Abstract
aidB is one of four genes of E. coli that is induced by alkylating agents and regulated by Ada protein. Three genes (ada, alkA, and alkB) encode DNA repair proteins that remove or repair alkylated bases. However, the role of AidB remains unclear despite extensive efforts to determine its function in cells exposed to alkylating agents. The E. coli AidB protein was identified as a component of the protein complex that assembles at strong promoters. We demonstrate that AidB protein preferentially binds to UP elements, AT rich transcription enhancer sequences found upstream of many highly expressed genes, several DNA repair genes, and housekeeping genes. AidB allows efficient transcription from promoters containing an UP element upon exposure to a DNA methylating agent and protects downstream genes from DNA damage. The DNA binding domain is required to target AidB to specific genes preferentially protecting them from alkylation damage. However, deletion of AidB’s DNA binding domain does not prevent its antimutagenic activity, instead this deletion appears to allow AidB to function as a cytoplasmic alkylation resistance protein. Our studies identify the role of AidB in alkylating agent exposed cells and suggest a new cellular strategy in which a subset of the genome is preferentially protected from damage by alkylating agents.
Keywords: AidB protein, alkylating agents, DNA protection, UP element
Introduction
The E. coli aidB gene is one of four genes of the adaptive response to alkylation damage and is regulated by Ada protein (For review see: [1,2]). Ada protein is a methyltransferase that functions as a transcriptional activator after transfer of a methyl group from DNA to a cysteine residue in its amino terminal domain. The alkylation of Ada is stable and activates it to function as a transcriptional activator that induces expression of the ada-alkB operon, the alkA and aidB genes. Ada, AlkA and AlkB are enzymes that repair different alkyl lesions in DNA. Ada removes alkyl groups from O6alkylguanine, O4 alkylthymine by transferring them to a cysteine residue in its C-terminal domain [3]. Its amino terminal domain is also a methyltransferase that repairs one stereoisomer of alkylated phosphates by transferring them to a cysteine residue in its N-terminal domain [4,5]. AlkA is a glycosylase that removes 6 different types of alkylated bases from DNA [6] and AlkB is an a-ketoglutarate-Fe(II)-dependent DNA dioxygenase that repairs 1-alkyladenine and 3-alkylcytosine lesions by oxidizing the alkyl groups to unstable derivatives that spontaneously decay restoring the bases to their original state [7,8].
The role for AidB in alkylated cells has remained an unsolved problem. AidB has similarity to the acyl-CoA dehydrogenase family of metabolic enzymes and has weak isovaleryl CoA-dehydrogenase activity [9,10]. AidB was also shown to be a flavoprotein that binds nonspecifically to double stranded DNA. This observation led to the suggestion that it might be a repair enzyme [9]. The recent crystal structure of AidB revealed that its flavin binding site lies within an interior channel, while its DNA binding site is accessible only from the exterior of the protein and is spatially distant from its flavin binding region. Based on these observations, it was suggested that AidB might instead bind and protect DNA by inactivating alkylators before they are able to react with DNA.
In this study we demonstrate that AidB has sequence specific DNA binding activity that targets AidB to UP element-containing genes. We propose the gene specific targeting of AidB protein to be a new cellular strategy that results in preferential protection from alkylation damage and counteracts transcription inhibition by alkylating agents at a subset of the genome, i.e., at genes controlled by promoters with UP elements.
Materials and Methods
Bacterial strains and plasmids
The bacterial strains and plasmids used in this work are listed in Table 1.
Table 1.
Bacterial strains and plasmids
| Strains/plasmids | Description | Reference or source |
|---|---|---|
| Strains | ||
| MG1655 | wild-type; F− λ− ilvG rfb50 rph1 | [38] |
| MV5924 | aidBΔ::TetR derivative of MG1655 in which the aidB gene is replaced by a tetracycline resistance cassette using the methods of Murphy and Campellone [39] | [11] |
| MV6774 | ada-alkBΔ25::CmR alkA1 tag-1 aidBΔ35::TetR derivative of MV1161 [13] | This study |
| MV6780 | ada-alkBΔ25::CmR alkA1 tag-1 aidBΔ35::TetR/pTrc99A) | This study |
| MV6782 | ada-alkBΔ25::CmR alkA1 tag-1 aidBΔ35::TetR/pMV435 (pTrc99A-AidB+) | This study |
| MV6790 | ada-alkBΔ25::CmR alkA1 tag-1 aidBΔ35::TetR/pMV1526 (pTrc99A-AidBΔ440– 451) | This study |
| Plasmids | ||
| pET22b(+) | carries an N-terminal pelB signal sequence for potential periplasmic localization, plus an optional C-terminal His-tag sequence | This study |
| pET22b-aidB | pET22bΔ(NdeI-HindIII)Ω(aidB gene) | This study |
| pET22b-lacZ | pET22bΔ(HindIII-XhoI)Ω(lacZ gene) | This study |
| pET22b-PrrnB(+UP)-lacZ | pET22b-lacZΔ(SphI-HindIII)ΩPrrnB(+UP) | This study |
| pET22b-PrrnB(−UP)-lacZ | pET22b-lacZΔ(SphI-HindIII)ΩPrrnB(−UP) | This study |
| pET22b-PleuA-lacZ | pET22b-lacZΔ(SphI-HindIII)ΩPleuA | This study |
| pET22b-PompF-lacZ | pET22b-lacZΔ(SphI-HindIII)ΩPompF | This study |
| pTrc99A | E. coli expression vector | [10] |
| pMV435 | pTrc99A-AidB+ | [10] |
| pMV1526 | pTrc99A-AidBΔ440–451 | This study |
Cloning of the aidB gene
The E. coli aidB gene was amplified from the bacterial chromosome by PCR using the primers listed in Table 2. The amplification product was digested with NdeI and HindIII (underlined in Table 2) and cloned into the pET22b (+) vector (Novagen) creating the plasmid pET22b-aidB. The resulting expression vector contains a 6X histidine tag to allow protein purification by Ni2+ affinity chromatography. Plasmid construction was verified by automated DNA sequencing. The recombinant AidB protein was produced and purified as described previously [11].
Table 2.
Oligonucleotides
| aidB Fw | 5'-ATACATATGGTGCACTGGCAAACTCA-3' |
| aidB Rv | 5'-ATAAAGCTTTAACACACACACTCCCC-3' |
| lacZ Fw | 5'-TGTAAGCTTATAACAATTTCACACAGGAA-3' |
| lacZ Rv | 5'-CGGCTCGAGTTATTTTTGACACCAGAC-3' |
| rrnB P1(+UP) Fw | 5'-TAAAGCATGCTCAGAAAATTATTTTAAATTTC-3' |
| rrnB P1Rv | 5'-ATTAAGCTTAGGAGAACCCCGCTGA-3' |
| rrnB P1(−UP) Fw | 5'-ATTTGCATGCCCTCTTGTCAGGCC-3' |
| PleuA Fw | 5'-ATAAGCATGCGGGACGTTTTTATTGCG-3' |
| PleuA Rv | 5'-AAGAAGCTTGATAAAGCGAACGATGTG-3' |
| PompF Fw | 5'-ATTTGCATGCACAAAGTTCCTTAAATTTTA-3' |
| PompF Rv | 5'-TAAAAGCTTAATAAAAATTTACGGAACTATTG-3' |
| rrnB P1(+UP) Fw bio (emsa) | 5'-bio-AGAAAATTATTTTAAATTTCCTCTTGTCAGGCCGGAATAAC TC CCTATAAT-3' |
| rrnB P1(+UP) Rv (emsa) | 5'-ATTATAGGGAGTTATTCCGGCCTGACAAGAGGAAATTTAA AATAA TTTTCT-3' |
| rrnB P1(+UP) Fw (emsa) | 5’-AGAAAATTATTTTAAATTTCCTCTTGTCAGGCCGGAATAAC TCCCT ATAAT-3’ |
| rrnB P1(−UP) Fw (emsa) | 5’-CCTCTTGTCAGGCCGGAATAACTCCCTATAAT-3’ |
| rrnB P1(−UP) Rv (emsa) | 5’-ATTATAGGGAGTTATTCCGGCCTGACAAGAGG-3’ |
Electrophoretic mobility shift assay (EMSA)
EMSA experiments were performed using rrnB P1wt as the biotin-labeled DNA probe. Sense and antisense oligonucleotides (Table 2) were annealed by incubation at 95°C for 5 min and successive gradual cooling to room temperature. Purified recombinant AidB was incubated with 20 ng of biotinylated DNA rrnB P1wt for 20 min at room temperature in 20 µl of buffer Z (25 mM HEPES pH 7.6, 50 mM KCl, 12.5 mM MgCl2, 1 mM DTT, 20% glycerol, 0.1% triton). Protein-DNA complexes were separated on 5% native polyacrylamide gel (29:1 cross-linking ratio) in 0.5x TBE (45 mM Tris pH 8.0, 45 mM boric acid, 1 mM EDTA) at 200 V (20 V/cm) at room temperature. Afterwards, electrophoretic transfer to a nylon membrane was carried out in 0.5× TBE at 380 mA for 45 min, and the transferred DNA was cross-linked to the membrane with UV light. After incubation in blocking buffer for 1 h at room temperature, the membrane was incubated with streptavidin-HRP conjugate (Sigma) for 30 min at room temperature. The membrane was washed and visualized with SuperSignal chemiluminescence reagent (Pierce).
