Abstract
A Roche 454 cDNA deep sequencing experiment was performed on a developing corm of Amorphophallus konjac—also known as voodoo lily. The dominant storage polymer in the corm of this plant is the polysaccharide glucomannan, a hemicellulose known to exist in the cell walls of higher plants and a major component of plant biomass derived from softwoods. A total of 246 mega base pairs of sequence data was obtained from which 4,513 distinct contigs were assembled. Within this voodoo lily expressed sequence tag collection genes representing the carbohydrate related pathway of glucomannan biosynthesis were identified, including sucrose metabolism, nucleotide sugar conversion pathways for the formation of activated precursors as well as a putative glucomannan synthase. In vivo expression of the putative glucomannan synthase and subsequent in vitro activity assays unambiguously demonstrate that the enzyme has indeed glucomannan mannosyl- and glucosyl transferase activities. Based on the expressed sequence tag analysis hitherto unknown pathways for the synthesis of GDP-glucose, a necessary precursor for glucomannan biosynthesis, could be proposed. Moreover, the results highlight transcriptional bottlenecks for the synthesis of this hemicellulose.
Electronic supplementary material
The online version of this article (doi:10.1007/s00425-011-1422-z) contains supplementary material, which is available to authorized users.
Keywords: Glucomannan, 454 deep sequencing, Cellulose synthase-like family A (CSLA), Amorphophallus konjac
Introduction
The most abundant components of plant biomass are the lignocellulosics composed of microfibrillar cellulose, a variety of hemicelluloses, and the polyphenol lignin (Pauly and Keegstra 2008). The hemicelluloses are thought to cross-link and tether the cellulose microfibrils in muro, thus providing a load-bearing structure (Fry 1989). In the case of gymnosperms, glucomannan—a polymer consisting of β-1,4-linked d-glucose and d-mannose residues that can be O-acetylated—is the major hemicellulose in the secondary cell wall, where it is also believed to have a structural function (Maeda et al. 2000). The non-crystalline glucomannans consist entirely of yeast fermentable hexoses and hence they present a useful feedstock source for the generation of biofuels from lignocellulosic plant materials (Pauly and Keegstra 2008; Mortimer et al. 2010). Thus, understanding the biosynthesis and metabolism of glucomannan and the regulation of these processes might lead to increasing the abundance of this relatively easily digestible and fermentable polymer in plant feedstocks improving its utility as a resource for the production of biofuels and/or other commodity chemicals.
The first identified gene involved in mannan biosynthesis was a galactosyltransferase (Edwards et al. 1999) resulting in the substitution of the mannan backbone rendering the polymer more soluble. A β-mannan synthase involved in the backbone formation also has been identified in guar seeds (Dhugga et al. 2004) and later studies could demonstrate in vitro activity (Liepman et al. 2005; Suzuki et al. 2006; Liepman et al. 2007). Glucomannan in the cell wall of the model species Arabidopsis thaliana is synthesized by members of the cellulose synthase-like family A (CSLA) as indicated by a reverse genetics approach (Goubet et al. 2009). The polymer is synthesized from the activated nucleotide sugars GDP-d-mannose and GDP-d-glucose (Liepman et al. 2005). While the nucleotide sugar interconversion enzymes involved in the pathway from sucrose, a product of the carbon fixation, to GDP-d-mannose have been identified, the necessary enzymes for the generation of GDP-d-glucose have not (for review see Reiter 2008). Additionally, how plants regulate mannan biosynthesis and the bottlenecks in its synthesis are not known.
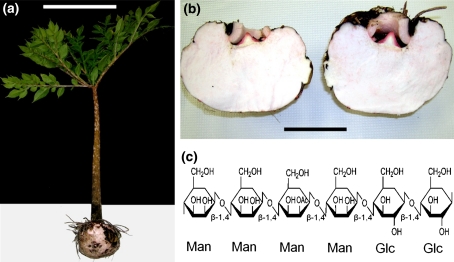
One approach to fill these gaps in our knowledge is to study a plant system that produces massive amounts of mannan (Dhugga et al. 2004; Pauly and Keegstra 2010). One example of such a plant system is the corm of Amorphophallus konjac, a specialized tissue which deposits large amounts glucomannan as a storage polymer (Fig. 1). Plants of the genus Amorphophallus (order of Alismatales, family of Araceae) are perennial plants with an underground stem in the form of a corm that have a highly dissected umbrella-shaped leaf blade (Chua et al. 2010). The known 170 species are mainly distributed in the tropics between West Africa to the east into Polynesia (Hetterscheid and Ittenbach 1996). Especially in the tropical and subtropical regions of Asia the diverse Amorphophallus species have been historically used as a food source as well as in traditional medicine. The most widely utilized species is Amorphophallus konjac K. Koch (Fig. 1a) that has been used in China for thousands of years (Chua et al. 2010). More recent interest in A. konjac—also known as voodoo lily or devil’s tongue—was spurred due its potential use as a dietary fiber in human nutrition with potential in the treatment of obesity (Kraemer et al. 2007). The corm of A. konjac (Fig. 1b) consists of 49–60% (w/w) water soluble glucomannan—referred to as konjac glucomannan (KGM)—as the main component and storage carbohydrate, 10–30% (w/w) starch, 2.6–7% (w/w) inorganic elements, 5–14% (w/w) protein, 3–5% (w/w) soluble sugars and small amounts of alkaloids (trigonelline) (Li et al. 2005). The accumulated glucomannan is stored in egg-shaped idioblasts that can reach up to ~650 μm in diameter within the parenchyma of the corm (Takigami et al. 1997). The content of KGM in the corm changes throughout development and is at its highest just before the leaf dies off, prior to the development of the flower or dormancy (Brown 2000). Konjac glucomannan consists of β-1,4-linked d-mannose and d-glucose residues in a molar ratio of about 1.6:1 (Cescutti et al. 2002; Fig. 1c). Additionally, the KGM chain can be O-acetylated, a substitution that is proposed to contribute to the solubility properties of the glucomannan polymer (Nishinari et al. 1992).
Fig. 1.
Seventeen-week-old Amorphophallus konjac plant with developing corm grown on soil and molecular structure of glucomannan. a 17-week-old A. konjac plant, bar 30 cm. b Morphology of the 17-week-old dissected corm, bar 10 cm (c). Structure of A. konjac glucomannan; Man mannose, Glc glucose, Ac acetate
Hence, one can expect that all the necessary components for glucomannan biosynthesis and metabolism can be found in the developing voodoo lily corm. Here, we present data from 454® cDNA deep sequencing and subsequent EST analysis from the tissue of the developing A. konjac corm to extend our knowledge concerning glucomannan biosynthesis.
