Abstract
Somatic embryogenesis is used for vegetative propagation of conifers. Embryogenic cultures can be established from zygotic embryos; however, the embryogenic potential decreases during germination. In Arabidopsis, LEAFY COTYLEDON (LEC) genes are expressed during the embryonic stage, and must be repressed to allow germination. Treatment with the histone deacetylase inhibitor trichostatin A (TSA) causes de-repression of LEC genes. ABSCISIC ACID3 (ABI3) and its Zea mays ortholog VIVIPAROUS1 (VP1) act together with the LEC genes to promote embryo maturation. In this study, we have asked the question whether TSA treatment in a conifer affects the embryogenic potential and the expression of embryogenesis-related genes. We isolated two conifer LEC1-type HAP3 genes, HAP3A and HAP3B, from Picea abies and Pinus sylvestris. A comparative phylogenetic analysis of plant HAP3 genes suggests that HAP3A and HAP3B are paralogous genes originating from a duplication event in the conifer lineage. The expression of HAP3A is high, in both somatic and zygotic embryos, during early embryo development, but decreases during late embryogeny. In contrast, the expression of VP1 is initially low but increases during late embryogeny. After exposure to TSA, germinating somatic embryos of P. abies maintain the competence to differentiate embryogenic tissue, and simultaneously the germination progression is partially inhibited. Furthermore, when embryogenic cultures of P. abies are exposed to TSA during embryo maturation, the maturation process is arrested and the expression levels of PaHAP3A and PaVP1 are maintained, suggesting a possible link between chromatin structure and expression of embryogenesis-related genes in conifers.
Electronic supplementary material
The online version of this article (doi:10.1007/s00425-011-1418-8) contains supplementary material, which is available to authorized users.
Keywords: Conifer embryogenesis, Embryogenic potential, HAP3 gene family, LEAFY COTYLEDON1, Trichostatin A, VIVIPAROUS1
Introduction
Somatic embryogenesis is the process of differentiation of embryos from somatic cells. This requires a signal that induces a somatic cell to dedifferentiate and gain embryogenic competence as well as the expression of an appropriate cellular environment for the response of the inductive signal (Braybrook and Harada 2008). The molecular mechanisms involved in this transition, from a differentiated vegetative cell to a cell with embryogenic competence, have been best described in model angiosperm species, but are largely uncharacterized in gymnosperms. This process is, however, the foundation for propagation of conifers through somatic embryos.
Seed development can be divided into distinct phases, an early morphogenesis phase and a late maturation phase followed by desiccation (West and Harada 1993; Goldberg et al. 1994). During morphogenesis, most cell divisions and differentiation occur and the basic body plan of the embryo is established. During the maturation phase, embryo morphogenesis is arrested and the embryo increases in size by cell elongation. Furthermore, storage compounds are synthesized and at the end of the maturation phase the embryo becomes desiccation tolerant. Seed germination marks the end of the embryonic development, and rapid repression of embryonic genes has been observed with seed imbibition (Tai et al. 2005). Several transcriptional regulators that play critical roles in promoting expression of seed transcriptional programs have been identified in Arabidopsis thaliana (Arabidopsis) (Zhang and Ogas 2009; Le et al. 2010). The LEAFY COTYLEDON (LEC) genes LEAFY COTYLEDON1 (LEC1), LEAFY COTYLEDON2 (LEC2) and FUSCA3 (FUS3) are transcription factors that act as master regulators of embryogenesis and they have been used as tools to define the mechanisms that underlie the initiation of somatic embryogenesis (Braybrook and Harada 2008). All three LEC genes encode transcriptional activators that are primarily expressed in the seed (Santos-Mendoza et al. 2008). LEC1 encodes a HAP3 subunit of the CCAAT-box binding factor (CBF) (Lotan et al. 1998; Lee et al. 2003). LEC2 and FUS3 encode transcription factors of the plant-specific B3 family (Luerssen et al. 1998; Stone et al. 2001). During early embryogenesis, the LEC genes are required to maintain the embryonic fate and to specify cotyledon identity (reviewed by Santos-Mendoza et al. 2008). The activity of the LEC genes must be repressed post-embryonically to allow vegetative development to proceed (Braybrook and Harada 2008). Ectopic expression of all three LEC genes causes cells in vegetative and reproductive tissues to adopt characteristics of maturation phase embryos (reviewed by Braybrook and Harada 2008). ABSCISIC ACID3 (ABI3) is another master regulator that together with the LEC genes promotes maturation (Giraudat et al. 1992; Parcy et al. 1997; To et al. 2006). ABI3 is orthologous to VP1 from maize (Zea mays) (McCarty et al. 1991) and belongs to the same subfamily as FUS3 and LEC2, commonly jointly referred to with the acronym AFL genes (Suzuki and McCarty 2008 with references). The role of B3 domain transcription factors in the regulation of embryo maturation and ABA-regulated gene expression in seeds has been studied extensively (Gutierrez et al. 2007). Regulators of LEC1 and AFL genes include the VP1/ABI3-LIKE (VAL) family of B3 domain transcription factors, which forms a sister clade to the AFL family (Suzuki et al. 2007). Furthermore, LEC1 seems to act earlier and as a regulator of AFL genes, since ectopic expression of LEC1 also activates the expression of the AFL genes (Kagaya et al. 2005).
The intricate control of regulatory genes during development of the seed has in several studies been shown to involve altered histone modifications and epigenetic regulation (reviewed by Zhang and Ogas 2009). Inhibition of histone deacetylases (HDACs), by mutant analysis or treatment with a chemical inhibitor, affects development of the embryo as well as the expression of seed associated genes including transcription factors (Tai et al. 2005; Tanaka et al. 2008). In addition, it has been postulated that the CHD3-chromatin-remodeling factor PICKLE (PKL) is a regulator of the LEC genes in Arabidopsis acting to repress embryonic identity during germination (Dean Rider et al. 2003).
Mutant analysis has been used, most extensively in Arabidopsis, to elucidate the genetic regulation of embryo development in angiosperms (Laux et al. 2004). In gymnosperms, however, knowledge about the regulation of embryo development is limited. Molecular and fossil data suggest that extant seed plants (gymnosperms and angiosperms) share a common ancestor approximately 300 million years ago (Smith et al. 2010). Despite this, the complement of genes expressed during embryogenesis in both groups shares striking sequence similarity (Cairney and Pullman 2007). Furthermore, it has been shown that certain regulatory pathways controlling seed- and spore-specific gene expression are conserved across phylogenetically distant species, ranging from ferns through cycads, and gymnosperms to angiosperms (Schallau et al. 2008). This suggests that genes central to embryogenesis may exhibit a high degree of conservation between angiosperms and gymnosperms, despite the fact that patterning during embryo development is very different between gymnosperms and angiosperms (von Arnold et al. 2002).