Competition experiments were performed using increasing quantities (100×–500×) of either unlabelled rrnB P1wt, which contains its UP element used as a specific competitor or rrnB P1ΔUP is used as a non-specific competitor.
Construction of fusion plasmids for transcription assays
The lacZ gene was amplified from genomic DNA of E. coli by PCR using the primers listed in Table 2. The amplification product was digested with HindIII and XhoI (underlined in Table 2) and cloned into the pET22b (+) vector (Novagen) generating the plasmid pET22b-lacZ. The rrnB P1 promoter with (rrnB P1wt) and without its UP element (rrnB P1ΔUP), PleuA and PompF were amplified by PCR, digested with SphI and HindIII and inserted into pET22b-lacZ linearized with the same restriction enzymes. The resulting plasmids, designated as listed in Table 1, were verified by automated DNA sequencing.
In vivo transcription assays
MG1655 and MV5924 E. coli strains were individually transformed with pET22b-lacZ, pET22b-P rrnB P1wt-lacZ, pET22b-P rrnB P1ΔUP-lacZ, pET22b-PleuA-lacZ and pET22b-PompF-lacZ plasmids. These bacterial cultures grown overnight in LB medium at 30°C, were diluted 1:100 in fresh medium. At an A600 nm of 0.4, the cultures were divided in four aliquots: one was not supplemented and the other three aliquots were supplemented with MNNG (5µg/ml), ENNG (5µg/ml), MMS 0.04%, respectively. Cellular pellets were collected during the exponential growth phase. β-galactosidase activity from the promoters-lacZ fusions was determined by measuring ONPG-hydrolysis, as described by Miller [12] and was compared to the activity obtained using a promoterless lacZ gene.
Isolation of plasmid DNA and damage assay
The MG1655 and MV5924 E. coli strains bearing pET22b-lacZ were grown overnight in LB medium at 30°C; these bacterial cultures were then diluted 1:100 in fresh medium. At an A600 nm of 0.4, the cultures were divided in four aliquots: one was not supplemented and the other three aliquots were supplemented with MNNG (5µg/ml), ENNG (5µg/ml), MMS 0.04%, respectively. After addition of alkylating agent, the bacterial cells were allowed to grow for 3 h; the plasmid DNA was isolated and served as a probe for the estimation of alkylated bases. The plasmids were divided into 2 aliquots, one of which was treated with the E. coli AlkA (a kind gift from Patrick J. O’Brien) and AP Endo (NEB); the other aliquot did not receive further treatment (control). Treatment with AlkA was performed in 70 mM MOPS, pH 7.5, 1 mM EDTA, 1mM DTT, 5% glycerol for 30 min at 37°C, followed by treatment with AP Endo for 1 h at 37°C. Then the samples were subjected to electrophoresis in 0.8% agarose gel for ~1 h at 80 V using 40 mM Tris, pH 7.8, 1 mM EDTA buffer.
Determination of DNA damage in the lacZ gene
MG1655 and MV5924 E. coli strains were individually transformed with pET22b-lacZ, pET22b-P rrnB P1wt-lacZ, pET22b-P rrnB P1ΔUP-lacZ. These bacterial cultures grown overnight in LB medium at 30°C, were diluted 1:100 in fresh medium. At an A600 nm of 0.4, the cultures were divided in two aliquots, and one was supplemented with 0.04% MMS to activate the adaptive response. The bacterial cells were allowed to grow for 3h. Then, the plasmids under study were isolated from these bacterial cells and were digested with HindIII and XhoI to release the lacZ fragment. To estimate the presence of alkyl lesions, the DNA fragments were treated or not with the AlkA and AP Endo proteins. The samples were then subjected to electrophoresis on alkaline agarose gels in 30 mM NaOH, 1 mM EDTA, pH 8 buffer, at 60 V for 3 h at 25°C. The gel was neutralized by soaking in a solution containing 1.5 M NaCl and 1 M Tris-HCl, pH 7.6 for 1 h. Finally, the gel was stained in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.4) containing SYBR® Gold for 30 min at 25°C and the samples were then analysed for single-strand DNA breaks.
Cell survival and mutagenesis
Cell survival was tested by growing cells to a density of 1–3 × 108 cells/ml, treating with MNNG for 30 min, then diluting cells in phosphate buffered saline containing 4% Na2S2O3 to inactivate residual MNNG [13], then plating cells on LB plates [13]. Mutation frequencies were determined using DSEM plates [13]. Cultures grown to approximately 3 × 108 cells per ml then spread on alkylating agent containing plates and incubated for 3 days at 37°C and Arg+ mutant colonies counted. Since alkylating agents are relatively unstable, plates containing mutagens were made by first adding alkylators at volumes needed to attain the specified final concentration, then adding 25 ml cooled (50°C) DSEM medium. The plates were cooled for 20 min, then dried by incubation at 37°C for 20 min with covers removed and immediately inoculated. All mutagenesis measurements were made at sub-lethal doses of alkylators using strains deficient in most alkylation specific DNA repair mechanisms ada-alkBΔ25:: CmR alkA1 tag-1 aidBΔ35::TetR) carrying either the vector pTrc99A, or pTrc99A derivatives that express aidB alleles.
Results
To identify proteins that bind the upstream regions of strong promoters, we investigated the protein complex that assembles at the upstream elements of the rrnB P1 promoter (see Supplemental Data and Supplemental FigureS1) by comparing proteins that bind to a sequence containing the −35 region and the UP element, an A/T rich enhancer sequence that constitutes the upstream element of many genes [14–16],, but not to a similar sequence lacking the UP element. The presence of the E. coli AidB protein among these proteins was unexpected, and suggested a possible regulatory role for AidB in transcription.