Materials and methods
Plant material
Corms of Amorphophallus konjac were acquired from the online auction website http://ebay.com. The corms were planted in Baccto soil (Michigan Peat Company, Houston, TX, USA) and grown at 25–35°C under light conditions natural to central Michigan in late summer and fall of 2009. The plants were watered three times per week.
Corm growth was verified in regular time intervals to pinpoint the stage at which the corm was rapidly expanding. After 17 weeks the whole plant (Fig. 1a) including its corm was harvested. The corm material was immediately sliced (Fig. 1b), frozen in liquid nitrogen and stored at −80°C until further analyses.
Glucomannan extraction and compositional analysis
Frozen corm samples were ground to a fine powder using a ball mill. The powder was washed for 3 h in 85% aqueous ethanol at room temperature (RT) and subsequently filtered using filter papers (Whatman, UK). The flow through was discarded and the filtered alcohol insoluble residue (AIR) was dried. Glucomannan was extracted from the AIR by incubation in double distilled water at 37°C for 16 h and vertical agitation at 200 rpm. The solution was centrifuged for 20 min at 2,000 rpm and the glucomannan containing supernatant was separated from the pellet and freeze-dried.
The extracted glucomannan (KGM) was analyzed for its monosaccharide composition by hydrolyzing 1 mg of KGM extract with 2 M trifluoroacetic acid (TFA) followed by derivatization to alditol acetates and GC–MS analysis as described (York et al. 1985).
Determination of starch content
The total starch content of freeze-dried corm material and the warm-water-extracted polymer was determined using the Megazyme “Total Starch” kit (catalog #K-TSTA, Megazyme, Wicklow, Ireland) according to the manufacturer’s recommendations.
Determination of the glucomannan-bound acetate content
The acetate content of KGM was determined using the Megazyme “Acetic Acid Kit” (catalog #K-ACET, Megazyme, Wicklow, Ireland). The assay was downscaled and adapted to a 96-well format. A total amount of 300 μg of extracted KGM was solubilized in 100 μl water. The polymer-bound acetate was released by adding 100 μl NaOH (1 M) and incubating for 1 h at RT with shaking at 500 rpm. The samples were neutralized with 1 M HCl, centrifuged for 10 min at 14,000 rpm, 10 μl of the supernatant containing released acetate was transferred to a UV capable 96-well flat bottom assay plate and diluted with 94 μl water. The kit content was used as follows. Solution 1 and Solution 2 were mixed in a ratio of 2.5:1 (30 μl + 12 μl per sample) and 42 μl of the mixture were added to each sample, mixed and incubated at RT for 3 min. The absorption was read at 340 nm (A0). Solution 3 was diluted 1:10 in water; 12 μl was added to each sample, mixed and incubated at RT for 4 min. The absorption was read at 340 nm (A1). Solution 4 was diluted 1:10 in water, 12 μl was added each sample, mixed and incubated for 12 min at RT. The absorption was read at 340 nm (A2). The amount of acetate in the samples was calculated based on an acetic acid standard curve and according to the manufacturer’s recommendations.
1H-NMR analysis of glucomannan
The KGM extract was saturated with deuterium oxide (D2O) for 24 h and then freeze-dried. Approximately 5 mg of KGM was suspended in 0.7 ml D2O (D, 99.96%) with 0.01 mg/ml 3-(trimethylsilyl)-1-propanesulfonic acid sodium salt (DSS) as an internal standard. The suspension was centrifuged to remove the non-soluble polymer (starch). The remaining KGM solution was transferred in a 5 mm NMR tube. The 1H NMR spectrum was recorded in an 800 MHz Bruker DRX-800 NMR spectrometer at 338 K.
Total RNA extraction, construction of cDNA library and 454 sequencing
Frozen corm material was ground to a fine powder using a mortar and pestle. Total RNA was extracted from 4.18 g of ground tissue using the pine tree method published by Chang et al. (1993). DNA was digested and removed using Qiagen RNase free DNase I (catalog #29254, Qiagen, Hilden, Germany) according to the manufacturer’s recommendations. The RNA quality was checked and assured using Agilent Bioanalyzer RNA pico 6000 chip (Agilent, Santa Clara, USA).
First strand cDNA was synthesized using the ClonTech SMART cDNA library construction kit (catalog #63490, ClonTech, Mountain View, CA, USA) according to the manufactures instructions. A modified version of the CDS III/3′ primer was used (5′-TAG AGG CCG AGG CGG CCG ACA TGT TTT GTT TTT TTT TCT TTT TTT TTT VN-3′, Integrated DNA Technologies, Coralville, IA, USA). Reverse transcription was conducted using Invitrogen SuperScript II Reverse Transcriptase (catalog #18064-022, Invitrogen, Carlsbad, CA, USA).
The cDNA was amplified by ligation-dependent PCR (LD-PCR) using the ClonTech Advantage 2 Polymerase mix (catalog #639201, ClonTech) according to the manufacturer’s recommendations. The amplified cDNA was digested with SfiI, size fractionated and precipitated using ClonTech SMART cDNA library construction kit (catalog #63490, ClonTech) according to the manufacturer’s instructions.
A total of 10 μg cDNA was submitted for Roche® 454 GSFLX Sequencing. The 454 sequencing was carried out by the Michigan State University Research Technology Support Facility Genomics Core (Michigan State University, East Lansing, MI, USA).
EST processing, assembly and gene annotation
The obtained 454 reads were assembled using CLC Genomics Workbench version v3.7 (CLC bio, Cambridge, MA, USA). The assembled contig sequences were aligned to the A. thaliana genome using BLASTN 2.2.23+ (Altschul et al. 1997) on the nucleotide level. The results were exported to a text searchable html-based database. The blast results were searched for known genes encoding proteins involved the nucleotide sugar conversion pathway as well as in the biosynthesis of glucomannan in Arabidopsis. The blast hits and annotations of the encoded A. thaliana proteins were used to assign annotations to the corresponding A. konjac contigs. In the case of multiple contigs being assigned to a single Arabidopsis gene the EST counts were combined.
