the SμF or SμR construct in an mft1 mutant. Given the similar GC contents of the test sequences, the Mft1-related difference may reflect the role of the THO complex in promoting transcription through the repetitive Sμ sequences [51]. Second, whereas there were 5 - 10-fold more recombinants obtained with the SμF than with the SμR construct under high-transcription conditions in either RNase H- or Top1-deficient background, there were 2-fold more recombinants with the SμR than with the SμF construct in the mft1 mutant. A reason for this difference is not immediately obvious, but could reflect impaired transcription of Sμ sequences that is unrelated to R-loop formation, or more global changes in gene expression in the absence of the THO complex.
The yeast Pif1 helicase can unwind G4 DNA in vitro and its absence greatly increases recombination-dependent instability associated with the human CEB1 minisatellite [16]. Given that Sμ sequences also form G4 DNA in vitro, we anticipated that Pif1 loss would be associated with an orientation-dependent increase in Sμ-associated recombination. Although recombination was elevated in the pif1 background, the greater effect we expected to see with the pTET-lys2::SμF than with the pTET-lys2::SμR construct, especially under high-transcription conditions, was not observed. G4 DNA structures are known to vary depending on the primary sequence of the G-rich motifs [52], however, and it is possible that Pif1 is specific for the CEB1-associated structure. Alternatively, the disparity in the pif1 results could reflect inherent differences in the two assay systems. In the CEB1 system, the assay measures the interstitial deletion within repeats where the repair template was presumably the sister chromatid [16]. In contrast, the repair template in our Sμ system is on a nonhomologous chromosome. Also, the CEB1 sequence was embedded in a non-transcribed region and the formation of the associated G4 DNA was attributed to the process of replication. In our system, highly activated transcription is necessary to reveal the difference in stability between G-loop forming (SμF) and non-G-loop forming (SμR) sequences and, thus, G4 formation is clearly co-transcriptional.
In addition to the potential of Sμ sequences to form non-B DNA structures, these sequences are also targets of AID, which exhibits single-strand specific cytosine deaminase activity in vitro. During CSR, the uracils generated by AID activity are excised by uracil DNA glycosylases and then are further processed by base excision repair enzymes to produce recombination-initiating breaks in DNA [29, 40]. The uneven distribution of G’s and C’s between the two strands of the Sμ fragment used here (the C-rich strand contains 358 cytosines but only 117 guanines) allowed us to infer whether AID more efficiently targets Sμ-associated cytosines when they are on the nontranscribed as opposed to the transcribed strand of a highly expressed gene. As predicted by its preference for single-standed DNA, expression of hAID in yeast increased the recombination rate associated with the pTET-lys2-SμR allele 7- and 16-fold in WT and top1 backgrounds, repectively, but had no effect on recombination with the pTET-lys2-SμF allele in either background. As seen in prior experiments [45], the effect of AID on recombination was greatly amplified in an mft1 background. The heterologous expression of AID was recently reported to increase chromosomal translocations at both the physiologically and inversely oriented Sμ sequence fragments embedded in THO-deficient yeast strains [46]. Although the extent of instability observed by Ruiz et al. was higher for the physiologically oriented Sμ sequence, the reverse of the pattern reported here, this difference could reflect the larger size of the Sμ fragments used in our assays or basic differences between the two systems.