AidB preferentially binds DNA containing UP elements
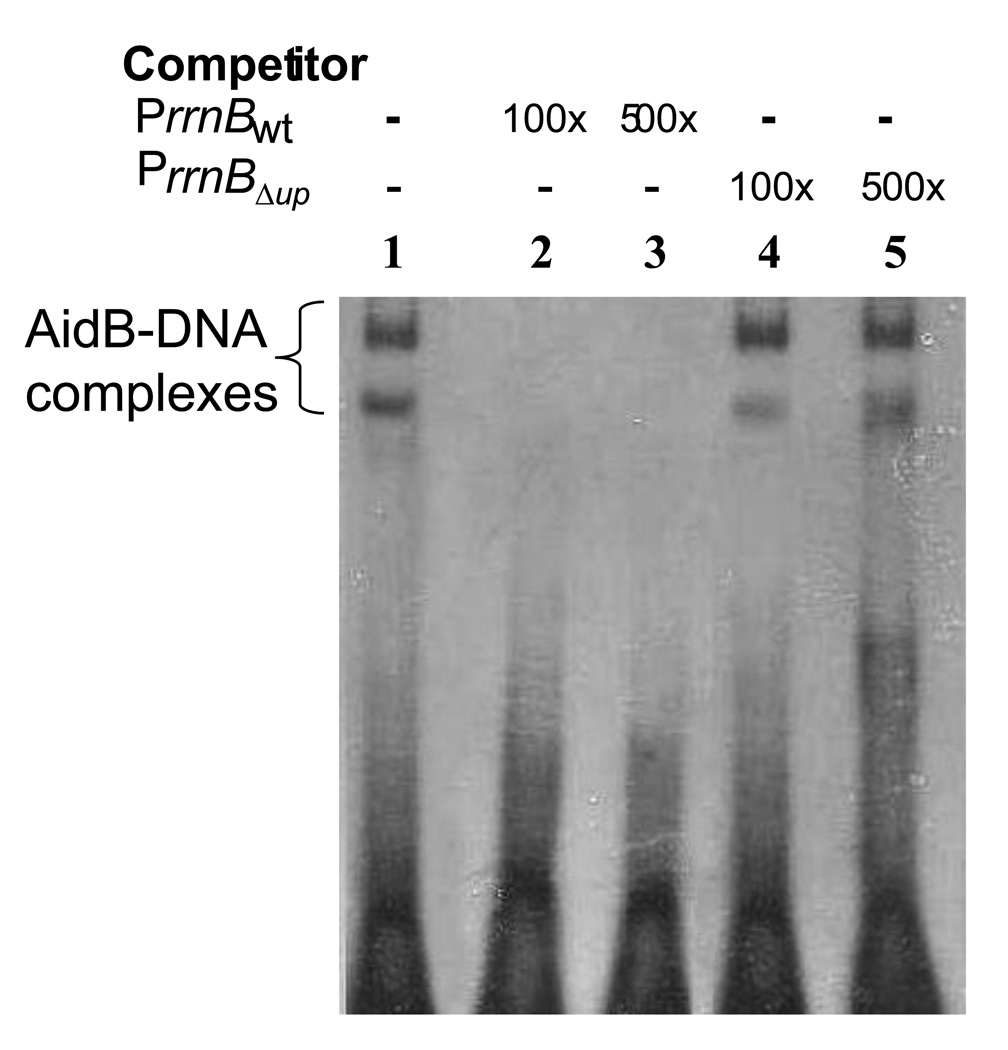
Figure 1 shows that AidB protein binds to DNA containing the rrnB P1wt promoter retarding the fragment in an electrophoretic mobility shift experiment (EMSA). When rrnB P1wt DNA is used as competitor, there is a rapid loss of binding to the labeled DNA. However, when the rrnB P1ΔUP promoter lacking the UP element is used as competitor, no inhibition of binding to the labeled rrnB P1wt sequence is seen even when it is added at a 500-fold excess. This indicates that AidB protein preferentially binds to rrnB P1 promoter only when the UP element is present. Similar results were also seen when random DNA containing the same base pair composition was used as competitor (see supplemental data). The preferential binding is not restricted to the rrnB P1wt promoter, since binding of AidB to its own promoter also requires the presence of the UP element [11].
Figure 1. Gel retardation.
Experiments were performed by incubating the AidB protein with rrnB P1wt; competitors were included as indicated. Lane 1: AidB protein incubated with rrnB P1wt. Lanes 2–3: Competition assay with rrnB P1wt (100×–500×) as specific competitor. Lanes 4–5: Competition assay with rrnB P1ΔUP as non specific competitor (100×–500×).
Functional analysis of AidB during transcription
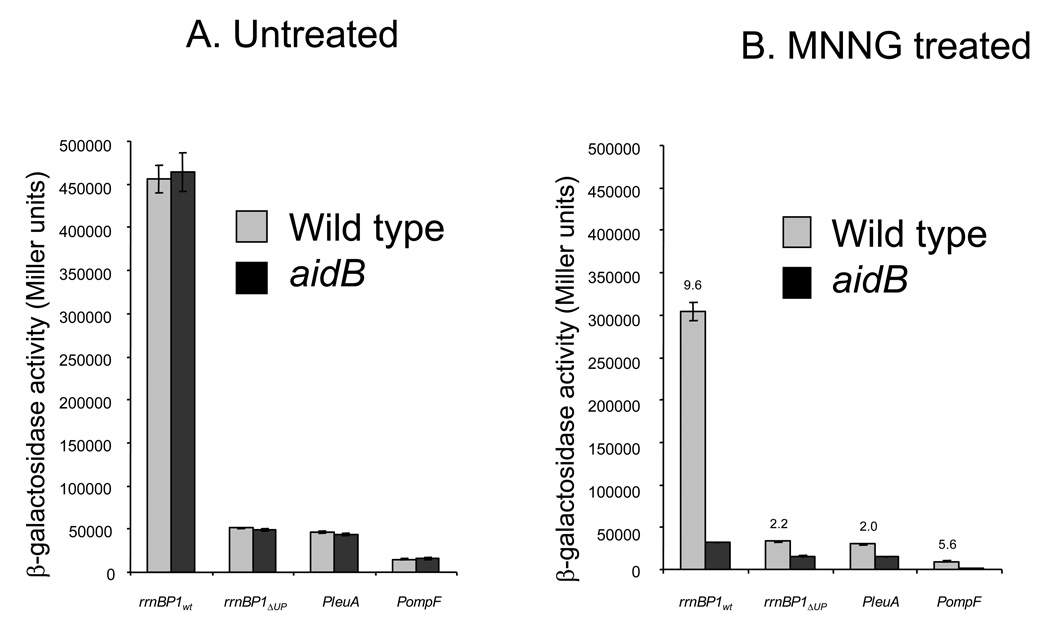
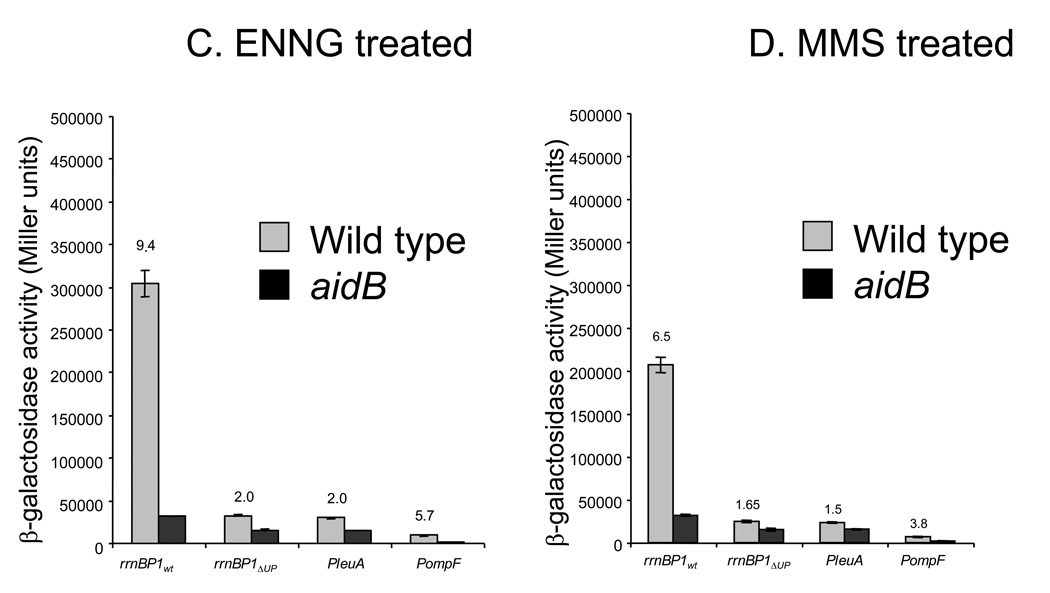
In order to determine whether the presence of AidB at the rrnB P1 promoter might be of biological significance, we tested its effect on transcription from this promoter by in vivo transcription assays. In addition, we tested other promoters that differ with respect to presence or absence of an UP element, namely: 1) the rrnB p1 promoter with its UP element (rrnBWT), 2) the rrnB promoter deleted of its UP element (rrnBΔUP), 3) PleuA, which lacks an UP element and 4) PompF, which has an UP element. All promoters were individually fused to a promoterless lacZ gene contained in the reporter plasmid pET22b-lacZ. Both MG1655 (wild type) and MV5924 (ΔaidB) E. coli strains were then transformed with the fusion plasmids and grown in LB medium, either in the absence or in the presence of alkylating agents (MMS, MNNG, ENNG). After 2 hours incubation in the presence or absence of the alkylating agent, β-galactosidase activity was measured during the exponential growth phase. As shown in Fig. 2A, wild type and aidB mutant strains not exposed to alkylators showed identical levels of β-galactosidase activity, indicating that the presence of AidB has no effect on transcription in untreated cells experiencing normal growth.