Production of recombinant CSLA proteins in Pichia pastoris
For production of NH3-terminal epitope-tagged AkCSLA3 protein, the coding sequence of the AkCSLA3 gene (Genbank accession number HQ833588) was fused in-frame and immediately downstream of the sequence encoding the T7 epitope tag. The sequence was synthesized and inserted into the pDONR vector by DNA2.0 (Menlo Park, CA, USA). This sequence was recombined from the pDONR vector into a Gateway-compatible pPICZ vector using LR clonase (Invitrogen) by standard procedures (Davis et al. 2010) to produce the pPICZ-AkCSLA3 vector. After linearization of the pPICZ-AkCSLA3 vector using PmeI, electroporation with a Gene Pulser (BioRad) was used for transformation of X-33 wild-type P. pastoris cells. Selection of transformed cells was achieved by culturing cells from transformation reactions for 2–3 days at 30°C on yeast extract peptone dextrose (YPD) plates supplemented with 1 M sorbitol and 100 μg/ml zeocin. PCR with the AkCSLA680F primer (5′-CTACCACTTCAAGGTGGAGCAG-3′) and AkCSLA1288R primer (5′-CTCCCGATCTCCAACAAACC-3′) was used to verify the presence of the AkCSLA3 transgene in Pichia cells by using the method of Davis et al. (2010). The pPICZ-OsCSLA1 vector and Pichia cells containing the OsCSLA1 transgene were produced and validated using similar procedures.
To induce protein expression, PCR-validated transgenic Pichia cultures (and X-33 control cells) were patched on YPD plates supplemented with ampicillin (50 μg/ml) to suppress bacterial contamination. After growth for 1 day at 30°C, patches were expanded by using a sterile toothpick to cover the entire plate surface; plates were incubated for 2 additional days at 30°C to obtain a lawn. Using a spatula, cells were collected and transferred directly into 250 ml baffled flasks containing 33 ml buffered methanol complex medium (BMMY) supplemented with ampicillin (50 μg/ml) as described in Davis et al. (2010). Cultures were grown at 28°C with orbital shaking at 250 rpm for 16–20 h, supplemented with 1% methanol, and cultured for 2–4 additional hours before collection of cells.
Preparation of microsomal membrane fractions from Pichia pastoris cells
Pichia cells were harvested from cultures and washed by using the procedure described by Davis et al. (2010). Washed cell pellets either were stored frozen at −80°C or used immediately for preparation of microsomal membranes as described by Cocuron et al. (2007).
Glucomannan synthase activity assays
In vitro glucomannan synthase activity assays were conducted as previously described (Liepman et al. 2005), except that 5 mM MnCl2 was present in the assay buffer. Two different types of glucomannan synthase assays, differing in the [14C]-labeled nucleotide sugar donor, were conducted: Glc*ManS (GDP-[14C]-glucose), and GlcMan*S (GDP-[14C]-mannose). GDP-glucose and GDP-mannose were each present at a total concentration of 20 μM in both types of assays. For Glc*ManS assays 3 μM GDP-[14C]-glucose was included, and for GlcMan*S assays 4 µM GDP-[14C]-mannose was included.
Results
Phenotypic identification of Amorphophallus konjac
An acquired dormant A. konjac corm was grown for 17 weeks in the greenhouse. The plant developed a solitary leaf about 60 cm in length with a smooth light pink petiole that was marbled with dark-green to brown spots (Fig. 1a). The leaf blades carried green, pointed, elliptical leaflets about 5–10 cm long. These observations are in agreement with the botanical description of A. konjac K. Koch (Brown 2000).
Carbohydrate composition of the developing corm
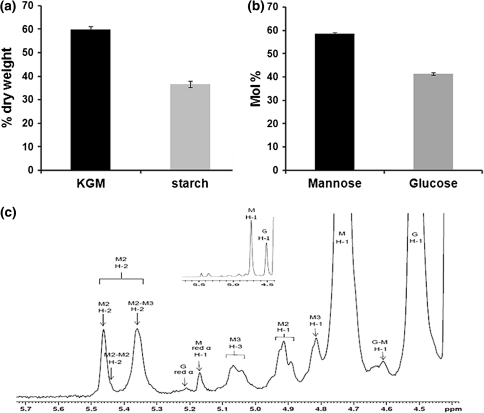
The corm of a 17-week-old plant was harvested and analyzed for its carbohydrate composition. The sliced corm was uniformly filled with a light pink, hard and moist tissue (Fig. 1b). Alcohol insoluble residue (AIR) was prepared from the corm tissue and its composition was determined. The extracted AIR consisted mainly of warm-water-extractable polymers (59.9% of the dry weight) and starch (36.7% of the dry weight) (Fig. 2a). The warm-water-extracted polymers were assayed for starch as well as monosaccharide composition, revealing a residual starch content of 8.1% of the dry weight of this fraction and a molar ratio of 1.4:1 mannose:glucose (Fig. 2b). Factoring for the starch content, the main polymer of the extract, glucomannan, had a mannose:glucose ratio of 1.8:1, which closely matches previous analysis of this of this polymer in A. konjac (Cescutti et al. 2002). The alkali-released acetate content of the warm-water extract amounted to 15.7 μg acetic acid per mg extract (±0.5 μg/mg). Taking the starch content of the warm water-extracted polymer into account, this indicates a 1.7% O-acetyl substitution of the extracted corm glucomannan.
Fig. 2.
Compositional analyses of the developing A. konjac corm. a Composition of a 17-week-old A. konjac corm. Amount of warm-water-extractable konjac glucomannan (KGM) and starch. The error bars represent the standard deviation, n = 3. b Monosaccharide composition of the KGM extract. The error bars represent the standard deviation, n = 6. c 1H-NMR spectrum of the KGM extract. See footnote in Table 1 for designation of signals
The extract also was characterized by 1H-NMR (Fig. 2c). Chemical shifts (Table 1) could be assigned based on published data derived from spruce, aspen and birch glucomannan (Lundqvist et al. 2002; Teleman et al. 2003). Integration of the anomeric signals (H1—M, M2, M3, M red: H1—G) revealed a mannose to glucose ratio of 1.78:1 in good agreement with the analysis presented above. Based on the observed reducing ends (H1—M, M2, M3, G/H1—M red) an average degree of polymerization of the konjac glucomannan of 53 could be estimated in the extract. Signals also were observed consistent with the O-acetylation of the polymer, particular on the O-2 and O-3 position of the mannosyl residues with a ratio of 1:1 (H1—M2: H-1—M3). No O-acetyl substituents were observed on the glucosyl residues. The degree of mannose O-acetylation of the extract was determined to be 11% (H1—M2, M3/H-1M), which based on the mass percent of the total polymer would represent an acetate content of 1.7% of the whole glucomannan polymer, which is in agreement with the data obtained above on the content of alkali released acetic acid.