In this study, we provide insights into the molecular regulation of the transition from the embryonic to the vegetative stage during embryo development in conifers. Both Norway spruce (Picea abies) and Scots pine (Pinus sylvestris) are included, since the developmental pattern of the zygotic embryos differs between the two species. We have isolated two LEC1-type conifer HAP3A genes, PaHAP3A and PsHAP3A, as well as the Scots pine ABI3 homolog, PsVP1 (PaVP1 has been reported earlier by Footitt et al. 2003). A comparative phylogenetic analysis of plant HAP3 genes suggests that HAP3A and HAP3B are paralogous genes originating from a duplication event in the conifer lineage. The expression levels of both PsHAP3A and PsVP1 are similar during development of zygotic and somatic embryos. In addition, changes in the expression levels of the HAP3A and VP1 genes during somatic embryogenesis show similar trends in the two conifer species and resemble that of homologous genes in angiosperms. Treatment of germinating embryos of Norway spruce with an HDAC inhibitor partially inhibits the progression of germination and maintains the embryogenic potential. Furthermore, we show that HDAC inhibitor treatment during embryo maturation arrests maturation and affects the expression of PaHAP3A and PaVP1, suggesting a possible link between chromatin structure and expression of embryogenesis-related genes in conifers.
Materials and methods
Plant material and growth conditions
Embryogenic cell lines 06.21.00 and 06.28.05 of Norway spruce (Picea abies L. Karst.) and 12:12 of Scots pine (Pinus sylvestris L.) were used in this study. The embryogenic cultures were treated as described previously for Norway spruce (von Arnold and Clapham 2008) and Scots pine (Burg et al. 2007), except that the cultures of Norway spruce were proliferated on solidified medium. Briefly, the cultures proliferated as proembryogenic masses (PEMs) on proliferation medium, i.e., medium supplemented with the plant growth regulators (PGRs), auxin (2,4-dichlorophenoxyacetic acid) and cytokinin (N6-benzyladenine). To stimulate differentiation of early somatic embryos from PEMs, the cultures were transferred to pre-maturation medium, i.e., medium lacking PGRs for 1 week (Norway spruce) or 2 weeks (Scots pine). Maturation of embryos was stimulated as cultures were transferred to maturation medium containing abscisic acid (ABA). Cotyledonary somatic embryos were formed after 4–8 weeks of maturation treatment. Before germination, the embryos were partially desiccated (von Arnold and Clapham 2008).
New embryogenic cultures of Norway spruce were initiated from mature cotyledonary somatic embryos and from germinating somatic embryos incubated on proliferation medium containing PGRs for 4–5 weeks.
Zygotic embryos of Scots pine representing eight developmental stages were isolated from seeds collected every second to the third day during late June to late July 2009, from trees growing around Uppsala, Sweden. Somatic embryos of Scots pine at corresponding developmental stages as the zygotic embryos were isolated from embryogenic cultures during proliferation, pre-maturation and maturation. PEMs, early-, late- and cotyledonary embryos were collected from embryogenic cultures of Norway spruce after 2 weeks on proliferation medium, 1 week on pre-maturation medium, 1 week on maturation medium and 5 weeks on maturation medium. After sampling, the embryos were stored at −70°C until future use.
TSA treatments
The inhibitor of histone deacetylases, trichostatin A (TSA) (Sigma-Aldrich, St. Louis, MO, USA) was dissolved in DMSO to obtain a stock solution of 10 mM. Initially mature zygotic embryos of Norway spruce were screened for their potential of initiating embryogenic cultures on media containing 0.01, 0.1, 1 and 10 μM TSA. The highest initiation frequency was obtained on media containing 1 and 10 μM TSA (data not shown). In this study, we have used 10 μM TSA. A medium lacking TSA was used as control in each experiment. Since no effect of DMSO alone was observed, we did not include data from DMSO controls.
To assess the effects of TSA on embryo maturation and gene expression, the cultures were exposed to TSA during the whole maturation phase. Tissues for gene expression analyses were sampled weekly. The presented data are based on three biological replicates.
To elucidate if the embryogenic potential in germinated embryos was influenced by inhibition of histone deacetylases, TSA was added to the germination medium for 10 days. Thereafter, the germinated embryos were transferred to proliferation medium containing PGRs but lacking TSA. The embryogenic potential of germinated embryos was determined 4–5 weeks after transfer to the proliferation medium. The data presented are based on three biological replicates with each replicate including at least 100 embryos per treatment. To examine the possibility of recovering the embryogenic potential after germination, embryos were first germinated for 10 days under standard germination conditions and then transferred to germination medium supplemented with TSA for an additional 5 days, before incubation on proliferation medium.
RNA isolation
Total RNA was isolated from zygotic embryos of Scots pine using the RNAqueous Micro Kit (Ambion-Applied Biosystems, Austin, TX, USA) and somatic embryos of Scots pine and Norway spruce with the Spectrum Plant Total RNA kit (Sigma-Aldrich). Total RNA was subjected to DNase digestion, according to the manufacturer’s instruction (DNA-free, Ambion). cDNA was prepared from 0.5 to 1 μg total RNA using the Quanta cDNA synthesis kit (Quanta Biosciences, Gaithersburg, MD, USA).
Isolation and sequencing of PaHAP3A, PsHAP3A and PsVP1
Putative LEC1 homologs from the HAP3 gene family in Norway spruce and Scots pine as well as the VP1/ABI3 gene from the B3 gene superfamily in Scots pine were isolated from cDNA derived from embryogenic cultures. PaVP1 has previously been isolated in Norway spruce (Footitt et al. 2003). Primers for isolating conifer homologs of LEC1 and ABI3 were designed from loblolly pine (Pinus taeda) and white spruce (Picea glauca) expressed sequence tags (ESTs) and the PaVP1 gene (Supplementary Table 1). PCR amplifications were performed in 50 μl reactions using the Phusion enzyme (Finnzymes Oy, Espoo, Finland) according to the manufacturer’s protocol. The products were separated on a 1% TBE-gel and bands were excised and purified using the Gel purification kit (Fermentas-Thermo Fischer Scientific, Burlington, Canada). Purified products were sent to GATC Biotech, Konstanz, Germany, for sequence verification.
Sequence alignment and analysis
Additional HAP3 and VP1/ABI3 sequences were selected based on previously published data and BLAST searches (Altschul et al. 1997; for references and accession numbers, see Supplementary Table 2). In cases where the database contained identical or nearly identical sequences from the same taxon, only one representative was included.