Figure 2. In vivo transcription of lacZ fused to different promoters.
The pET22b-P rrnB P1wt-lacZ, pET22b-P rrnB P1ΔUP-lacZ, pET22b-PleuA-lacZ and pET22b-PompF-lacZ plasmids were individually introduced into MG1655 (wild type) and MV5924 (ΔaidB) E. coli strains and the specific activity of β-galactosidase was determined in the absence (A) and in the presence of MNNG (5µg/ml) (B), ENNG (5µg/ml) (C), MMS 0.04% (D. The activities of promoters are reported in Miller units; the activity obtained using a promoterless lacZ gene was subtracted. Numbers above bars refer to the ratio of the β-galactosidase activity of the promoter measured in the wild type cells to the activity of that same promoter in the aidB mutant strain. Means and standard deviations have been calculated from four independent assays.
When MG1655 cells are treated with MMS, MNNG, or ENNG, transcription is reduced by roughly 2-fold for all promoters tested. In contrast, the aidB mutant showed a much more severe reduction in transcription, especially at the rrnBWT and PompF, the two UP element containing promoters (Fig. 2). This suggests that the interaction of AidB protein with this class of promoters is of functional significance and that AidB prevents transcription block by alkylation stress. The smaller effect of the aidB deletion on the two promoters lacking UP elements is consistent with preferential binding of AidB to this region (Fig. 1). Taken together, these data strongly suggest that AidB is required for high levels of transcription during alkylation stress and that it has a more pronounced effect on transcription from promoters containing an upstream UP element sequence. While it is formally possible that AidB is a transcriptional regulator of UP element containing genes when alkylating agents are present, a more likely explanation, based on its role as part of an alkylation inducible DNA repair response, is that AidB prevents or repairs DNA damage in specific regions of the genome, preferentially preserving the coding capacity of genes transcribed from UP element containing promoters.
AidB reduces the level of alkylation damage in DNA
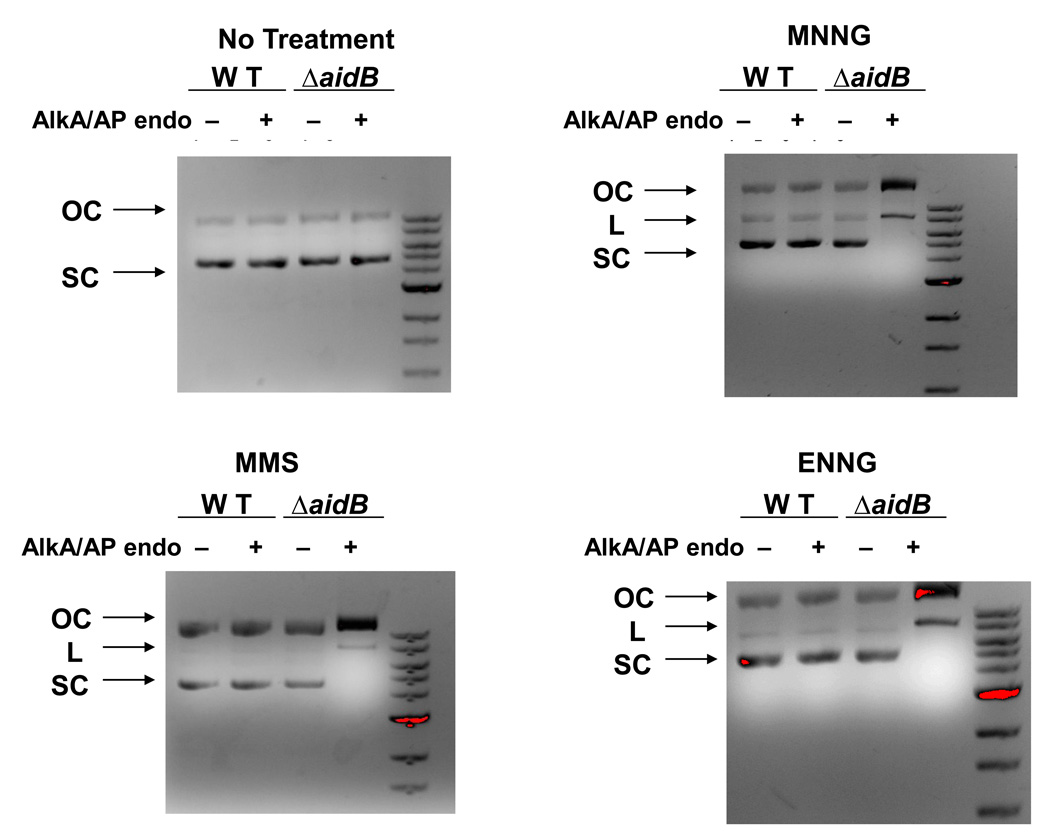
To test directly if AidB might be able to prevent or repair alkylation damage to DNA the pET22b-lacZ plasmid was isolated from wild type and aidB mutant cells grown either in the absence or in the presence of alkylators (MMS, MNNG, ENNG) and served as a probe for the estimation of alkylated bases in DNA. The plasmids were divided into 2 aliquots, one was treated with E. coli AlkA (a gift from Patrick J. O’Brien) and AP Endonuclease IV (AP Endo) (New England Biolabs); the other aliquot did not receive further treatment and served as a control. The AlkA glycosylase recognizes and removes a wide variety of alkylated bases converting them to abasic sites [6] and AP Endo is an apurinic/apyrimidinic (AP) endonuclease that converts the abasic sites to nicks [17,18]. The combined action of these two enzymes on a damage1d plasmid results in the conversion of the covalently closed circular (supercoiled) DNA to open circular and, if lesions are closely spaced, linear forms. AlkA treated and untreated plasmids were then subjected to electrophoresis on agarose gels and tested for conversion of the supercoiled form to open circular and linear forms. As shown in Fig. 3A, alkyl lesions were not detected in plasmids isolated from bacteria grown in LB medium without the addition of alkylating agents, indicating there is no detectable endogenous damage or non-specific cleavage by these enzymes in vitro. When plasmid DNA isolated from wild type cells exposed to alkylating agents was analyzed (Fig. 3B–D), treatment with AlkA and AP Endo did not result in nicking (Lane 2) indicating a lack of DNA damage, but when DNA isolated from the alkylating agent treated aidB mutant was analyzed, the supercoiled fraction was completely absent after AlkA/AP Endo treatment and there was an increase in both open circular and linear forms (Lanes 4). These results indicate that the presence of AidB reduces the level of alkylation damage in plasmid DNA. Moreover, AidB protects DNA from all three alkylating agents tested, although they differ in the nature of DNA lesions they produce. Indeed, MNNG methylates and ENNG ethylates DNA more effectively at O6-G than MMS. In contrast MMS methylates double stranded DNA primarily at N7-G and N3-A sites and in single stranded DNA regions it also methylates N1-A and N3-C more efficiently than MNNG [19].