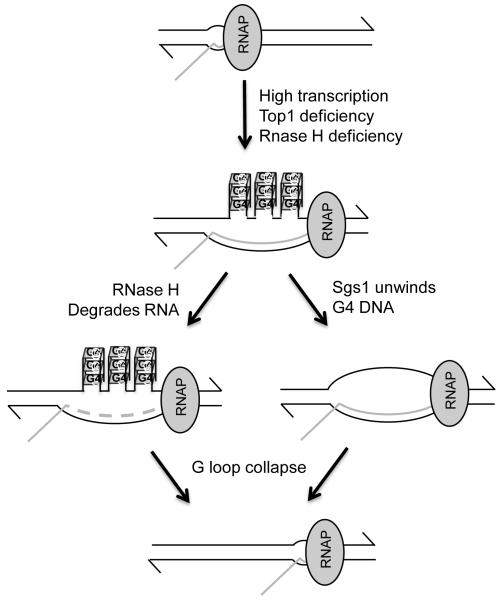
The genome-destabilizing potential of repeat-containing sequence motifs is closely linked to their ability to assume non-B DNA structures in vivo. Using yeast as a model system, we have demonstrated that mitotic instability associated with the murine Sμ sequence is increased by transcription and exhibits strong orientation dependence. Our data point to two possible factors that can promote G-loop formation – highly activated transcription and the associated accumulation of DNA superhelical stresses in the absence of Top1 (Figure 2). Once formed, RNase Hs function to destabilize G-loops by degradation of RNA, which is consistent with the formation of RNA:DNA hybrids and co-transcriptional G-loop structures. G-loops can also be destabilized by Sgs1 helicase, which presumably unwind G quartets on the non-transcribed strand. Although this system appears to recapitulate some physiological features associated with Sμ sequences, it is not intended to model the process of CSR. The assay system used here, however, will be useful in investigating whether the collision between transcription and replication machineries can influence the stability of G-loop forming sequences and also in elucidating additional helicases and nucleases that might function to collapse G-loop structures. The general approach outline in this study can also be used to study the requirements of maintaining other types of repetitive sequences that have been associated with genetic instability.
Figure 2.
Regulation of G-loop formation in highly transcribed DNA.
See Discussion for details.
Highlights.
We have developed a yeast genetic assay system to analyze genomic instability conferred by the repetitive G-rich immunoglobulin switch Mu region (Sμ) sequence.
Activated transcription of Sμ sequence embedded in yeast genome leads to orientation-specific instability with the higher instability being observed when G-rich strand is on the non-transcribed strand mimicking the physiological orientation.
Under conditions in which extended R-loop formation likely occurs – in the absence of Top1 or the loss of RNase H and Sgs1 helicase – we observed further increase in the instability of Sμ sequence and greater orientation bias.
Acknowledgements
The authors thank F. Alt for the gift of pT7-Sμ, Y. Pavlov for pESC-LEU-hAIDsc, and T. Petes for the critical reading of the manuscript. This work was supported by NIH grant GM38464 to SJ-R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimersm that apply to the journal pertain.
Conflict of interest statement
The authors declare that we have no conflict of interest.
References
- [1].Bacolla A, Wojciechowska M, Kosmider B, Larson JE, Wells RD. The involvement of non-B DNA structures in gross chromosomal rearrangements. DNA Repair (Amst) 2006;5:1161–1170. doi: 10.1016/j.dnarep.2006.05.032. [DOI] [PubMed] [Google Scholar]
- [2].Gordenin DA, Resnick MA. Yeast ARMs (DNA at-risk motifs) can reveal sources of genome instability. Mutat Res. 1998;400:45–58. doi: 10.1016/s0027-5107(98)00047-5. [DOI] [PubMed] [Google Scholar]
- [3].Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- [4].Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat Struct Mol Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- [5].Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- [6].Duquette ML, Huber MD, Maizels N. G-rich proto-oncogenes are targeted for genomic instability in B-cell lymphomas. Cancer Res. 2007;67:2586–2594. doi: 10.1158/0008-5472.CAN-06-2419. [DOI] [PubMed] [Google Scholar]
- [7].Willis TG, Dyer MJ. The role of immunoglobulin translocations in the pathogenesis of B-cell malignancies. Blood. 2000;96:808–822. [PubMed] [Google Scholar]
- [8].Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Schawalder J, Paric E, Neff NF. Telomere and ribosomal DNA repeats are chromosomal targets of the bloom syndrome DNA helicase. BMC Cell Biol. 2003;4:15. doi: 10.1186/1471-2121-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Weitao T, Budd M, Campbell JL. Evidence that yeast SGS1, DNA2, SRS2, and FOB1 interact to maintain rDNA stability. Mutat Res. 2003;532:157–172. doi: 10.1016/j.mrfmmm.2003.08.015. [DOI] [PubMed] [Google Scholar]