Full-length amino acid alignments of 28 HAP3 genes were initially compiled using the MUSCLE and ClustalX algorithms in the program Geneious 5.0 (Drummond et al. 2010). Both alignments were then compared for discrepancies using the AltaVist alternative alignment visualization tool v. 1.0 (Morgenstern et al. 2003) before manual refinement and back-translation using Geneious. The nucleotide alignment was used for phylogenetic analyses, while the equivalent amino acid alignment was used only to identify shared sequence characters and sequence motifs. Bayesian phylogenetic analyses of the HAP3 genes were based on the conserved B-domain of the sequences, using the program MrBayes v3.1.2 (Ronquist and Huelsenbeck 2003). Owing to the long evolutionary distances of the HAP3 genes, a test for substitution saturation was performed using the software Dambe v4.5.56 (Xia et al. 2003). The variable third position was found to be saturated and was omitted in subsequent analyses. The model of evolution selected was GTR + I + G, which assumes a general time reversibility (GTR), a certain proportion of invariable sites (I) and a gamma approximation of the rate variation among sites (G). We ran four heated chains of the Markov chain Monte Carlo in parallel, sampling one tree every 500 generations for 1,500,000 generations starting with a random tree. The search reached stationarity after ~110,000 generations. The first 110,000 generations were considered the “burn-in” period and were discarded from generating the consensus phylogeny. Parsimony analysis was performed using PAUP* 4.0 (Swofford, 2001). Trees were generated using heuristic search replicates with 1,000 random stepwise taxon additions with a Tree Bisection–Reconnection branch-swapping algorithm and MulTrees ON. Gaps were treated as missing data and third position nucleotides were assigned zero weight. Bootstrap support for nodes (Felsenstein 1985) was estimated with 1,000 heuristic search replicates, using the same settings as the original search, with 1,000 random stepwise additions for each bootstrap replicate.
The analyses of PaVP1 and PsVP1 were made using all B3 family genes from Arabidopsis and the VP1 gene from maize. The conserved B3 region was used for alignment and phylogenetic analyses. Arabidopsis B3 protein sequences were collected using domain search (http://www.sanger.ac.uk/). The amino acids were then back-translated to nucleotide sequences using the Protogene server (Moretti et al. 2006). Duplicate sequences were omitted before sequence alignment. Alignments and Bayesian phylogenetic analyses were performed as described for the HAP3 genes. Starting with a random tree, one tree was sampled every 500 generations for 11,500,000 generations. The search reached stationarity after about 1,150,050 generations, and the “burn-in” period was set to the first 1,150,050 generations. The consensus tree showed a clear separation of known subgroups in the phylogeny of B3 genes, i.e., the AFL (ABI3/FUS3/LEC2) genes and the VAL (VP1/ABI3-LIKE) embryonic repressor genes (VAL1, VAL2 and VAL3), as suggested in Swaminathan et al. (2008); Romanel et al. (2009). From the consensus tree, a subsample of 35 sequences were selected for the final analysis, which was run for 2,000,000 generations. The search reached stationarity after 200,000 generations and the “burn-in” period was set to 200,000 generations.
Quantitative real-time PCR
Quantitative real-time PCR (qRT-PCR) was performed using the iQ5 Real-Time Detection System in iCycler iQ 96-well PCR plates with adhesive seals (Bio-Rad Laboratories, Hercules, CA, USA). Primers used to quantify expression levels are presented in Supplementary Table 3. The expression data of each gene was normalized against the expression of three reference genes, PHOSPHOGLUCOMUTASE, CELL DIVISION CYCLE 2 (CDC2) and ELONGATION FACTOR1-α (EF1α), previously selected based on their stability during embryo development (Vestman et al. 2011) using the geNorm software (Vandesompele et al. 2002). Amplifications were carried out using the DyNAmo Flash Sybr Green qPCR kit (Finnzymes). PCR cycling conditions were as advised by the manufacturer, with annealing and extension at 60°C for 30 s. The reactions were run for 40 cycles and at the end of each run melt curves were generated to ensure product uniformity. Three independent biological replicate samples were assessed and samples were added to the plates in triplicates. In all studies, inter-run connector samples were included to correct for the use of multiple plates. All calculations and normalizations were done using the iQ5 software (Bio-Rad).
Statistical methods
Data from the qRT-PCR were analyzed using a mixed model approach, see e.g., Littell et al. (2007), as implemented in the mixed procedure of the SAS (2008) system. The data from the zygotic and somatic embryo comparison in Scots pine were assessed with one-way ANOVA analyses, followed by post hoc Tukey’s HSD comparison tests. For the data from TSA-treated Norway spruce embryos, the model included time, treatment and the interaction between time and treatment. An unstructured covariance matrix was used to model the within-treatment covariances over time. Different genes were analyzed separately. Differences of P < 0.05 were regarded as significant.
Results
The embryogenic potential decreases during germination, but can be maintained with a histone deacetylase inhibitor
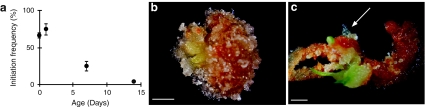
Embryogenic cultures of Norway spruce are routinely established from mature non-desiccated zygotic embryos (von Arnold and Clapham 2008). The potential to initiate embryogenic cultures in conifers decreases successively as the embryos germinate (Klimaszewska et al. 2010b). Initially, we analyzed this decrease in embryogenic potential during germination in Norway spruce (Fig. 1a). About 70% of the cotyledonary embryos and embryos germinated for 1 day differentiated embryogenic tissue within 5 weeks on proliferation medium. When embryogenic tissue was induced on cotyledonary embryos, usually the whole explant developed protruding embryogenic tissue (Fig. 1b). The embryogenic potential decreased drastically during germination (Fig. 1a). Furthermore, when germinating embryos were used as explants, embryogenic tissue differentiated from fewer more localized regions on the explant (Fig. 1c).
Fig. 1.
The embryogenic potential decreases during germination of Norway spruce. a Cotyledonary somatic embryos germinated for 0–14 days were stimulated to differentiate embryogenic tissue. The proportion of embryos that had initiated embryogenic tissue within 5 weeks is presented. Frequencies are based on three biological replicates with 30 embryos per treatment. b Embryogenic tissue differentiated from a mature cotyledonary embryo. c Embryogenic (arrow) and non-embryogenic tissue differentiated from a 2 week-old germinated embryo. Bars, 2 mm
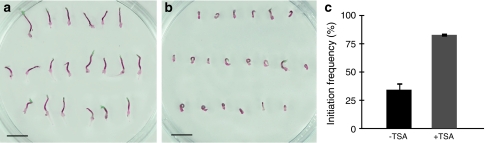
To establish whether treatment with an HDAC inhibitor affects the embryogenic potential, cotyledonary somatic embryos were germinated for 10 days on medium supplemented with TSA and thereafter transferred to proliferation medium lacking TSA to stimulate differentiation of embryogenic tissue. The germination progression was partially inhibited when the embryos were exposed to TSA (Fig. 2a, b). Among untreated embryos, the frequency of initiating embryogenic tissue was on average only 35%. However, when embryos were TSA treated during germination, the average frequency of initiation of embryogenic tissue was high, 85% (Fig. 2c). This is similar to the initiation frequency from cotyledonary embryos (Fig. 1a). This shows that the embryogenic potential of embryos (both cotyledonary embryos and embryos germinated for 10 days) was maintained when they were transferred to medium containing TSA. To test if treatment with TSA could also affect the embryogenic potential of already germinated embryos, we first germinated embryos for 10 days and then exposed them to TSA for 5 days before transfer to the proliferation medium. The initiation frequency was only 5% in untreated control embryos germinated for 15 days. In contrast, this 5-day treatment of already germinated embryos resulted in an initiation frequency of 22%, which was significantly higher (P < 0.05, Fisher’s exact test) than for untreated control embryos. The embryogenic tissue from TSA-treated germinating embryos differentiated from localized regions on the explant, similar to the patterns seen on germinated control embryos (Fig. 1c). Embryogenic cultures derived from TSA-treated embryos proliferated and differentiated into cotyledonary embryos as in the original embryogenic culture (data not shown).