Table 1.
Assignment of 1H NMR chemical shifts of the KGM extract (see also Fig. 2c)
| Residuea | 1H chemical shifts in ppmb | ||||
|---|---|---|---|---|---|
| H-1 | H-2 | H-3 | H-4 | H-5, H-6, H-6′ | |
| M red α | 5.17 | 3.99 | 3.90 | n.d. | n.d. |
| M red β | 4.89 | 4.01 | n.d. | n.d. | n.d. |
| M | 4.74 | 4.10 | n.d. | n.d. | n.d. |
| M2 | 4.92 | 5.46 | 3.99 | n.d. | n.d. |
| M2 (M2–M3) | 4.88 | 5.36 | 3.99 | n.d. | n.d. |
| M3 | 4.81 | 4.17 | 5.06 | n.d. | n.d. |
| G red α | 5.21 | n.d. | n.d. | n.d. | n.d. |
| G | 4.51 | 3.35 | 3.64 | 3.54 | n.d. |
| G (G-M) | 4.61 | 3.41 | 3.65 | n.d. | n.d. |
aThe following abbreviations are used (Teleman et al. 2003): M red non-acetylated Man reducing end, M non-acetylated Man, M2 2-O-acetylated Man, M3 3-O-acetylated Man, G non-acetylated Glc, M2 (M2–M3) M2 within the sequences→4)[2-O-Ac]-β-d-Manp-(1→4)[2-O-Ac]-β-d-Manp-(1→, G (G–M) Glc within the sequences→4)-β-d-Manp-(1→4)-β-d-Glcp-(1→
bRelative to internal DSS standard; n.d. not determined
In summary, consistent with data found in the literature (Li et al. 2005; Brown 2000) these results indicate that the major carbohydrate components in the developing A. konjac corm at this growth stage are acetylated glucomannan and starch.
cDNA deep sequencing and EST contig generation
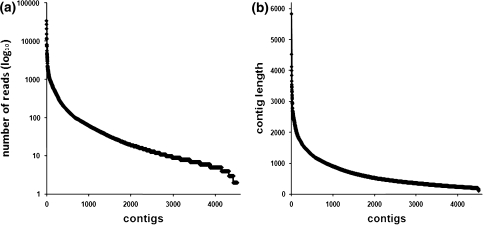
The material of the 17-week-old corm was used to create a cDNA library for Roche 454® deep sequencing. The deep sequencing run yielded 1,735,537 reads with an average read length of 327 bp. Of these 754,784 reads were of high quality, yielding a total of 246,640,187 high-quality base pairs from this experiment. The sequencing data has been uploaded to the NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/sra) under SRS150964. The high-quality ESTs were assembled into 4,513 contigs with an average length of 658 bp and an average coverage of 125 reads (Fig. 3). The contigs were annotated by aligning them to the A. thaliana nucleotide collection using BLASTN (Altschul et al. 1997). The contigs with ≥1,100 EST reads are shown in Supplemental Table 1. Contigs ranged from representation by 34,001 ESTs (putative ion transmembrane transporter) to single EST representation (191,027 singletons, 25.3% of total ESTs).
Fig. 3.
Number of reads and length of the assembled contig sequences. a The 454® deep sequencing reads were assembled into 4,513 contig sequences with an average coverage of 124.86 reads per contig and b an average length of 658 bp
Identification of genes involved in glucomannan and starch biosynthesis
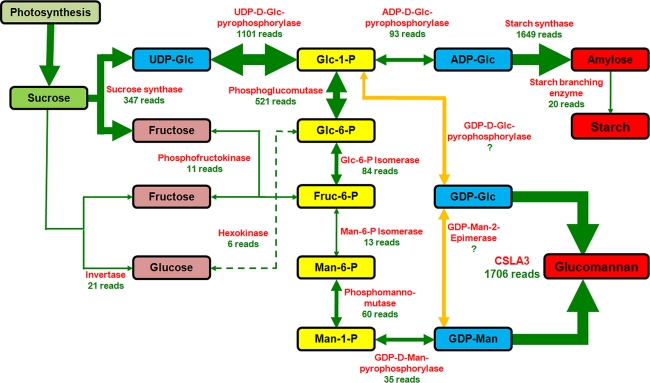
Our focus was to identify genes involved in glucomannan biosynthesis. Hence, the annotated contigs were searched for genes necessary for the de novo biosynthesis of glucomannan as well as starch, the other major polymer in the corm. Both polymers are presumably generated from sucrose originating from carbon assimilation in the leaf tissue. Based on contig orthologs from A. thaliana (Table 2) transcripts of all enzymes known to play a role in the glucomannan biosynthesis pathway were found in the EST database (Fig. 4). The assembled contig sequences of the genes encoding these proteins are presented in Supplemental Table 2. However, they were represented by different EST levels. For the glucomannan and starch biosynthesis pathways the highest number of ESTs (1,706 reads) was found for a transcript most similar (E value 2e−140, Table 2) to a glucomannan synthase AtCSLA3; this sequence was among the 40 most abundant contigs found in the database (Supplemental Table 1). Hence, it can be assumed that the harvested corm tissue was in a developmental stage during which it actively synthesized glucomannan.
Table 2.