Figure 3. Plasmid damage assay.
The pET22b-lacZ DNA was isolated from wild type (Lanes 1–2) and ΔaidB (Lanes 3–4) E. coli strains grown in the absence (A) or in the presence of MNNG (5µg/ml) (B), MMS 0.04% (C), ENNG (5µg/ml) (D), digested (Lanes 2, 4) or not (Lanes 1, 3) with AlkA and AP Endo and subjected to agarose gel electrophoresis. Lane 5, 1 Kb DNA marker (NEB). OC: open circular; L: linear; SC: supercoiled.
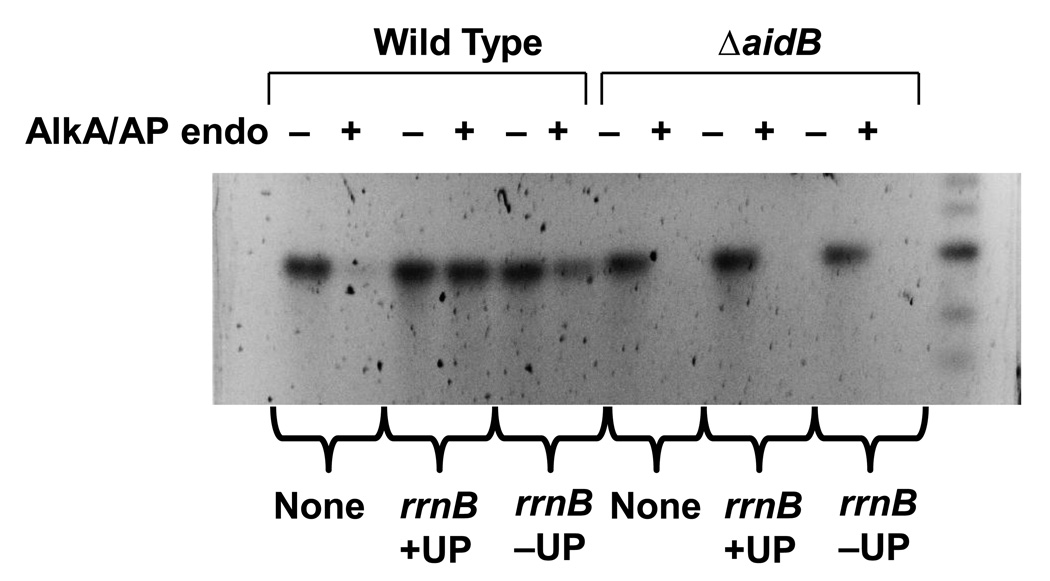
AidB preferentially protects DNA regions downstream of an UP element
Since AidB appears to protect DNA from alkylating agents very effectively (Figure 3), the lack of an alkylation sensitivity phenotype of aidB mutants remains a puzzle (see Figure 5 and [13,20]). The observation that AidB protein allows more effective transcription of genes with UP element promoters in the presence of an alkylating agent (Fig. 2) and has a higher affinity for UP element containing promoters [11], suggests that AidB may not protect the entire genome equally and may show a preference for UP element containing regions of DNA. To test this possibility, we analyzed the effect of AidB on alkylation damage in vivo in the lacZ fragment. In this experiment, we used three plasmids, each carrying the lacZ gene fused to three different upstream sequences: the rrnBWT promoter, the rrnBΔUP promoter, and a third plasmid carrying a promoterless lacZ gene. We investigated whether the presence of AidB might affect the content of alkyl lesions within the lacZ sequences. Wild type and aidB mutant cells containing these plasmids were first treated with MMS. After isolation, the plasmids were digested with restriction enzymes to release the lacZ fragment. The lacZ fragment was then purified by agarose gel electrophoresis and divided into two aliquots. One aliquot was treated with the AlkA/AP Endo to nick the DNA at the lesion sites. The samples were then subjected to electrophoresis on alkaline agarose gel to denature DNA and to separate nicked from full-length ssDNA fragments. Since only undamaged strands will run as full-length molecules, the fraction of strands containing lesions can be estimated by comparing the AlkA/AP Endo treated samples with controls not treated with AlkA/AP Endo. Figure 4 shows that the aidB mutant cells are not able to protect the lacZ gene regardless of the upstream sequence present and all DNA samples are equally sensitive to AlkA/AP Endo treatment (Lanes 8, 10,12). In Wild type cells, essentially all DNA from the rrnBWT bearing plasmid exposed to AlkA/AP Endo remains as full length (Fig. 4, compare Lanes 3 and 4). When lacZ is fused to the rrnBΔUP promoter, the sample treated with AlkA/AP Endo (Lane 6) shows a clear decrease in the amount of full-length fragments compared with the control DNA not treated with AlkA/AP Endo (Lane 5). Treatment of lacZ from the promoterless plasmid with AlkA/AP Endo resulted in an almost complete loss of full-length DNA fragments indicating a higher level of damage (Compare Lanes 1 and 2), thus confirming that the presence of AidB is required for the protection against alkyl damage. It also suggests that transcription itself cannot be solely responsible for damage prevention, since transcription at the onset of damage is identical in wild type and the aidB mutant (Fig. 2). Additionally, there is no detectable difference in damage levels when DNA samples isolated from the aidB mutant are compared with one another despite the markedly higher level of transcription of the lacZ gene transcribed from the rrnB P1wt element versus the promoterless lacZ. By contrast, plasmids from MMS treated wild type cells show a clear difference in their levels of protection from alkylation damage. Figure 4 shows that lacZ fused to the UP element containing the rrnB P1wt promoter is well protected from MMS exposure when compared, either to the samples from the aidB mutant, or lacZ fused to the rrnB promoter that lacks the UP element, or has no promoter.
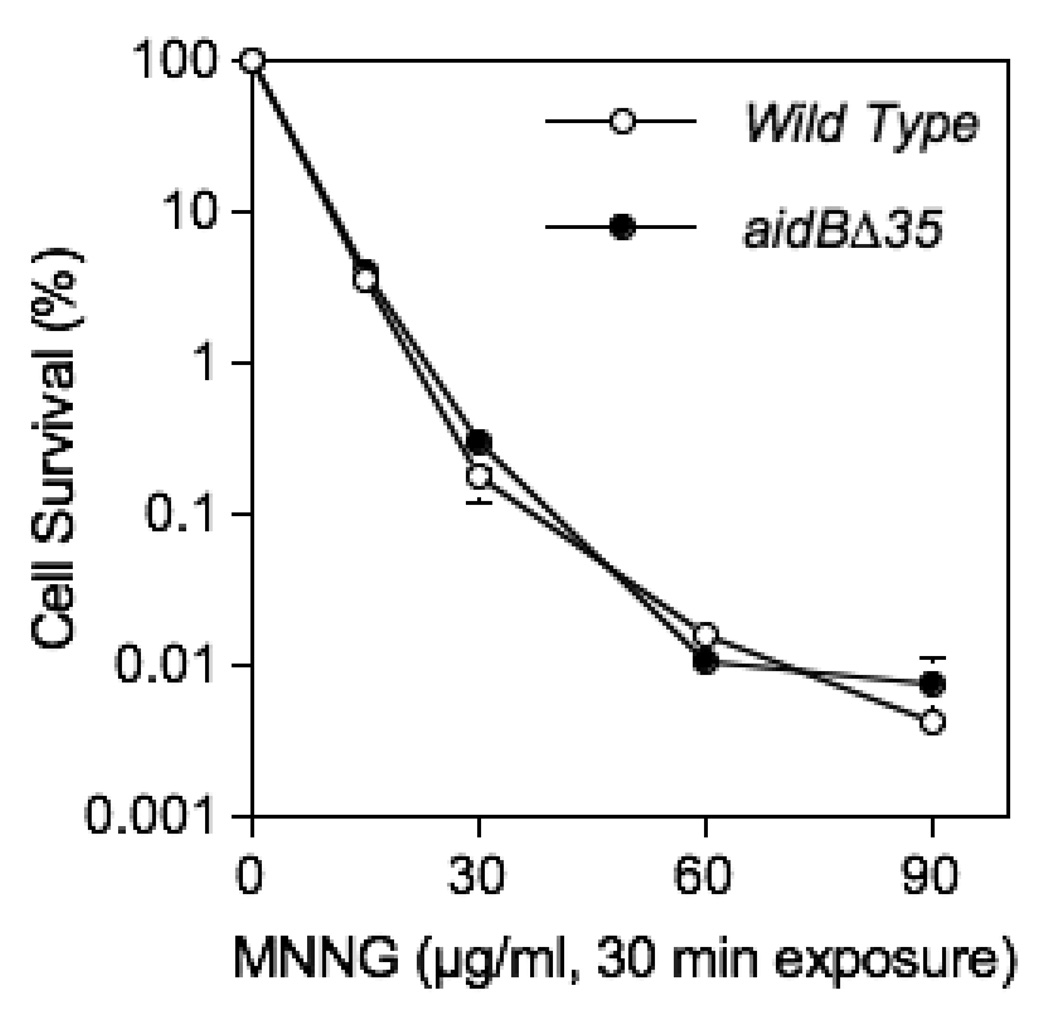
Figure 5. Alkylating agent sensitivity of strains deficient in adaptive response genes.