Fig. 2.
The HDAC inhibitor TSA inhibits post-germination growth and maintains the embryogenic potential in germinating somatic embryos of Norway spruce. a Control embryos germinated for 10 days and b embryos germinated on medium containing 10 μM TSA for 10 days. Bars, 10 mm. c After 10 days germination on medium lacking (−TSA) or supplemented (+TSA) with TSA, the embryos were incubated on proliferation medium for 5 weeks to stimulate initiation of embryogenic tissue. The frequency of initiation is based on three biological replicates with 100 embryos per treatment. The difference in initiation frequency between −TSA and +TSA was significant, P < 0.001, Fisher’s exact test
Isolation and phylogenetic analysis of LEC1 and VP1/ABI3 homologs from Norway spruce and Scots pine
We isolated two HAP3 genes from both Norway spruce and Scots pine using primers designed from publicly available homologous conifer sequences (Supplementary Table 1). The genes were annotated PaHAP3A (accession number JF280794), PaHAP3B, PsHAP3A (accession number JF280795) and PsHAP3B. Complete putative coding sequences were obtained for both HAP3A genes comprising 540 bp of coding sequence corresponding to translated regions of 180 amino acids. Both peptides, generated from the nucleotide sequences, are highly similar to those of the two LEC1-type Arabidopsis genes LEC1 and LEAFY COTYLEDON1-LIKE (L1L). The conserved B-domain of PaHAP3A shares 83 and 89% of the amino acids with LEC1 and L1L, respectively, and PsHAP3A shares 80 and 86% of translated amino acid identity. Both PaHAP3A and PsHAP3A contain all characteristic amino acids that define the LEC1-type HAP3 genes (Kwong et al. 2003), except that the glutamic acid (77) is replaced by asparagine in PsHAP3A. So far, only partial transcripts of the HAP3B genes have been sequenced, including the conserved B-domain.
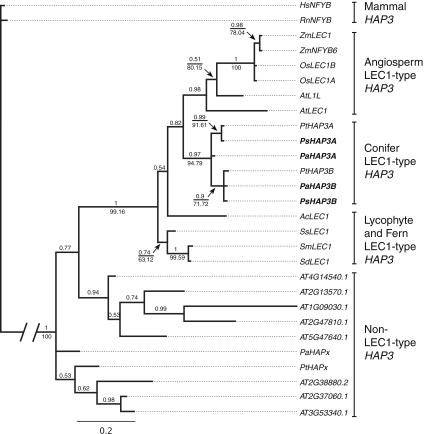
To analyze the relationship between the conifer HAP3 genes and HAP3 genes from other species, phylogenetic analyses based on nucleotide sequences from the conserved B-domain were made (Fig. 3). The phylogenetic analyses of HAP3 genes using Bayesian inference or heuristic searches of most parsimonious trees are in general agreement with Kwong et al. (2003) and Xie et al. (2008), supporting that plant HAP3 genes can be subdivided into LEC1-type and non-LEC1-type HAP3 genes (Fig. 3). Lycophyte and fern LEC1-type genes group at the base of the seed plant LEC1-type HAP3 genes. Both seed plant lineages harbor paralogous LEC1-type HAP3 genes, as exemplified by the presence of two LEC1-type HAP3 genes in rice (Oryza sativa) and Arabidopsis, as well as in conifers (HAP3A and HAP3B).
Fig. 3.
The conifer HAP3A and HAP3B genes are grouped within the plant-specific LEC1-type HAP3 genes. The figure shows a 50% majority rule tree derived from trees sampled after “burn-in” using Bayesian inference. Numbers above branches represent the posterior probabilities and numbers below branches represent parsimony bootstrap support values for branches that agreed between the estimates. Major clades are indicated to the right of the tree. Genes in boldface type were identified in this study. The root branch has been decreased to save space as indicated with the two leaning vertical lines. Names and accession numbers are found in Supplementary Table 2
ABI3 belongs to the large B3 family of plant-specific transcription factors (Giraudat et al. 1992; Suzuki and McCarty 2008) The full-length PaVP1 cDNA (accession number AF175576), which has been reported earlier (Footitt et al. 2003), contains all described conserved regions (A1, B1–B3) reported for ABI3 andVP1 (McCarty et al. 1991; Giraudat et al. 1992). The putative Scots pine VP1/ABI3 homolog PsVP1 was isolated using primers designed from a loblolly pine EST and PaVP1 (Supplementary Table 1). The partial PsVP1 cDNA, starting in the A1 region and extending beyond the conserved B3 region, included 1,939 nucleotides and shares 78% similarity with PaVP1.
To analyze the correspondence and evolution of PaVP1 and PsVP1, a Bayesian phylogenetic analysis was performed using nucleotide sequences from the B3 domains of the two conifer genes, the maize VP1 gene and all 118 B3 genes of Arabidopsis. The consensus tree showed a clear separation of known subgroups in the phylogeny of B3 genes, i.e., the AFL genes and the embryonic repressor genes (VAL1, VAL2 and VAL3), as suggested previously (Swaminathan et al. 2008; Romanel et al. 2009). From the consensus tree, a subsample of 35 genes was selected for a final analysis (Supplementary Fig. 1) The conifer genes were positioned closest to the maize VP1 gene and ABI3 from Arabidopsis, confirming a close relationship between the conifer and angiosperm ABI3/VP1 genes.
Expression levels of the conifer HAP3A and VP1 genes during development of zygotic and somatic embryos
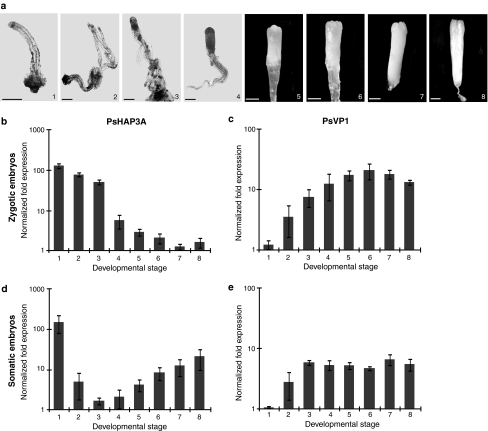
The expression of PsHAP3A and PsVP1 during development of zygotic and somatic embryos in Scots pine was analyzed by quantitative RT-PCR. The differences in expression levels between various developmental stages (Fig. 4a) were significant (Supplementary Table 4). PsHAP3A was highly expressed during early zygotic embryogenesis and a significant decrease in expression occurred at stage 4 (when the subordinate embryos were degraded), whereafter the expression remained low (Fig. 4b). In contrast, the expression of PsVP1 was initially low, but increased significantly at stage 3 (after cleavage polyembryogeny), whereafter it remained high (Fig. 4c). We also analyzed the expression of PsHAP3A and PsVP1 during development of somatic embryos of Scots pine at developmental stages corresponding to stages 2–8 in zygotic embryos. Stage 1 zygotic and somatic embryos are not comparable. Stage 1 zygotic embryos represent single, zygote-derived embryos, while stage 1 somatic embryos represent proliferating PEMs. PsHAP3A was highly expressed during proliferation (stage 1), but decreased significantly during embryo development (Fig. 4d). The expression of PsVP1 was low during proliferation (stage 1) and increased significantly at stage 3 (Fig. 4e). Our results show a similar trend in expression levels of both PsHAP3A and PsVP1 during the development of zygotic and somatic embryos.