A. thaliana orthologs based on the A. konjac ESTs potentially involved in the biosynthesis of glucomannan and starch (Fig. 4)
| Description | A. thaliana genetic locus | 454 contig length | EST reads | E value | Annotation in pathway (Fig. 4) |
|---|---|---|---|---|---|
| AkCSLA3 (cellulose synthase-like A3) | At1g23480 | 2,014 | 1,706 | 2e−140 | CSLA3 |
| Ak starch synthase | At1g32900 | 2,186 | 1,649 | 7e−171 | Starch synthase |
| Ak UDP-d-glucose pyrophosphorylase (UGP2) | At5g17310 | 2,606 | 1,101 | 0.0 | UDP-d-Glc-pyrophosphorylase |
| Ak phosphoglucomutase | At1g23190 | 2,702 | 521 | 0.0 | Phosphoglucomutase |
| Ak SUS2 (sucrose synthase 2) | At5g49190 | 2,943 | 347 | 0.0 | Sucrose synthase |
| Ak ADP-glucose pyrophosphorylase | At5g19220 | 2,360 | 93 | 0.0 | ADP-d-Glc-pyrophosphorylase |
| Ak PGIC (glucose-6-phosphate isomerase) | At5g42740 | 1,477 | 84 | 0.0 | Glc-6-P isomerase |
| Ak PMM (phosphomannomutase) | At2g45790 | 1,062 | 60 | 1e−129 | Phosphomannomutase |
| Ak CYT1 (GDP-d-mannose pyrophosphorylase) | At2g39770 | 801 | 35 | 5e−50 | GDP-d-Man-pyrophosphorylase |
| Ak CINV1 (cytosolic invertase 1) | At1g35580 | 782 | 21 | 4e−10 | Invertase |
| Ak SBE2.2 (starch branching enzyme 2.2) | At5g03650 | 642 | 20 | 1e−10 | Starch branching enzyme |
| Ak DIN9 (mannose-6-phosphate isomerase) | At1g67070 | 392 | 13 | 8.5 | Man-6-P isomerase |
| Ak phosphofructokinase | At1g12000 | 469 | 11 | 1e−97 | Phosphofructokinase |
| Ak HXK1 (hexokinase 1) | At4g29130 | 253 | 6 | 3e−08 | Hexokinase |
The assembled contig sequences of the A. konjac genes; see Supplemental Table 2
Fig. 4.
Proposed glucomannan and starch biosynthesis pathway in the A. konjac corm based on the established EST database. Presented is the nucleotide sugar interconversion pathway from sucrose to GDP-mannose and GDP-glucose, the two building blocks of glucomannan as well as the starch biosynthesis pathway. The necessary converting enzymes found in the EST data are given in red with the number of obtained reads in green. The thickness of the arrows is representative for the number of reads. The highest number of ESTs for this pathway was found to be from CSLA3—a putative glucomannan synthase. UDP-Glc UDP-d-glucose, Glc-1-P glucose-1-phosphate, Glc-6-P glucose-6-phosphate, Fruc-6-P fructose-6-phosphate, Man-6-P mannose-6-phosphate, Man-1-P mannose-1-phosphate, GDP-Man GDP-d-mannose, GDP-Glc GDP-d-glucose, ADP-Glc ADP-d-glucose, CSLA3 cellulose synthase-like family A member 3
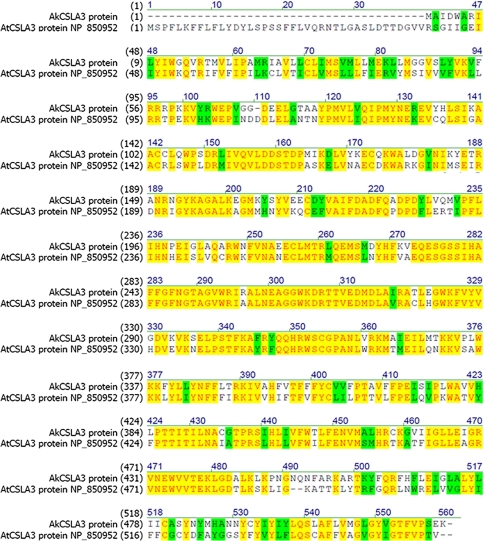
The full coding sequence of the presumptive AtCSLA3 ortholog in A. konjac—AkCSLA3—was assembled from the ESTs and deposited in NCBI under HQ833588. The coding sequence was translated into a protein sequence and aligned to the AtCSLA3 protein sequence (accession number NP_850952) (Fig. 5). The two proteins shared 59% identity and 71% similarity. Analysis with TargetP v1.1 (Emanuelsson et al. 2007) predicts that AkCSLA3 is most likely to be targeted to the secretory pathway. Hence, the protein was analyzed for putative transmembrane domains using TMHMM server 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/, Krogh et al. 2001). This analysis predicted the presence of five putative transmembrane domains, similar to the predicted topology of AtCSLA3 (Supplemental Fig. 1).
Fig. 5.
Protein alignment of the A. konjac glucomannan synthase AkCSLA3 and the A. thaliana mannan synthase AtCSLA3
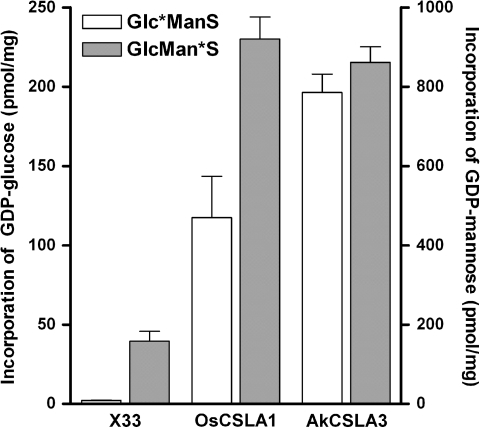
To determine whether the AkCSLA3 protein catalyzes glucomannan biosynthesis, the activity of recombinant AkCSLA3 protein and the previously described OsCSLA1 protein (Liepman et al. 2007) as a positive control, produced in the yeast P. pastoris was assayed. Microsomal membrane samples prepared from Pichia cells expressing the AkCSLA3 protein incorporated significantly more GDP-glucose and GDP-mannose into 70% ethanol-insoluble products than wild-type X-33 samples (Fig. 6). Relative to the positive control OsCSLA1, the AkCSLA3 protein incorporated glucose from the donor substrate GDP-glucose into polymeric products more readily, suggesting that it may have a higher capacity to incorporate glucose into glucomannans than the OsCSLA1 protein.
Fig. 6.
Glucomannan synthase activity assays of recombinant AkCSLA3 produced in Pichia pastoris. Microsomal membranes from untransformed Pichia cells (X33) or transgenic lines producing either AkCSLA3 or OsCSLA1 (positive control), were assayed for glucomannan synthase activity in vitro. The incorporation of glucose or mannose from the substrates GDP-glucose or GDP-mannose, respectively, into 70% ethanol-insoluble products is shown. For Glc*Man synthase assays, GDP-[14C]-glucose was used in the presence of non-radioactive GDP-man. For GlcMan* synthase assays, GDP-[14C]-mannose was used in the presence of non-radioactive GDP-glucose. Error bars represent standard deviation, n = 3
Another of the most abundant transcripts was a putative starch synthase (1,649 EST reads) (Table 2), consistent with the large abundance of starch in the analyzed developing corm tissue. The lowest abundance in terms of EST reads for genes involved in the biosynthesis of glucomannan and starch was found for a putative hexokinase (6 EST reads), which supports that this enzyme may not play a major role in mannan and/or starch biosynthesis.