- [11].Smith S, Banerjee S, Rilo R, Myung K. Dynamic regulation of single-stranded telomeres in Saccharomyces cerevisiae. Genetics. 2008;178:693–701. doi: 10.1534/genetics.107.081091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Stavropoulos DJ, Bradshaw PS, Li X, Pasic I, Truong K, Ikura M, Ungrin M, Meyn MS. The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum Mol Genet. 2002;11:3135–3144. doi: 10.1093/hmg/11.25.3135. [DOI] [PubMed] [Google Scholar]
- [13].Killen MW, Stults DM, Adachi N, Hanakahi L, Pierce AJ. Loss of Bloom syndrome protein destabilizes human gene cluster architecture. Hum Mol Genet. 2009;18:3417–3428. doi: 10.1093/hmg/ddp282. [DOI] [PubMed] [Google Scholar]
- [14].Sun H, Bennett RJ, Maizels N. The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucleic Acids Res. 1999;27:1978–1984. doi: 10.1093/nar/27.9.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sun H, Karow JK, Hickson ID, Maizels N. The Bloom’s syndrome helicase unwinds G4 DNA. J Biol Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- [16].Ribeyre C, Lopes J, Boule JB, Piazza A, Guedin A, Zakian VA, Mergny JL, Nicolas A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kruisselbrink E, Guryev V, Brouwer K, Pontier DB, Cuppen E, Tijsterman M. Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective C. elegans. Curr Biol. 2008;18:900–905. doi: 10.1016/j.cub.2008.05.013. [DOI] [PubMed] [Google Scholar]
- [18].Sundararajan R, Gellon L, Zunder RM, Freudenreich CH. Double-strand break repair pathways protect against CAG/CTG repeat expansions, contractions and repeat-mediated chromosomal fragility in Saccharomyces cerevisiae. Genetics. 2010;184:65–77. doi: 10.1534/genetics.109.111039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Balakumaran BS, Freudenreich CH, Zakian VA. CGG/CCG repeats exhibit orientation-dependent instability and orientation-independent fragility in Saccharomyces cerevisiae. Hum Mol Genet. 2000;9:93–100. doi: 10.1093/hmg/9.1.93. [DOI] [PubMed] [Google Scholar]
- [20].Kim HM, Narayanan V, Mieczkowski PA, Petes TD, Krasilnikova MM, Mirkin SM, Lobachev KS. Chromosome fragility at GAA tracts in yeast depends on repeat orientation and requires mismatch repair. EMBO J. 2008;27:2896–2906. doi: 10.1038/emboj.2008.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lobachev KS, Stenger JE, Kozyreva OG, Jurka J, Gordenin DA, Resnick MA. Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 2000;19:3822–3830. doi: 10.1093/emboj/19.14.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tian M, Alt FW. Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleases in vitro. J Biol Chem. 2000;275:24163–24172. doi: 10.1074/jbc.M003343200. [DOI] [PubMed] [Google Scholar]
- [23].Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Freedman JA, Jinks-Robertson S. Genetic requirements for spontaneous and transcription-stimulated mitotic recombination in Saccharomyces cerevisiae. Genetics. 2002;162:15–27. doi: 10.1093/genetics/162.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- [26].Mayorov VI, Rogozin IB, Adkison LR, Frahm C, Kunkel TA, Pavlov YI. Expression of human AID in yeast induces mutations in context similar to the context of somatic hypermutation at G-C pairs in immunoglobulin genes. BMC Immunol. 2005;6:10. doi: 10.1186/1471-2172-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- [28].Kim N, Abdulovic AL, Gealy R, Lippert MJ, Jinks-Robertson S. Transcription-associated mutagenesis in yeast is directly proportional to the level of gene expression and influenced by the direction of DNA replication. DNA Repair (Amst) 2007;6:1285–1296. doi: 10.1016/j.dnarep.2007.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Spell RM, Jinks-Robertson S. Determination of mitotic recombination rates by fluctuation analysis in Saccharomyces cerevisiae. Methods Mol Biol. 2004;262:3–12. doi: 10.1385/1-59259-761-0:003. [DOI] [PubMed] [Google Scholar]