Cells were grown to a density of approximately 1–3 × 108 cells/ml and treated with MNNG as indicated. Wild type (MG1655), aidBΔ35::TetR (MV5924). Each data point represents 3 (15, 90 µg/ml) or 6 repetitions (0, 30 and 60 µg/ml). Standard errors of the mean are shown where visible beyond the data point.
Figure 4. AidB preferentially protects DNA regions containing an UP element.
The pET22b-lacZ, pET22b-P rrnB P1wt-lacZ and pET22b-P rrnB P1ΔUP-lacZ plasmids were isolated from wild type (Lanes 1–6) and ΔaidB (Lanes 7–12) E. coli strains grown in the presence of MMS 0.04% and digested to release and purify the lacZ fragment. The lacZ containing DNA fragments were untreated (Lanes 1, 3, 5, 7, 9, 11) or treated (Lanes 2, 4, 6, 8, 10, 12) with AlkA and AP Endo and subjected to electrophoresis on alkaline agarose gel. Lanes 1, 2, 7, 8: lacZ lacking a promoter; Lanes 3, 4, 9, 10: lacZ fused to the rrnB P1wt promoter with its UP element; Lanes 5, 6, 11, 12: lacZ fused to the rrnB P1ΔUP promoter without its UP element; Lane 13: 1Kb DNA marker (NEB).
These results demonstrate that AidB preferentially protects the DNA of genes transcribed from UP element-containing promoters to a greater extent than DNA fragments bearing promoters lacking an UP element.
AidB does not confer cellular resistance to alkylation damage
The aidB mutant strain MV5924was tested for its sensitivity to MNNG damage. Figure 5 shows that even a complete deletion of aidB results in little or no sensitivity to MNNG when compared with its isogenic wild type. The aidB mutation also does not affect MMS sensitivity (Figure S3, supplementary material). These results are consistent with previous observations that mutants carrying insertions in the aidB gene show no increase in sensitivity to alkylating agents [13]. A lack of alkylation sensitivity of the aidB deletion mutant is inconsistent with a general DNA damage prevention mechanism, since the ability to prevent damage throughout the genome should result in increased resistance. However, protection of only some DNA regions would prevent damage to only those genes that are targets for AidB protein and is unlikely to have a major effect on overall survival.
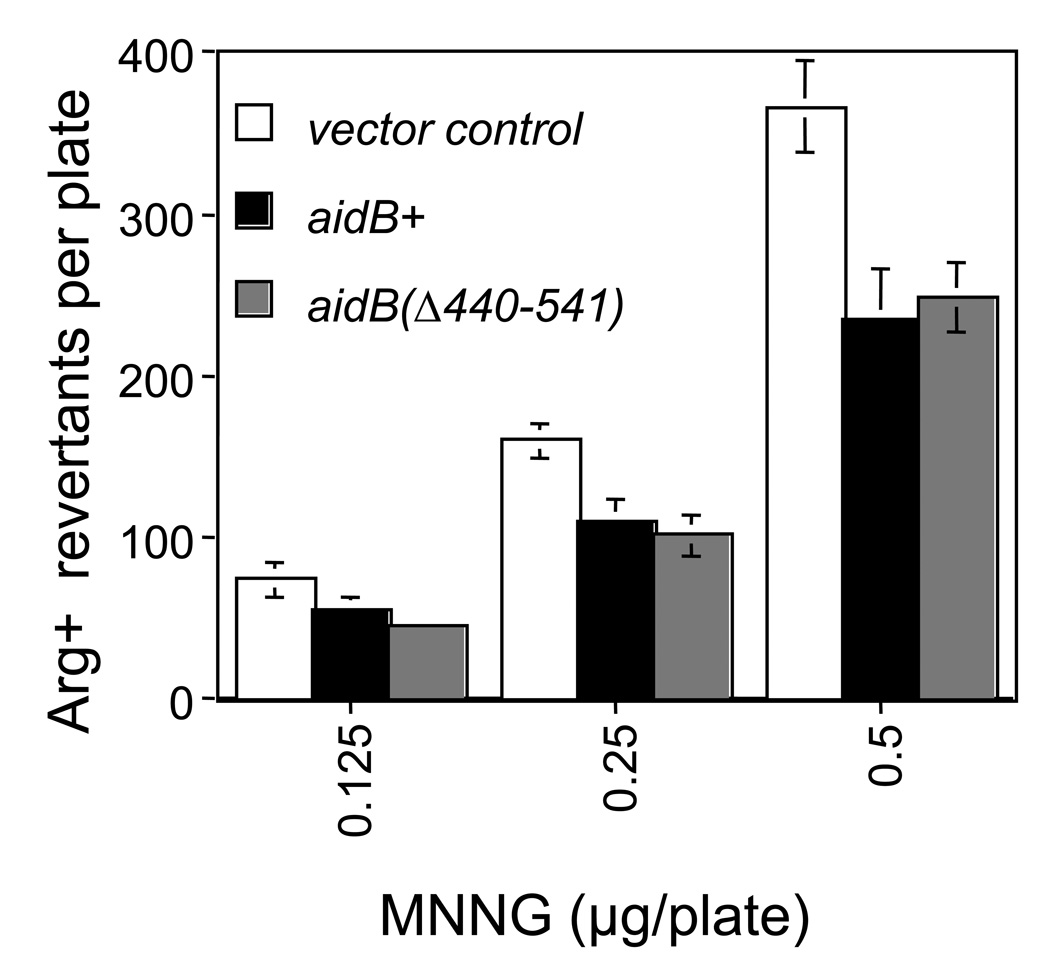
Based on the 3-dimensional structure of AidB protein, it has been suggested that AidB is unlikely to function as a DNA repair protein. Instead it may bind DNA and enzymatically inactivate alkylators as they approach the DNA. This notion is based on the observation that the dehydrogenase active site of AidB is spatially distant from its DNA binding face and accessible only from the exterior of the protein [23]. Since a DNA repair protein predicts that the DNA binding domain will be required for activity, we constructed an AidB mutant that lacks the entire DNA binding domain (aidBΔ440–541). This mutant has previously been shown to have IVD activity identical to the wild type, but no detectable DNA binding activity [11]. Since wild type AidB protein functions as an antimutator when cells are grown in the presence of MNNG, we tested if the DNA binding deficient AidB mutant protein retains the antimutator activity. Figure 6 shows that the aidBΔ440–541 mutant allele is as active as the wild type allele in the antimutator assay, indicating that DNA binding is not required for the alkylation resistance function of AidB and suggesting DNA binding instead serves to target AidB to specific genes.