Discussion
A 17-week-old corm of A. konjac actively produces glucomannan
Carbohydrate analysis of a developing 17-week-old corm of A. konjac revealed that the main polymeric carbohydrates in the corm are starch and a glucomannan with a mannose to glucose ratio of 1.4–1.8:1, whereby 11% of the mannosyl residues are O-acetylated equally at position O-2 and O-3 (Fig. 2c). The observed KGM mannose to glucose ratio is in agreement of 1.6:1 for purified KGM (Nishinari et al. 1992). Detailed analysis of the fine structure of the konjac glucomannan revealed that the substitutions of the glucomannan backbone are the same as those found in various wood species, underscoring the usefulness of the voodoo lily corm system as a relevant model system for glucomannan biosynthesis.
Together with the corm compositional data, the finding of a high abundance of ESTs (1,706 reads) aligning to AtCSLA3 (a glucomannan synthase) and an almost equally high abundance of 1,649 reads aligned to a gene encoding an A. thaliana starch synthase indicates highly active KGM and starch biosynthetic processes in the corm at this stage.
Identification of a mannan synthase in A.konjac
From the ESTs 1,706 reads aligned to the gene encoding for the A. thaliana cellulose synthase-like A3 (AtCSLA3) protein—this number is among the top 40 most abundant ESTs in the database. Proteins of the family CSLA belong to the glycosyltransferase family 2 (GT2) (http://www.cazy.org/, Cantarel et al. 2009). The identified protein from the developing corm shares a 71% similarity with AtCSLA3 (Fig. 5) and hence was designated AkCSLA3 on the basis of amino acid sequence similarity. Goubet et al. (2009) demonstrated that AtCSLA3 is involved in glucomannan biosynthesis in secondary cell walls of Arabidopsis. Based on the high similarity to AtCSLA3, the high abundance of the AkCSLA3 transcript in the developing corm, and the ability of recombinant AkCSLA3 to catalyze the synthesis of glucomannans in vitro, we provided conclusive evidence that AkCSLA3 is the glucomannan synthase responsible for the synthesis of storage glucomannan in corms of A. konjac using the substrates GDP-d-mannose and GDP-d-glucose. Both proteins—AkCSLA3 and AtCSLA3—contain five predicted transmembrane domains and also are thought to share a very similar topology (Supplemental Fig. 1). Recently, the topology of AtCSLA9—another Arabidopsis glucomannan synthase with the same topology as AtCSLA3—was demonstrated (Davis et al. 2010). This study provided compelling evidence that AtCSLA9 is localized to the Golgi apparatus and that the active site of this enzyme faces the Golgi lumen. Due to the very similar topology this also can be assumed for AtCSLA3. Interestingly, when the identified A. konjac glucomannan synthase, which is somewhat truncated on the N-terminus compared to AtCSLA03, is analyzed with a transmembrane topology prediction algorithm (http://www.cbs.dtu.dk/services/TMHMM-2.0/, Krogh et al. 2001), the active site is predicted to face the cytosol, in direct contrast to the Arabidopsis ortholog AtCSLA3 (Supplemental Fig. 1). One reason for this difference in the predicted orientation may be the limits of the prediction algorithm. On the other hand, the glucomannan produced in the voodoo lily corm is deposited in idioblasts in the parenchyma of the corm (Takigami et al. 1997) and not in the apoplast as in Arabidopsis. This difference in the deposition of the product might prompt such an orientation of the active site in AkCSLA3. However, without experimental evidence of the topology of AkCSLA3 these points are highly speculative.
Identification of a comprehensive pathway for the production of glucomannan in A. konjac
Based on annotation of the contigs to Arabidopsis genes the metabolic pathway from sucrose to glucomannan and starch could be re-constructed (Fig. 4) based on our current knowledge of the relevant portions of this pathway. On a transcriptional level all necessary enzymes are transcribed in the corm at the selected developmental stage (Table 2; Fig. 4).
Assuming that photo-assimilates originating from the A. konjac leaf are transported to the corm in the form of sucrose, multiple pathways are possible to convert this disaccharide into the hexose phosphate pool. Sucrose can be converted either by an invertase, a sucrose synthase or via phosphorylation followed by the action of a sucrose-phosphate synthase (for review see Winter and Huber 2000). Based on the observed transcript abundance of sucrose synthase (347 EST reads), converting sucrose to fructose and UDP-d-glucose seems to be the dominant pathway (Fig. 4). Sucrose hydrolysis to glucose and fructose followed by phosphorylation of the monosaccharides by a hexokinase seems most likely to play a minor, insignificant role (21 and 6 EST reads, respectively). The identified putative mannose-6-phosphate isomerase exhibits only an insignificant E value of 8.5 (Table 2). One reason might be that the retrieved EST contig assigned to this gene is quite short (320 bp) and in low abundance as indicated by the EST read number.
It is of interest to note that only a low abundance (11 EST reads) of phosphofructokinase transcripts was found; this enzyme is responsible for feeding fructose into the phosphate sugar pool for further conversion into glucose-6-phosphate and/or mannose-6-phosphate (Fig. 4). These data indicate that fructose may only play a minor role in this pathway. The fate of the fructose derived from the sucrose hydrolysis in the corm is unknown at this point. In contrast, there is a high abundance of the UDP-d-glucose pyrophosphorylase transcript (1,101 EST reads) feeding glucose into the hexose phosphate pool. Glucose-1-phosphate is further converted into ADP-d-glucose, which is utilized by the starch synthetic enzymes to produce starch. The transcripts of all these enzymes were observed with the highest transcript level in the last steps of starch synthesis.
The transcript of phosphoglucomutase, an equilibrium enzyme, which is necessary to ensure diversion of glucose-1-phosphate from the starch biosynthetic pathway to the synthesis of GDP-d-mannose was also shown to be abundant in a relative high abundance (521 EST reads). The transcripts of the enzymes for the conversion of glucose-6-phosphate via several intermediates to GDP-d-mannose are present, albeit in relatively low abundance (Fig. 4).
Hypothesis for the synthesis of GDP-d-glucose
It has been shown that one precursor for glucomannan biosynthesis is GDP-d-glucose, which is accepted by CSLA mannan synthases in addition to GDP-d-mannose (Liepman et al. 2005). This dual donor substrate specificity of CSLAs was confirmed here with the AkCSLA3. However, how GDP-d-glucose is synthesized from the hexose phosphate pool has not been described in plants and the corresponding genes have not been cloned (Reiter 2008).