Fig. 4.
Development of embryos in Scots pine and the expression of PsHAP3A and PsVP1 at sequential developmental stages. a Stages during development of a zygotic embryo; 1 a single zygote-derived early embryo, 2 four equal-sized embryos formed after cleavage, 3 a dominant embryo and several subordinate embryos, 4 a dominant embryo, and the subordinate embryos have been eliminated, 5 and 6 maturing embryos with developing cotyledon primordia, 7 and 8 fully mature cotyledonary embryos. Bars, 0.5 mm. Quantitative real-time PCR analysis of PsHAP3A (b, d) and PsVP1 (c, e) during the development of zygotic (b, c) and somatic (d, e) embryos. Samples were taken at the stages presented in a. Expression values are presented as relative values to the sample with lowest expression in each replicate. Expression levels are mean values of three biological replicates. Error bars indicate ± SE of biological replicates. The significance of the differences in expression levels between various developmental stages are presented in Supplementary Table 4
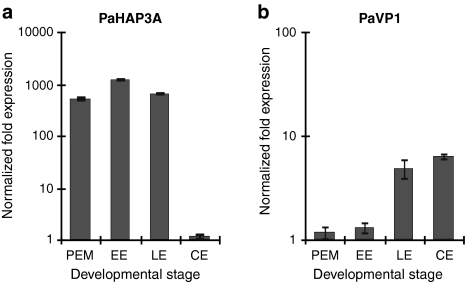
Expression of PaHAP3A and PaVP1 in Norway spruce was assessed at four different time points during the development of somatic embryos, i.e., proliferating PEMs on proliferation medium, early embryos 1 week after withdrawal of PGRs, late embryos after 1 week on maturation medium and cotyledonary embryos after 5 weeks on maturation medium (Fig. 5). The selected stages corresponded to somatic embryo stages 1, 3, 4 and 8 of Scots pine (Fig. 4a). PaHAP3A expression was high in proliferating PEMs and low in cotyledonary embryos (Fig. 5a). In contrast, the expression of PaVP1 was low in PEMs and high in cotyledonary embryos (Fig. 5b). The expression of PaVP1 correlates well with our earlier studies, where the expression of PaVP1 is highest around the early cotyledonary stage (Footitt et al. 2003).
Fig. 5.
Expression of PaHAP3A and PaVP1 during the development of somatic embryos in Norway spruce. Somatic embryos collected from embryogenic cultures of Norway spruce were taken at four developmental stages, i.e., proliferating PEMs on proliferation medium, early embryos (EE) 1 week after withdrawal of PGRs, late embryos (LE) after 1 week on maturation medium and cotyledonary embryos (CE) after 5 weeks on maturation medium. Quantitative real-time PCR analysis of PaHAP3A (a) and PaVP1 (b). Expression values are presented as relative values to the sample with lowest expression in each replicate. Expression levels are mean values of two biological replicates assessed three times each. Error bars indicate ±SE of biological replicates
TSA affects the expression of PaVP1 and PaHAP3A during embryo development
To test whether HDAC inhibition affects the expression of PaHAP3A and PaVP1, expression analyses were initially carried out on 10 day-old germinated somatic embryos of Norway spruce treated with TSA. However, the expression levels of both genes were very low, under a reliable detection limit, in both control and TSA-treated embryos.
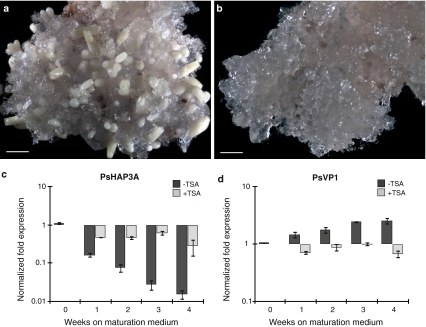
The effect of TSA treatment on the expression of PaHAP3A and PaVP1 was therefore analyzed during the maturation phase of somatic embryos. In control cultures, many cotyledonary embryos had developed after 4 weeks on maturation medium (Fig. 6a). In contrast, cultures developing on maturation medium supplemented with TSA continued to proliferate and maturation was arrested (Fig. 6b). However, if TSA was excluded from the maturation medium after 4 weeks, the embryos continued to mature and develop into cotyledonary embryos. In control cultures, PaHAP3A expression decreased during maturation. This decrease in PaHAP3A expression was significantly inhibited in TSA-treated cultures (Fig. 6c). The expression of PaVP1 increased in control cultures during maturation. The increase in expression of PaVP1 was significantly inhibited in TSA-treated cultures (Fig. 6d).
Fig. 6.
The HDAC inhibitor TSA blocks maturation and affects the expression of PaHAP3A and PaVP1 during the development of somatic embryos in Norway spruce. Embryogenic cultures incubated for 4 weeks on maturation medium lacking TSA (a) or containing 10 μM TSA (b). Bars, 2 mm. Quantitative real-time PCR analysis of PaHAP3A (c) and PaVP1 (d) in embryogenic cultures treated with TSA (light bars) and control cultures without TSA (dark bars). Samples were collected during differentiation of early somatic embryos (0 weeks) and after 1–4 weeks on maturation medium. Expression values are presented as relative values to one of the samples from untreated early embryogenic cultures (0 weeks). Expression levels are shown as mean values of three biological replicates assessed three times each. Error bars indicate ±SE of biological replicates. The expression levels of TSA-treated samples (+TSA) significantly differed from the untreated control samples (−TSA) at each developmental stage for both genes, P < 0.05
Discussion
Stimulation of somatic embryogenesis in germinating embryos by TSA
Embryogenic cultures of conifers are routinely established from immature or mature zygotic embryos. The initiation frequency deceases dramatically when the embryos start germinating (Bonga et al. 2010; Klimaszewska et al. 2010b, and this work). Here, we demonstrate that after exposure to the HDAC inhibitor TSA, germinating embryos of Norway spruce maintain the competence to differentiate into embryogenic tissue at the same time as the germination progression is partially inhibited. It has previously been demonstrated that seed germination in Arabidopsis is inhibited by TSA. Simultaneously, the expression of embryogenesis-related genes, including the LEC genes and ABI3, are activated and embryo-like structures differentiate on the true leaves after withdrawal of TSA (Tanaka et al. 2008). Our results imply that TSA affects the embryogenic potential and germination in a similar way in conifers. To analyze if TSA also affects gene expression in conifers, we isolated embryogenesis-related genes from Norway spruce and Scots pine.