Figure 6. Antimutagenic activity of wild type and AidB(Δ440–541).
Cells were grown to a density of approximately 3 × 108 cells per ml and undiluted cultures were spread on DSEM plates ± alkylating agents and mutation frequencies determined. All mutagenesis experiments were conducted at sublethal doses of alkylating agents using strains deficient in most alkylation specific DNA repair mechanisms: aidB+ (MV6782, ada-alkBΔ25:: CmR alkA1 tag-1 aidBΔ35::TetR/pTrc99A-AidB+); aidBΔ440–541 (MV6790, ada-alkBΔ25::CmR alkA1 tag-1 aidBΔ35::TetR/pTrc99A-AidBΔ440–451); vector control (MV6780, ada-alkBΔ25::CmR alkA1 tag-1 aidBΔ35::TetR/pTrc99A).
Discussion
The biological role of AidB has long been uncertain. Our data demonstrate that AidB prevents DNA damage by alkylating agents and counteracts the block to transcription that results upon exposure to alkylating agents, especially in genes that are transcribed from promoters containing UP elements. These effects are seen after treatment with MMS, MNNG and ENNG, three alkylating agents that produce different DNA lesions or damage spectra [24]. The result that ENNG damage is also prevented is especially interesting since ENNG lesions are repaired not only the by adaptive response repair system, but also by nucleotide excision repair in E. coli [25].
The result that AidB can prevent DNA damage seems inconsistent with the result that loss of AidB function by a complete deletion has little or no effect on MMS or MNNG sensitivity of the mutant strain (Figure 5 and S3). However, effects of an aidB mutation on DNA damage and mutagenesis were seen at sublethal doses of alkylating agents (Figures 3–6). Since Ada dependent aidB induction is relatively weak compared to that of other adaptive response genes [13,20], it is possible that AidB protein levels are too low to provide adequate protection against lethal doses of alkylating agents. The primary function of AidB may be to protect DNA from the low levels of alkylators that are produced as by-products of stationary phase metabolism [26–29], a possibility that is consistent with the observation that aidB is induced and expressed at elevated levels in stationary phase [10,30,31].
The result that the AidB protein specifically binds to DNA sequences that include the UP element [11] (see also Fig. 1), suggests that the lack of increased sensitivity to high levels of alkylating agents in the aidB mutant (Fig. 5) may also be due to the fact that AidB only protects a subset of the genome, leaving other genes, including essential ones, exposed to DNA damage. The aidB mutant phenotype is consistent with targeted repair or damage prevention and is analogous to the effect seen in strains that lack the ability to carry out transcription-coupled repair (TCR) of UV damage, the only other gene specific repair or damage prevention system currently known. A TCR deficient mfd mutant shows only a modest decrease in cellular resistance to UV, but a dramatic reduction in the rate at which repair of active genes occurs [32–34]. Thus, the AidB prevention mechanism appears to be a cellular strategy to preferentially protect a subset of genes. In this case the genes include ones important for basic metabolic processes and key DNA repair genes. AidB is targeted towards genes whose promoters have upstream UP elements. This includes genes such as most of the ribosomal RNA genes and many tRNA genes as well as several key DNA repair genes required for recovery from alkylation damage such as recA, polA, sulA, recN the ada-alkB operon, and aidB itself [14,35,36].
The presence of a functional aidB gene protects UP element genes from alkylation damage and results in more efficient transcription in the presence of alkylating agents. lacZ fused to the two UP element containing rrn and ompF promoters are transcribed 10- and 6- fold more efficiently in the presence of an alkylating agent than lacZ fused to an rrn promoter whose UP element has been deleted, or the leuA promoter, which has no UP element. Although it is possible that AidB has regulatory effects on these genes, a lower level of template damage should clearly contribute to the transcription efficiency.
Promoters lacking an UP element, and thus not efficiently bound by AidB protein still show a slightly higher level of transcription in wild type versus aidB mutants upon alkylation (2.2 and 2-fold enhancement for rrnBΔUP and Pleu, Fig. 2). It is unclear if this aidB-dependent enhancement of transcription in the presence of an alkylating agent represents some direct protection by aidB, or is an indirect effect of the elevated levels of ribosomes, tRNAs and possibly other components of the translational machinery that are transcribed at a higher levels in the aidB+ strain under these conditions. The observation that the protection of lacZ fused to the rrn promoter lacking an UP element and the observation that plasmid DNA shows better protection in wild type than in an aidB mutant strain (Fig. 4), suggests that there may be some general protection resulting from the presence of aidB, especially when it is highly expressed, or induced for a long period of time as in these experiments. Under these conditions AidB may initially protect the genes preferentially targeted, followed by other parts of the genome if AidB protein accumulates to sufficiently high levels. The precise mechanism of action of AidB remains to be determined, though it is possible that it provides protection of DNA adjacent to its preferred binding site, either by simply inactivating alkylating agents and reducing the local concentration, or by polymerizing into multimers that extend from the initial binding site. In the latter case, it is likely to protect both by shielding the DNA and by inactivating alkylators.
However, the MNNG resistance resulting from expression of the DNA binding deficient aidB mutant protein, AidB(Δ440–541) indicates that the mutant lacking DNA binding activity still functions to prevent alkylation mutagenesis. This observation makes it unlikely that AidB functions by simply binding and coating the DNA, thus preventing access by alkylators. The ability of the DNA binding defective AidB protein to prevent mutagenesis suggests that AidB is not a DNA repair protein, since DNA repair would be inhibited by lack of DNA binding activity. Instead, AidB is more likely to function to prevent damage by detoxifying alkylating agents, which could reduce DNA alkylation even in the absence of DNA binding activity by reducing the intracellular concentration of active alkylators. A role for AidB in alkylating agent detoxification is also consistent with earlier work on AidB and analysis of the structural features of the protein [10,23]. Determination of the precise mechanism by which AidB may inactivate alkylating agents requires further work to examine the chemistry of the hypothetical process.
It is unclear how widespread preferential damage prevention mechanisms such as AidB are, if other prokaryotes and eukaryotes have similar damage prevention proteins, or if the strategy of preferential DNA protection extends to mechanisms that prevent damage by other agents. In E. coli the dps gene is highly expressed in stationary phase and prevents oxidative DNA damage. Unlike AidB, however, this protein is produced at very high levels and appears to function as a genome wide protective protein. It is unclear if it may also have a preference for specific sequences when it is expressed at lower levels [37].
Highlights.
In this manuscript we demonstrate that aidB binds preferentially to UP element containing genes and preferentially protects them from alkylation damage. Additionally, the reduction in transcription normally seen in the presence of alkylators is much greater in the aidB mutant than in wild type, indicating that transcription capacity is preserved when aidB is functional. We further demonstrate that aidB overexpression reduces mutagenesis and that this does not require the DNA binding domain, suggesting that it does not repair DNA, but prevents damage from occurring.