In theory, one could envision two pathways (Fig. 4). The first pathway would entail the conversion of glucose-1-phosphate to GDP-d-glucose via a GDP-d-glucose pyrophosphorylase (Reiter 2008). Such an enzyme has been found in E. coli where a GDP-mannose pyrophosphorylase apparently has a dual donor substrate specificity and also was able to synthesize GDP-d-glucose from glucose-1-phosphate and GTP (Yang et al. 2005). Currently, there are three genetic loci in Arabidopsis annotated as GDP-d-mannose pyrophosphorylase (At2g39770, At3g55590 and At4g30570) of which only for one (At2g39770) activity has been demonstrated (Conklin et al. 1997). Hence, there is the possibility that this enzyme also has dual donor substrate specificity and is able to generate GDP-d-glucose in addition to GDP-d-mannose or that one of the other two genes encodes an enzyme with GDP-d-glucose pyrophosphorylase activity. However, in the database obtained from the developing corm described, we were unable to identify orthologs to the two Arabidopsis genes currently annotated as GDP-d-mannose pyrophosphorylase At3g55590 and At4g30570—only a putative ortholog to At2g39770 (CYT1) (Table 2) was identified. This supports the theory that an enzyme with dual donor substrate specificity acts within this pathway.
The second pathway would entail a GDP-d-mannose-2-epimerase converting GDP-d-mannose into GDP-d-glucose (Reiter 2008). Indeed, the EST database does contain two contigs aligning to Arabidopsis genetic loci with putative aldose epimerase activities (At5g66530, E value 1e−06, 58 EST reads and At3g17940, E value 1e−03, 20 EST reads). No activity for either of these proteins has been demonstrated; hence, their involvement in the conversion of GDP-d-mannose to GDP-d-glucose is speculative.
Transcriptional bottlenecks in the synthesis of glucomannan in A. konjac
The developing corm of A. konjac is a tissue that actively deposits glucomannan and starch. The biological function of this tissue is to convert as much assimilates as possible into these storage polymers. If one assumes that this process is partly under transcriptional control the established expression level of genes encoding the various enzymes necessary for the synthesis of these polymers can give hints about rate limiting steps. Based on high EST abundance, these steps include the very last stage of polymer synthesis (mannan synthase and starch synthase), as well as the very early stages of the pathway (sucrose synthase, UDP-glucose pyrophosphorylase and to some extent phosphoglucomutase). Ectopic expression of one or multiple of these enzymes in transgenic plants is predicted to increase the kinetics of the processes and thus might increase the abundance of glucomannan and/or starch in plants. Indeed, overexpression of sucrose synthase and UDP-glucose pyrophosphorylase led to an increase in cell wall material in poplar (Coleman et al. 2009). In addition, overexpression of various Arabidopsis CSLAs led to an accumulation of mannans in stem tissue (Goubet et al. 2009) confirming that transcript levels of some of these genes represent transcriptional bottlenecks.
However, it is expected that the synthesis of these polymers is also regulated by posttranslational modifications and thus a proper flux analysis of the intermediates is needed to identify rate limiting steps. Moreover, additional cellular components not investigated in this study might be necessary for the proper synthesis and deposition of glucomannan. These might include sugar phosphate and nucleotide sugar transporters or the vesicular machinery necessary to deposit glucomannan.
In conclusion, we created an EST collection from a cDNA library prepared from a developing A. konjac tuber. The results obtained demonstrate the usefulness of this system to identify metabolic pathways related to polysaccharide biosynthesis and underscore the opportunity to develop novel hypotheses concerning their biosynthesis. Using some of the highly expressed genes as targets in transgenic or marker-selected breeding approaches should result in an enrichment of glucomannan in plant feedstocks allowing for a more economical and sustainable production of biofuels or other chemicals.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
Funding for this study was provided by the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, US Department of Energy (award no. DE-FG02-91ER20021) and the Energy Biosciences Institute (grant # OO0G01). A.H.L. acknowledges Ms. Adewunmi Adetayo for assistance with manipulations of the AkCSLA3 gene, and the EMU Provost’s office for financial support (Faculty Research Fellowship). A.H.L. also acknowledges Dr. Kenneth Keegstra (Michigan State University, Plant Research Lab) for generous provision of reagents for glucomannan synthase assays, and Dr. Yan Wang for assistance with assays.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- AIR
Alcohol insoluble residue
- AtCSLA3
Arabidopsis thaliana cellulose synthase-like family A member 3
- AkCSLA3
Amorphophallus konjac cellulose synthase-like family A member 3
- cDNA
Complementary DNA
- CSLA
Cellulose synthase-like family A
- D2O
Deuterium oxide
- EST
Expressed sequence tag
- GC
Gas chromatography
- GDP
Guanosine diphosphate
- GT
Glycosyltransferase
- UDP
Uridine diphosphate
- KGM
Konjac glucomannan
- MS
Mass spectrometry
- NMR
Nuclear magnetic resonance
- RT
Room temperature
- TFA
Trifluoroacetic acid
- UV
Ultraviolet
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3398–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. Aroids. Plants of the Arum Family. Portland, Oregon: Timber Press; 2000. [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:233–238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cescutti P, Campa C, Delben F, Rizzo R. Structure of the oligomers obtained by enzymatic hydrolysis of the glucomannan produced by the plant Amorphophallus konjac. Carbohydr Res. 2002;337:2505–2511. doi: 10.1016/S0008-6215(02)00332-4. [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Bio Rep. 1993;11:113–116. doi: 10.1007/BF02670468. [DOI] [Google Scholar]
- Chua M, Baldwin TC, Hocking TJ, Chan K. Traditional uses and potential health benefits of Amorphophallus konjac K. Koch ex NEBr. J Ethnopharmacol. 2010;128:268–278. doi: 10.1016/j.jep.2010.01.021. [DOI] [PubMed] [Google Scholar]