Conifer homologs of AtLEC1 and AtABI3 are expressed during embryo development
To isolate putative conifer homologs of LEC1, LEC2, FUS3 and ABI3, we initially blasted the Arabidopsis sequences against conifer EST databases. Only genes highly similar to LEC1 and ABI3 were found. At present, no full genome sequence of any gymnosperm is available and therefore it is too early to exclude the possibility that there are conifer homologs to LEC2 and FUS3 as well.
We isolated two conifer LEC1-type HAP3 genes in both Norway spruce and Scots pine, PaHAP3A, PaHAP3B, PsHAP3A and PsHAP3B. Both conifer HAP3A and HAP3B genes encode proteins that are highly similar to LEC1-type HAP3 subunits of the CCAAT binding transcription factor (CBF, or NF-Y) (Lotan et al. 1998; Lee et al. 2003; Kwong et al. 2003). Furthermore, they all contain nearly all amino acids characteristic for LEC1-type HAP3 subunits in Arabidopsis, including the important amino acid asp 55, which confer DNA binding specificity to the CBF complex (Lee et al. 2003). Phylogenetic analyses position both conifer HAP3A and HAP3B genes among LEC1-type genes. Within the LEC1-type genes there are well-supported clades separating angiosperms, gymnosperms and lycophytes (Fig. 4). Lycophyte LEC1-type genes group basal to the angiosperm and gymnosperm clade suggesting that an ancestral LEC1-type gene was present in the last common ancestor of all extant seed plants. Both angiosperms and gymnosperms seem to have undergone separate duplications within the LEC1-type lineage. Maize has been suggested to harbor up to three copies of LEC1-type HAP3 genes, although the accessions are not publicly available (Suzuki et al. 2008). Similarly, the conifer LEC1-type HAP3A and HAP3B genes are most likely the result of a separate duplication event within the gymnosperm lineage.
The Norway spruce homolog to ABI3, PaVP1, has previously been characterized (Footitt et al. 2003). In this study we isolated its Scots pine homolog, PsVP1. The protein is highly similar to that of Norway spruce and sequence analysis revealed close relationship of both conifer genes to ABI3 and its maize homolog VP1, within the group of plant-specific B3 transcription factors.
During early conifer embryogenesis, the expression of PaHAP3A and PsHAP3A is initially high, in agreement with recently reported expression of a putative LEC1-type HAP3 gene in lodgepole pine (Pinus contorta) (Park et al. 2010). Later, during development of the embryos, the expression of PaHAP3A and PsHAP3A decreases dramatically and remains low throughout maturation (Figs. 4, 5). In contrast, the expression of the conifer PaVP1 and PsVP1 genes increases early during embryo development and peaks at the time point when the cotyledons emerge (Figs. 4, 5). These results are analogous with what has been reported in Arabidopsis, where LEC1 is expressed at higher levels during early embryo development than in maturing embryos (Lotan et al. 1998; Baumbusch et al. 2004). Furthermore, the expression of ABI3 can first be detected at the globular stage and persists throughout maturation of the embryo (Parcy et al. 1997; To et al. 2006).
Preliminary expression studies of PaHAP3B indicate a comparable low overall expression, and in contrast to PaHAP3A the expression increases at later maturation stages (data not shown). Duplication within a gene lineage could be indicative of functional divergence leading to subfunctionalization, and/or neofunctionalization according to Irish and Litt 2005, which may be reflected by differential expression during development. Hence, the data suggest that the conifer HAP3 genes might have gone through subfunctionalization, and/or neofunctionalization, with PaHAP3A showing the highest similarity in activity to extant angiosperm LEC1 genes. However, we cannot exclude that the HAP3B gene might have important roles and partly overlapping functions with HAP3A during embryogenesis in conifers, as suggested for LEC1 and L1L in Arabidopsis (Kwong et al. 2003; Yamamoto et al. 2009).
The LEC genes (LEC1, LEC2 and FUS3) have been shown necessary to confer embryonic cell fate and specification of cotyledon identity early during embryogenesis (Meinke 1992; Bäumlein et al. 1994; Keith et al. 1994; Meinke et al. 1994; West et al. 1994; Lotan et al. 1998; Luerssen et al. 1998; Stone et al. 2001; Gazzarrini et al. 2004). Mutant analysis has indicated that LEC1 might act upstream of LEC2 and FUS3 (Meinke et al. 1994). The expression of FUS3 and ABI3 is regulated by LEC1 and LEC2 as well as by themselves. Furthermore, the spatial expression seems to be very important, i.e., it has been demonstrated that ABI3 is regulated by a complex network that involves LEC1 in the cotyledons, but not in the embryo axis (To et al. 2006). Molecular data has also shown that ectopic LEC1 expression induces expression of ABI3 (Kagaya et al. 2005). Later during embryogenesis, ABI3, together with the LEC genes, is responsible for the initiation and maintenance of the maturation phase. To what extent the conifer HAP3A and VP1 genes interact is presently not known, but the fact that the expression profiles of the genes are each other’s opposite during embryo development might indicate a direct or indirect feedback regulation or at least a common regulatory machinery.
Ectopic expression of CHAP3A (a putative LEC1-type HAP3 gene from Picea mariana) under an inducible promoter was recently reported in germinating somatic embryos of white spruce (Klimaszewska et al. 2010b). However, the ectopic expression of CHAP3A did not induce the expression of other embryogenesis-related genes, neither was the development of the embryos affected. This is in contrast to that previously shown in Arabidopsis, where ectopic post-embryonic expression of the LEC1 gene in vegetative cells can induce the expression of embryo-specific genes and stimulate formation of embryo-like structures (Lotan et al. 1998). Further studies on the functions of the LEC1-type HAP3A and HAP3B genes are necessary to understand their roles during conifer embryogenesis.
Expression of embryo-specific transcription factors is affected by TSA
We have shown that treatment of Norway spruce embryos with TSA during germination partially inhibits germination and maintains the embryogenic potential (Figs. 2, 3). However, we could not show whether the TSA treatment during germination affects the expression of PaHAP3A or PaVP1, since both genes were expressed at very low levels. Of importance is that inhibition of HDAC activity during embryo germination stimulates the initiation of embryogenic tissues in a localized non-uniform pattern. Recently, Klimaszewska et al. (2010a) reported initiation of embryogenic tissue from buds of 10 year-old trees regenerated from somatic embryos of white spruce. The induction of embryogenic tissue was shown to be associated with formation of nodules on the surface of the needle. The nodules differentiate in a similar pattern as we have shown for protruding embryogenic tissue on germinating embryos (Fig. 1c). Interestingly, Klimaszewska et al. (2010a) showed a high expression of CHAP3A in embryogenic tissue, but could not detect any expression in the needles. The results from the Klimaszewska group and results presented in this study suggest that the expression of genes stimulating the initiation of embryogenic competence are localized to a limited number of cells in the explant, not detectable in total RNA from the whole explant. We assume that some cells in the germinating embryo still possess the intracellular environment needed for embryogenic competence and that application of the HDAC inhibitor TSA enables transcription of embryogenesis-related genes and transcription factors needed for generating embryogenic tissue from these cells.