Supplementary Material
Acknowledgements
We thank Patrick J. O’Brien (University of Michigan) for AlkA protein. This work was funded in part by NIH Grant CA100122 to MRV, Italian MIUR PRIN 2008 and FIRB 2007 Italian Human ProteoNet grants to AD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Volkert MR, Landini P. Transcriptional responses to DNA damage. Curr. Opin. Microbiol. 2001;4:178–185. doi: 10.1016/s1369-5274(00)00186-7. [DOI] [PubMed] [Google Scholar]
- 2.Landini P, Volkert MR. Regulatory responses of the adaptive response to alkylation damage: a simple regulon with complex regulatory features. J. Bacteriol. 2000;182:6543–6549. doi: 10.1128/jb.182.23.6543-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCarthy TV, Karran P, Lindahl T. Inducible repair of O-alkylated DNA pyrimidines in Escherichia coli. EMBO J. 1984;3:545–550. doi: 10.1002/j.1460-2075.1984.tb01844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teo I, Sedgwick B, Kilpatrick MW, McCarthy TV, Lindahl T. The intracellular signal for induction of resistance to alkylating agents in E. coli. Cell. 1986;45:315–324. doi: 10.1016/0092-8674(86)90396-x. [DOI] [PubMed] [Google Scholar]
- 5.Nakabeppu Y, Sekiguchi M. Regulatory mechanisms for induction of synthesis of repair enzymes in response to alkylating agents: Ada protein acts as a transcriptional regulator. Proc. Natl. Acad. Sci. USA. 1986;83:6297–6301. doi: 10.1073/pnas.83.17.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Brien PJ, Ellenberger T. The Escherichia coli 3-methyladenine DNA glycosylase AlkA has a remarkably versatile active site. J Biol Chem. 2004;279:26876–26884. doi: 10.1074/jbc.M403860200. [DOI] [PubMed] [Google Scholar]
- 7.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 8.Falnes PO, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 9.Rohankhedkar MS, Mulrooney SB, Wedemeyer WJ, Hausinger RP. The AidB component of the Escherichia coli adaptive response to alkylating agents is a flavin-containing, DNA-binding protein. J Bacteriol. 2006;188:223–230. doi: 10.1128/JB.188.1.223-230.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landini P, Hajec LI, Volkert MR. Structure and transcriptional regulation of the Escherichia coli adaptive response gene aidB. J. Bacteriol. 1994;176:6583–6589. doi: 10.1128/jb.176.21.6583-6589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rippa V, Amoresano A, Esposito C, Landini P, Volkert M, Duilio A. Specific DNA binding and regulation of its own expression by the AidB protein in Escherichia coli. J Bacteriol. 2010;192:6136–6142. doi: 10.1128/JB.00858-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratories, Cold Spring Harbor; 1972. [Google Scholar]
- 13.Volkert MR, Nguyen DC. Induction of specific Escherichia coli genes by sublethal treatments with alkylating agents. Proc. Natl. Acad. Sci. USA. 1984;81:4110–4114. doi: 10.1073/pnas.81.13.4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gourse RL, Ross W, Gaal T. UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol Microbiol. 2000;37:687–695. doi: 10.1046/j.1365-2958.2000.01972.x. [DOI] [PubMed] [Google Scholar]
- 15.Estrem ST, Gaal T, Ross W, Gourse RL. Identification of an UP element consensus sequence for bacterial promoters. Proc. Natl. Acad. Sci. USA. 1998;95:9761–9766. doi: 10.1073/pnas.95.17.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ross W, Gosink KK, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse RL. A third recognition element in bacterial promoters: DNA binding by the a subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 17.Ljungquist S, Lindahl T. Relation between Escherichia coli endonucleases specific for apurinic sites in DNA and exonuclease III. Nucleic Acids Res. 1977;4:2871–2879. doi: 10.1093/nar/4.8.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ljungquist S. A new endonuclease from Escherichia coli acting at apurinic sites in DNA. J Biol Chem. 1977;252:2808–2814. [PubMed] [Google Scholar]
- 19.Singer B. The chemical effects of nucleic acid alkylation and their relation to mutagenesis and carcinogenesis. Prog Nucleic Acid Res Mol Biol. 1975;15:219–284. [PubMed] [Google Scholar]
- 20.Volkert MR, Nguyen DC, Beard KC. Escherichia coli gene induction by alkylation treatment. Genetics. 1986;112:11–26. doi: 10.1093/genetics/112.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Volkert MR. Adaptive response of Escherichia coli to alkylation damage. Environ. Molec. Mutagen. 1988;11:241–255. doi: 10.1002/em.2850110210. [DOI] [PubMed] [Google Scholar]
- 22.Lindahl T, Sedgwick B, Sekiguchi M, Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu. Rev. Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- 23.Bowles T, Metz AH, O'Quin J, Wawrzak Z, Eichman BF. Structure and DNA binding of alkylation response protein AidB. Proc Natl Acad Sci U S A. 2008;105:15299–15304. doi: 10.1073/pnas.0806521105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singer B, Grunberger D. Molecular Biology of Mutagens and Carcinogens. New York: Plenum Press; 1983. [Google Scholar]
- 25.Bonanno K, Wyrzykowski J, Chong W, Matijasevic Z, Volkert MR. Alkylation resistance of E. coli cells expressing different isoforms of human alkyladenine DNA glycosylase (hAAG) DNA Rep. 2002;1:507–517. doi: 10.1016/s1568-7864(02)00051-4. [DOI] [PubMed] [Google Scholar]
- 26.Rebeck GW, Samson L. Increased spontaneous mutation and alkylation sensitivity of Escherichia coli strains lacking the ogt O6-methylguanine DNA repair methyltransferase. J. Bacteriol. 1991;173:2068–2076. doi: 10.1128/jb.173.6.2068-2076.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sedgwick B. Nitrosated peptides and polyamines as endogenous mutagens in O6-alkylguanine-DNA alkyltransferase deficient cells. Carcinogenesis. 1997;18:1561–1567. doi: 10.1093/carcin/18.8.1561. [DOI] [PubMed] [Google Scholar]
- 28.Taverna P, Sedgwick B. Generation of edogenous methylating agents by nitrosation in Escherichia coli. J. Bacteriol. 1996;178:5105–5111. doi: 10.1128/jb.178.17.5105-5111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sedgwick B. Oxidation of methylhydrazines to mutagenic methylating derivatives and inducers of the adaptive response of Escherichia coli to alkylation damage. Cancer Res. 1992;52:2693–2697. [PubMed] [Google Scholar]
- 30.Landini P, Hajec LI, Nguyen LH, Burgess RR, Volkert MR. The leucine-responsive protein (Lrp) acts as a specific repressor for ss-dependent transcription of the Escherichia coli aidB gene. Molec. Microbiol. 1996;20:947–955. doi: 10.1111/j.1365-2958.1996.tb02536.x. [DOI] [PubMed] [Google Scholar]
- 31.Volkert MR, Hajec LI, Matijasevic Z, Fang F, Prince R. Induction of the Escherichia coli aidB gene under oxygen limiting conditions requires a functional rpoS (katF) gene. J. Bacteriol. 1994;176:7638–7645. doi: 10.1128/jb.176.24.7638-7645.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selby CP, Sancar A. Mechanisms of transcription-repair coupling and mutation frequency decline. Microbiol Rev. 1994;58:317–329. doi: 10.1128/mr.58.3.317-329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selby CP, Sancar A. Molecular Mechanism of Transcription-Repair Coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 34.Selby CP, Witkin E, Sancar A. Escherichia coli mfd mutant deficient in "mutation frequency decline" lacks strand-specific repair: in vitro complementation with purified coupling factor. Proc. Natl. Acad. Sci. USA. 1991;88 doi: 10.1073/pnas.88.24.11574. 11574-11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ross W, Aiyar SE, Salomon J, Gourse RL. Escherichia coli promoters with UP elements of different strengths: modular structure of bacterial promoters. J Bacteriol. 1998;180:5375–5383. doi: 10.1128/jb.180.20.5375-5383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landini P, Volkert MR. RNA polymerase a subunit binding site in positively controlled promoters: a new model for RNA polymerase/promoter interaction and transcriptional activation in the E. coli ada and aidB genes. EMBO J. 1995;14:4329–4335. doi: 10.1002/j.1460-2075.1995.tb00107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez A, Kolter R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J. Bacteriol. 1997;179:5188–5194. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jensen KF. The Escherichia coli K-12 "wild types" W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J Bacteriol. 1993;175:3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy KC, Campellone KG. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol Biol. 2003;4:11. doi: 10.1186/1471-2199-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volkert MR. Altered induction of the adaptive response to alkylation damage in Escherichia coli recF mutants. J. Bacteriol. 1989;171:99–103. doi: 10.1128/jb.171.1.99-103.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.