- Cocuron JC, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG. A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proc Natl Acad Sci USA. 2007;104:8550–8555. doi: 10.1073/pnas.0703133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman H, Yan J, Mansfield S. Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proc Natl Acad Sci USA. 2009;106:13118–13123. doi: 10.1073/pnas.0900188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PL, Pallanca JE, Last RL, Smirnoff N. l-Ascorbic acid metabolism in the ascorbate-deficient Arabidopsis mutant vtc1. Plant Physiol. 1997;115:1277–1285. doi: 10.1104/pp.115.3.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J, Brandizzi F, Liepman AH, Keegstra K. Arabidopsis mannan synthase CSLA9 and glucan synthase CSLC4 have opposite orientations in the Golgi membrane. Plant J. 2010;64:1028–1037. doi: 10.1111/j.1365-313X.2010.04392.x. [DOI] [PubMed] [Google Scholar]
- Dhugga KS, Barreiro R, Whitten B, Stecca K, Hazebroek J, Randhawa GS, Dolan M, Kinney AJ, Tomes D, Nichols S, Anderson P. Guar seed beta-mannan synthase is a member of the cellulose synthase super gene family. Science. 2004;303:363–366. doi: 10.1126/science.1090908. [DOI] [PubMed] [Google Scholar]
- Edwards ME, Dickson CA, Chengappa S, Sidebottom C, Gidley MJ, Reid JSG. Molecular characterisation of a membrane-bound galactosyltransferase of plant cell wall matrix polysaccharide biosynthesis. Plant J. 1999;19:601–697. doi: 10.1046/j.1365-313x.1999.00566.x. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Fry SC. The structure and functions of xyloglucan. J Exp Bot. 1989;40:1–11. doi: 10.1093/jxb/40.1.1. [DOI] [Google Scholar]
- Goubet F, Barton CJ, Mortimer JC, Yu XL, Zhang ZN, Miles GP, Richens J, Liepman AH, Seffen K, Dupree P. Cell wall glucomannan in Arabidopsis is synthesised by CSLA glycosyltransferases, and influences the progression of embryogenesis. Plant J. 2009;60:527–538. doi: 10.1111/j.1365-313X.2009.03977.x. [DOI] [PubMed] [Google Scholar]
- Hetterscheid WLA, Ittenbach S. Everything you always wanted to know about Amorphophallus but were afraid to stick your nose into. Aroideana. 1996;19:7–129. [Google Scholar]
- Kraemer WJ, Vingren JL, Silvestre R, Spiering BA, Hatfield DL, Ho JY, Fragala MS, Maresh CM, Volek JS. Effect of adding exercise to a diet containing glucomannan. Metabolism. 2007;56:1149–1158. doi: 10.1016/j.metabol.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Li B, Xia J, Wang Y, Xie BJ. Grain-size effect on the structure and antiobesity activity of konjac flour. J Agric Food Chem. 2005;53:7404–7407. doi: 10.1021/jf050751q. [DOI] [PubMed] [Google Scholar]
- Liepman AH, Wilkerson CG, Keegstra K. Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc Natl Acad Sci USA. 2005;102:2221–2226. doi: 10.1073/pnas.0409179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman AH, Nairn CJ, Willats WGT, Sorensen I, Roberts AW, Keegstra K. Functional genomic analysis supports conservation of function among cellulose synthase-like a gene family members and suggests diverse roles of mannans in plants. Plant Physiol. 2007;143:1881–1893. doi: 10.1104/pp.106.093989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundqvist J, Teleman A, Junel L, Zacchi G, Dahlman O, Tjerneld F, Stalbrand H. Isolation and characterization of galactoglucomannan from spruce (Picea abies) Carbohydr Polym. 2002;48:29–39. doi: 10.1016/S0144-8617(01)00210-7. [DOI] [Google Scholar]
- Maeda Y, Awano T, Takabe K, Fujita M. Immunolocalization of glucomannans in the cell wall of differentiating tracheids in Chamaecyparis obtuse. Protoplasma. 2000;213:148–156. doi: 10.1007/BF01282152. [DOI] [Google Scholar]
- Mortimer JC, Miles GP, Brown DM, Zhang ZN, Segura MP, Weimar T, Yu XL, Seffen KA, Stephens E, Turner SR, Dupree P. Absence of branches from xylan in Arabidopsis gux mutants reveals potential for simplification of lignocellulosic biomass. Proc Natl Acad Sci USA. 2010;107:17409–17414. doi: 10.1073/pnas.1005456107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishinari K, Williams PA, Phillips GO. Review of the physicochemical characteristics and properties of konjac mannan. Food Hydrocolloid. 1992;6:199–222. doi: 10.1016/S0268-005X(09)80360-3. [DOI] [Google Scholar]
- Pauly M, Keegstra K. Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J. 2008;54:559–568. doi: 10.1111/j.1365-313X.2008.03463.x. [DOI] [PubMed] [Google Scholar]
- Pauly M, Keegstra K. Plant cell wall polymers as precursors for biofuels. Curr Opin Plant Biol. 2010;13:305–312. doi: 10.1016/j.pbi.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Reiter WD. Biochemical genetics of nucleotide sugar interconversion reactions. Curr Opin Plant Biol. 2008;11:236–243. doi: 10.1016/j.pbi.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Li L, Sun YH, Chiang VL. The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol. 2006;142:1233–1245. doi: 10.1104/pp.106.086678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takigami S, Takiguchi T, Phillips GO. Microscopical studies of the tissue structure of konjac tubers. Food Hydrocolloid. 1997;11:479–484. doi: 10.1016/S0268-005X(97)80046-X. [DOI] [Google Scholar]
- Teleman A, Nordstrom M, Tenkanen M, Jacobs A, Dahlman O. Isolation and characterization of O-acetylated glucomannans from aspen and birch wood. Carbohydr Res. 2003;338:525–534. doi: 10.1016/S0008-6215(02)00491-3. [DOI] [PubMed] [Google Scholar]
- Winter H, Huber SC. Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes. CRC Crit Rev Plant Sci. 2000;19:31–67. doi: 10.1016/S0735-2689(01)80002-2. [DOI] [PubMed] [Google Scholar]
- Yang YH, Kang YB, Lee KW, Lee TH, Park SS, Hwang BY, Kim BG. Characterization of GDP-mannose pyrophosphorylase from Escherichia coli O157:H7 EDL933 and its broad substrate specificity. J Mol Catal B Enzym. 2005;37:1–8. doi: 10.1016/j.molcatb.2005.08.002. [DOI] [Google Scholar]
- York WS, Darvill AG, McNeil M, Albersheim P. Structure of plant-cell walls. 16.3-deoxy-d-manno-2-octulosonic acid (Kdo) is a component of rhamnogalacturonan-II, a pectic polysaccharide in the primary-cell walls of plants. Carbohydr Res. 1985;138:109–126. doi: 10.1016/0008-6215(85)85228-9. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.