To examine if TSA affects the expression of PaHAP3A and PaVP1, we analyzed the expression of the genes during the maturation phase. The expression of PaHAP3A decreases during maturation, while that of PaVP1 increases. When embryogenic cultures of Norway spruce are exposed to TSA during the maturation phase, the cultures continue to proliferate and maturation is arrested (Fig. 6a, b). Simultaneously, the expression of PaVP1 remains low while the expression of PaHAP3A remains high throughout the maturation treatment (Fig. 6c, d). Assuming that TSA affects histone acetylation in conifers, our results indicate a connection between changes in acetylation patterns and the levels of embryogenesis-related gene expression in Norway spruce.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors would like to thank Ulf Olsson for statistical consultancy. This work was supported by the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (Formas) and the Royal Swedish Academy of Agriculture and Forestry. Silvia Valladares was supported by an Angeles Alvariño postdoctoral fellowship from Xunta de Galicia (Spain).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- ABI3
ABSCISIC ACID INSENSITIVE3
- HDAC
Histone deacetylase
- LEC1
LEAFY COTYLEDON1
- TSA
Trichostatin A
- VP1
VIVIPAROUS1
Footnotes
J. F. Sundström and A. Sundås-Larsson contributed equally to this work.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbusch LO, Hughes DW, Galau GA, Jakobsen KS. LEC1, FUS3, ABI3 and Em expression reveals no correlation with dormancy in Arabidopsis. J Exp Bot. 2004;55:77–87. doi: 10.1093/jxb/erh014. [DOI] [PubMed] [Google Scholar]
- Bäumlein H, Miséra S, Luerßen H, Kölle K, Horstmann C, Wobus U, Müller AJ. The FUS3 gene of Arabidopsis thaliana is a regulator of gene expression during late embryogenesis. The Plant J. 1994;6:379–387. doi: 10.1046/j.1365-313X.1994.06030379.x. [DOI] [Google Scholar]
- Bonga JM, Klimaszewska KK, von Aderkas P. Recalcitrance in clonal propagation, in particular of conifers. Plant Cell Tiss Org. 2010;100:241–254. doi: 10.1007/s11240-009-9647-2. [DOI] [Google Scholar]
- Braybrook SA, Harada JJ. LECs go crazy in embryo development. Trends Plant Sci. 2008;13:624–630. doi: 10.1016/j.tplants.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Burg K, Helmersson A, Bozhkov P, von Arnold S. Developmental and genetic variation in nuclear microsatellite stability during somatic embryogenesis in pine. J Exp Bot. 2007;58:687–698. doi: 10.1093/jxb/erl241. [DOI] [PubMed] [Google Scholar]
- Cairney J, Pullman GS. The cellular and molecular biology of conifer embryogenesis. New Phytol. 2007;176:511–536. doi: 10.1111/j.1469-8137.2007.02239.x. [DOI] [PubMed] [Google Scholar]
- Dean Rider S, Henderson JT, Jerome RE, Edenberg HJ, Romero-Severson J, Ogas J. Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J. 2003;35:33–43. doi: 10.1046/j.1365-313X.2003.01783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Heled J, Kearse M, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A (2010) Geneious v5.1, available from http://www.geneious.com [DOI] [PMC free article] [PubMed]
- Felsenstein J. Confidence-limits on phylogenies—an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Footitt S, Ingouff M, Clapham D, von Arnold S. Expression of the viviparous 1 (Pavp1) and p34cdc2 protein kinase (cdc2 Pa) genes during somatic embryogenesis in Norway spruce (Picea abies [L.] Karst) J Exp Bot. 2003;54:1711–1719. doi: 10.1093/jxb/erg178. [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P. The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev Cell. 2004;7:373–385. doi: 10.1016/j.devcel.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. Isolation of the ArabidopsisABI3 gene by positional cloning. Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB, de Paiva G, Yadegari R. Plant embryogenesis: zygote to seed. Science. 1994;266:605–614. doi: 10.1126/science.266.5185.605. [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Van Wuytswinkel O, Castelain M, Bellini C. Combined networks regulating seed maturation. Trends Plant Sci. 2007;12:294–300. doi: 10.1016/j.tplants.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Irish VF, Litt A. Flower development and evolution: gene duplication, diversification and redeployment. Curr Opin Genet Dev. 2005;15:454–460. doi: 10.1016/j.gde.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Kagaya Y, Toyoshima R, Okuda R, Usui H, Yamamoto A, Hattori T. LEAFYCOTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISICACIDINSENSITIVE3. Plant Cell Physiol. 2005;46:399–406. doi: 10.1093/pcp/pci048. [DOI] [PubMed] [Google Scholar]
- Keith K, Kraml M, Dengler NG, McCourt P. fusca3: a heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell. 1994;6:589–600. doi: 10.1105/tpc.6.5.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimaszewska K, Overton C, Stewart D, Rutledge RG. Initiation of somatic embryos and regeneration of plants from primordial shoots of 10 year-old somatic white spruce and expression profiles of 11 genes followed during the tissue culture process. Planta. 2010;233:635–647. doi: 10.1007/s00425-010-1325-4. [DOI] [PubMed] [Google Scholar]
- Klimaszewska K, Pelletier G, Overton C, Stewart D, Rutledge RG. Hormonally regulated overexpression of ArabidopsisWUS and conifer LEC1 (CHAP3A) in transgenic white spruce: implications for somatic embryo development and somatic seedling growth. Plant Cell Rep. 2010;29:723–734. doi: 10.1007/s00299-010-0859-z. [DOI] [PubMed] [Google Scholar]
- Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ. LEAFYCOTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell. 2003;15:5–18. doi: 10.1105/tpc.006973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T, Würschum T, Breuninger H. Genetic regulation of embryonic pattern formation. Plant Cell . 2004;16 Suppl:S190–S202. doi: 10.1105/tpc.016014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, Drews GN, Fischer RL, Okamuro JK, Harada JJ, Goldberg RB. Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA. 2010;107:8063–8070. doi: 10.1073/pnas.1003530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc Natl Acad Sci USA. 2003;100:2152–2156. doi: 10.1073/pnas.0437909100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littell RC, Milliken GA, Stroup WW, Wolfinger RD, Schabenberger O. SAS for mixed models. J Roy Statist Soc. 2007;170:257–258. doi: 10.1111/j.1467-985X.2006.00455_9.x. [DOI] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 1998;93:1195–1205. doi: 10.1016/S0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- Luerssen H, Kirik V, Herrmann P, Miséra S. FUSCA3 encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsisthaliana. Plant J. 1998;15:755–764. doi: 10.1046/j.1365-313X.1998.00259.x. [DOI] [PubMed] [Google Scholar]
