Abstract
Background
Fibrous hamartoma is the key pathology of congenital pseudarthrosis of the tibia (CPT), which was shown to have low osteogenicity and high osteoclastogenicity. This study further investigated the mechanism of impaired osteoblastic differentiation of fibrous hamartoma cells.
Methods
Fibroblast-like cells were obtained from enzymatically dissociated fibrous hamartomas of 11 patients with CPT associated with neurofibromatosis type I (NF1). Periosteal cells were also obtained from the distal tibial periosteum of 3 patients without CPT or NF1 as control. The mRNA levels of Wnt ligands and their canonical receptors, such as Lrp5 and β-catenin, were assayed using reverse transcriptase PCR (RT-PCR). Changes in mRNA expression of osteoblast marker genes by rhBMP2 treatment were assayed using quantitative real time RT-PCR. Changes in mRNA expression of transcription factors specifically involved in osteoblastic differentiation by rhBMP2 treatment was also assayed using quantitative real-time RT-PCR.
Results
Wnt1 and Wnt3a mRNA expression was lower in fibrous hamartoma than in tibial periosteal cells, but their canonical receptors did not show significant difference. Response of osteoblastic marker gene expression to rhBMP2 treatment showed patient-to-patient variability. Col1a1 mRNA expression was up-regulated in most fibrous hamartoma tissues, osteocalcin was up-regulated in a small number of patients, and ALP expression was down-regulated in most fibrous hamartoma tissues. Changes in mRNA expression of the transcription factors in response to rhBMP2 also showed factor-to-factor and patient-to-patient variability. Dlx5 was consistently up-regulated by rhBMP2 treatment in all fibrous hamartoma tissues tested. Msx2 expression was down-regulated by rhBMP2 in most cases but by lesser extent than control tissue. Runx2 expression was up-regulated in 8 out of 18 fibrous hamartoma tissues tested. Osterix expression was up-regulated in 2 and down-regulated in 3 fibrous hamartoma tissues.
Conclusions
Congenital pseudarthrosis of the tibia appears to be caused by fibrous hamartoma originating from aberrant growth of Nf1 haploinsufficient periosteal cells, which failed in terminal osteoblastic differentiation and arrested at a certain stage of this process. This pathomechanism of CPT should be targeted in the development of novel therapeutic biologic intervention.
Keywords: Congenital, Pseudarthrosis, Osteoblastic differentiation
Congenital pseudarthrosis of the tibia (CPT) is a very rare but specific condition characterized by anterolateral bowing at birth, and recurrent pathologic fractures of the tibia during early childhood. Fracture usually occurs spontaneously or after minor trauma and rarely heals without appropriate surgical intervention, and easily results in pseudarthrosis.
Fibrous hamartoma tissue is regarded as the key pathology of CPT,1) which occupies the site of pseudarthrosis, and is continuous with abnormally thickened periosteum in the adjacent bone segment.
CPT frequently develops in patients with neurofibromatosis type 1 (NF1). It was reported that association with NF1 was found in 40 to 60 per cent of CPT patients,2) and CPT develops in 1 to 4 percent of patients with NF1.3) NF1 is one of the most common autosomal dominant disorders, affecting approximately 1 in 3500 births.4) It is characterized by neurofibromas, cafe-au-lait spots, Lisch nodules and numerous skeletal manifestations, such as macrocephaly, short stature, kyphoscoliosis, sphenoid wing dysplasia, congenital bowing and congenital pseudarthrosis of the tibia.
NF1 is caused by a heterozygous mutation in the Nf1 gene located in chromosome 17q11.2.5) This gene encodes neurofibromin, which is expressed in a wide range of cells and tissues,6) including maturing chondrocytes, hypertrophic chondrocytes, osteoblasts, osteocytes, and osteoclasts.7) It is also expressed in tissues around CPT.8) Neurofibromin negatively regulates the activity of an intracellular signaling molecule p21ras (Ras), by functioning as a GTPase activating protein (Ras-GAP).9,10) Haploinsufficiency or complete deficiency in Nf1 gene function results in a dose-dependent elevation in Ras activity, which can activate the mitogen-activated protein kinase (MAPK) pathway and the phosphatidylinositol-3-phosphate kinase (PI-3K) pathway.11)
The Nf1 gene is classified as a tumor suppressor gene. Heterozygous Nf1+/- mice experienced tumors and premature death.12) Chimeric mice containing moderate levels of Nf1-/- deficient cells went on to form numerous neurofibromas, which progressed to malignancy with loss of the p53 tumor suppressor gene.13) Furthermore, the loss of heterozygosity (LOH) is seen in benign and malignant tumors, including neurofibroma, malignant peripheral nerve sheath tumor (MPNST) and astrocytoma.14,15)
Previous studies demonstrated that the phenotype of the fibrous hamartoma cells is consistent with the immunophenotype of mesenchymal lineage cells as that of tibial periosteal cells.16,17) However, these cells do not undergo osteoblastic differentiation in response to bone morphogenetic protein (BMP), and they are more osteoclastogenic than the tibial periosteal cells.16) In this study, we investigated further steps of osteoblastic differentiation in cells isolated from fibrous hamartoma, in order to delineate the mechanism of impaired osteoblastic differentiation of fibrous hamartoma cells.