- [31].Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annu Rev Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- [32].Belli G, Gari E, Aldea M, Herrero E. Functional analysis of yeast essential genes using a promoter-substitution cassette and the tetracycline-regulatable dual expression system. Yeast. 1998;14:1127–1138. doi: 10.1002/(SICI)1097-0061(19980915)14:12<1127::AID-YEA300>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- [33].Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Arudchandran A, Cerritelli S, Narimatsu S, Itaya M, Shin DY, Shimada Y, Crouch RJ. The absence of ribonuclease H1 or H2 alters the sensitivity of Saccharomyces cerevisiae to hydroxyurea, caffeine and ethyl methanesulphonate: implications for roles of RNases H in DNA replication and repair. Genes Cells. 2000;5:789–802. doi: 10.1046/j.1365-2443.2000.00373.x. [DOI] [PubMed] [Google Scholar]
- [35].Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- [36].Hickson ID. RecQ helicases: caretakers of the genome. Nat Rev Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- [37].Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- [38].Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].El Hage A, French SL, Beyer AL, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].French SL, Sikes ML, Hontz RD, Osheim YN, Lambert TE, El Hage A, Smith MM, Tollervey D, Smith JS, Beyer AL. Distinguishing the roles of Topoisomerases I and II in relief of transcription-induced torsional stress in yeast rRNA genes. Mol Cell Biol. 2011;31:482–494. doi: 10.1128/MCB.00589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Rondon AG, Jimeno S, Aguilera A. The interface between transcription and mRNP export: from THO to THSC/TREX-2. Biochim Biophys Acta. 2010;1799:533–538. doi: 10.1016/j.bbagrm.2010.06.002. [DOI] [PubMed] [Google Scholar]
- [42].Chavez S, Beilharz T, Rondon AG, Erdjument-Bromage H, Tempst P, Svejstrup JQ, Lithgow T, Aguilera A. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 2000;19:5824–5834. doi: 10.1093/emboj/19.21.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci U S A. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- [45].Gomez-Gonzalez B, Aguilera A. Activation-induced cytidine deaminase action is strongly stimulated by mutations of the THO complex. Proc Natl Acad Sci U S A. 2007;104:8409–8414. doi: 10.1073/pnas.0702836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ruiz JF, Gomez-Gonzalez B, Aguilera A. AID Induces Double-Strand Breaks at Immunoglobulin Switch Regions and c-MYC Causing Chromosomal Translocations in Yeast THO Mutants. PLoS Genet. 2011;7:e1002009. doi: 10.1371/journal.pgen.1002009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Roberts RW, Crothers DM. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science. 1992;258:1463–1466. doi: 10.1126/science.1279808. [DOI] [PubMed] [Google Scholar]
- [48].Roy D, Yu K, Lieber MR. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol Cell Biol. 2008;28:50–60. doi: 10.1128/MCB.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Tornaletti S, Park-Snyder S, Hanawalt PC. G4-forming sequences in the non-transcribed DNA strand pose blocks to T7 RNA polymerase and mammalian RNA polymerase II. J Biol Chem. 2008;283:12756–12762. doi: 10.1074/jbc.M705003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Drolet M, Phoenix P, Menzel R, Masse E, Liu LF, Crouch RJ. Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Natl Acad Sci U S A. 1995;92:3526–3530. doi: 10.1073/pnas.92.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Voynov V, Verstrepen KJ, Jansen A, Runner VM, Buratowski S, Fink GR. Genes with internal repeats require the THO complex for transcription. Proc. Nat. Acad. Sci. USA. 2006;103:14423–14428. doi: 10.1073/pnas.0606546103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Neidle S. The structures of quadruplex nucleic acids and their drug complexes. Curr Opin Struct Biol. 2009;19:239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]