- McCarty DR, Hattori T, Carson CB, Vasil V, Lazar M, Vasil IK. The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell. 1991;66:895–905. doi: 10.1016/0092-8674(91)90436-3. [DOI] [PubMed] [Google Scholar]
- Meinke DW. A homoeotic mutant of Arabidopsisthaliana with leafy cotyledons. Science. 1992;258:1647–1650. doi: 10.1126/science.258.5088.1647. [DOI] [PubMed] [Google Scholar]
- Meinke DW, Franzmann LH, Nickle TC, Yeung EC. LeafyCotyledon mutants of Arabidopsis. Plant Cell. 1994;6:1049–1064. doi: 10.1105/tpc.6.8.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti S, Reinier F, Poirot O, Armougom F, Audic S, Keduas V, Notredame C. PROTOGENE: turning amino acid alignments into bona fide CDS nucleotide alignments. Nucleic Acids Res. 2006;34(Web Server issue):W600–W603. doi: 10.1093/nar/gkl170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern B, Goel S, Sczyrba A, Dress A. AltAVisT: comparing alternative multiple sequence alignments. Bioinformatics. 2003;19:425–426. doi: 10.1093/bioinformatics/btf882. [DOI] [PubMed] [Google Scholar]
- Parcy F, Valon C, Kohara A, Miséra S, Giraudat J. The ABSCISICACID-INSENSITIVE3, FUSCA3, and LEAFYCOTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell. 1997;9:1265–1277. doi: 10.1105/tpc.9.8.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-Y, Klimaszewska K, Park J-Y, Mansfield SD. Lodgepole pine: the first evidence of seed-based somatic embryogenesis and the expression of embryogenesis marker genes in shoot bud cultures of adult trees. Tree Physiol. 2010;30:1469–1478. doi: 10.1093/treephys/tpq081. [DOI] [PubMed] [Google Scholar]
- Romanel EAC, Schrago CG, Couñago RM, Russo CAM, Alves-Ferreira M. Evolution of the B3 DNA binding superfamily: new insights into REM family gene diversification. PLoS ONE. 2009;4:e5791. doi: 10.1371/journal.pone.0005791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L. Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 2008;54:608–620. doi: 10.1111/j.1365-313X.2008.03461.x. [DOI] [PubMed] [Google Scholar]
- Schallau A, Kakhovskaya I, Tewes A, Czihal A, Tiedemann J, Mohr M, Grosse I, Manteuffel R, Bäumlein H. Phylogenetic footprints in fern spore- and seed-specific gene promoters. Plant J. 2008;53:414–424. doi: 10.1111/j.1365-313X.2007.03354.x. [DOI] [PubMed] [Google Scholar]
- Smith SA, Beaulieu JM, Donoghue MJ. An uncorrelated relaxed-clock analysis suggests an earlier origin for flowering plants. Proc Natl Acad Sci USA. 2010;107:5897–5902. doi: 10.1073/pnas.1001225107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ. LEAFYCOTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA. 2001;98:11806–11811. doi: 10.1073/pnas.201413498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, McCarty DR. Functional symmetry of the B3 network controlling seed development. Curr Opin Plant Biol. 2008;11:548–553. doi: 10.1016/j.pbi.2008.06.015. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Wang HH-Y, McCarty DR. Repression of the LEAFYCOTYLEDON1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACIDINSENSITIVE 3-LIKE B3 genes. Plant Physiol. 2007;143:902–911. doi: 10.1104/pp.106.092320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Latshaw S, Sato Y, Settles AM, Koch KE, Hannah LC, Kojima M, Sakakibara H, McCarty DR. The maize Viviparous8 locus, encoding a putative ALTERED MERISTEM PROGRAM1-like peptidase, regulates abscisic acid accumulation and coordinates embryo and endosperm development. Plant Physiol. 2008;146:1193–1206. doi: 10.1104/pp.107.114108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan K, Peterson K, Jack T. The plant B3 superfamily. Trends Plant Sci. 2008;13:647–655. doi: 10.1016/j.tplants.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*:Phylogenetic analysis using parsimony (*and other methods) Sunderland: Sinauer Associates; 2001. [Google Scholar]
- Tai HH, Tai GCC, Beardmore T. Dynamic histone acetylation of late embryonic genes during seed germination. Plant Mol Biol. 2005;59:909–925. doi: 10.1007/s11103-005-2081-x. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Kikuchi A, Kamada H. The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 2008;146:149–161. doi: 10.1104/pp.107.111674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F. A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell. 2006;18:1642–1651. doi: 10.1105/tpc.105.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034.1-0034.11 [DOI] [PMC free article] [PubMed]
- Vestman D, Larsson E, Uddenberg D, Cairney J, Clapham D, Sundberg E, Arnold S (2011) Important processes during differentiation and early development of somatic embryos of Norway spruce as revealed by changes in global gene expression. Tree Genet Genomes 7:347–362
- von Arnold S, Clapham D. Spruce embryogenesis. Methods Mol Biol. 2008;427:31–47. doi: 10.1007/978-1-59745-273-1_3. [DOI] [PubMed] [Google Scholar]
- von Arnold S, Sabala I, Bozhkov P, Dyachok J, Filonova L. Developmental pathways of somatic embryogenesis. Plant Cell Tiss Org. 2002;69:233–249. doi: 10.1023/A:1015673200621. [DOI] [Google Scholar]
- West M, Harada JJ. Embryogenesis in higher plants: an overview. Plant Cell. 1993;5:1361–1369. doi: 10.1105/tpc.5.10.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M, Yee KM, Danao J, Zimmerman JL, Fischer RL, Goldberg RB, Harada JJ. LEAFYCOTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell. 1994;6:1731–1745. doi: 10.1105/tpc.6.12.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Xie Z, Salemi M, Chen L, Wang Y. An index of substitution saturation and its application. Mol Phylogenet Evol. 2003;26:1–7. doi: 10.1016/S1055-7903(02)00326-3. [DOI] [PubMed] [Google Scholar]
- Xie Z, Li X, Glover BJ, Bai S, Rao G-Y, Luo J, Yang J. Duplication and functional diversification of HAP3 genes leading to the origin of the seed-developmental regulatory gene, LEAFYCOTYLEDON1 (LEC1), in nonseed plant genomes. Mol Biol Evol. 2008;25:1581–1592. doi: 10.1093/molbev/msn105. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Kagaya Y, Toyoshima R, Kagaya M, Takeda S, Hattori T. Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J. 2009;58:843–856. doi: 10.1111/j.1365-313X.2009.03817.x. [DOI] [PubMed] [Google Scholar]
- Zhang H, Ogas J. An epigenetic perspective on developmental regulation of seed genes. Molecular Plant. 2009;2:610–627. doi: 10.1093/mp/ssp027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.