METHODS
Primary Cell Culture
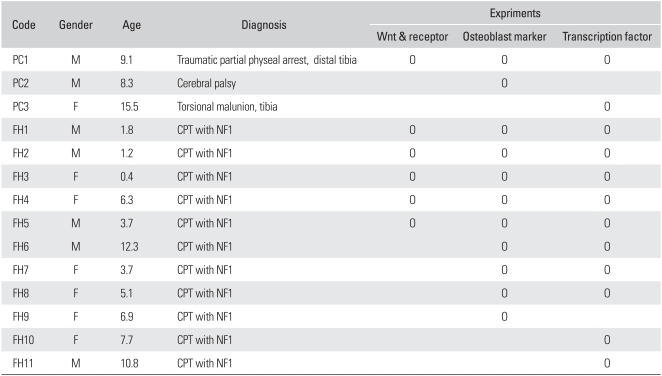
This study was approved by the institutional review board of Seoul National University Hospital. Harvested human tissues were investigated in experiments with informed consent. Fibrous hamartoma tissue was harvested from 11 patients (5 male and 6 female) with an atrophic type of CPT associated with NF1, during surgical procedures for osteosynthesis. Control tissue was obtained from the distal tibial periosteum. A strip of about 2 × 5 mm distal tibial periosteum was harvested during supramalleolar tibial derotation osteotomy from 3 patients without CPT or NF1 (Table 1). They underwent tissue culture. Fibrous hamartoma or tibial periosteum was enzymatically dissociated to obtain fibroblast-like cells, which were plated on a 100-mm dish, and cultured in DMEM containing 10% FBS and antibiotics at 37℃ in 5% CO2. When the second passage cells reached confluence, they were trypsinized, frozen, and stored in liquid nitrogen until required. The cells from fibrous hamartoma were denoted by the labels listed in Table 1 as FH1 through FH11, and cells from the tibial periosteum as PC1 through PC3. A vial of cells from one confluent dish were thawed and plated on 100-mm dish. When they reached confluence again, they were re-plated in 1:3 ratio, and rhBMP2 (R&D Systems, Minneapolis, MN, USA) treatment was commenced 48 hours after plating. As the amount of tissue harvested from patients was limited, not all tissues were used in the experiments (Table 1).
Table 1.
Tissue Samples Used in This Study
PC: periosteal cell, FH: fibrous hamartoma, CPT: congenital pseudarthrosis of the thibia, NF1: neurofibromatosis type 1.
Wnt Ligands and Receptors Expression
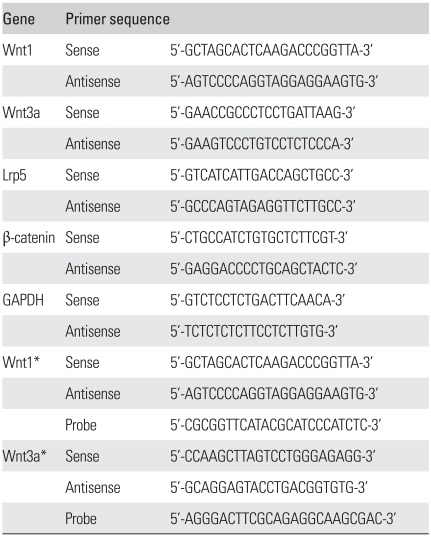
Total RNA was extracted from plated cells using Trizol reagent (Invitrogen, Carlsbad, CA, USA). The mRNA expressions of Wnt1, Wnt3a, LRP-5 and β-catenin were assayed by reverse transcription-polymerase chain reaction (Table 2), to investigate for endogenous Wnt signaling. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as internal control. Regular PCR was performed using AccuPower® HotStart PCR Premix (Bioneer, Daejeon, Korea) with primers for each gene (Table 2). Quantitative real-time PCR was performed for Wnt1 and Wnt3a as they showed different band intensity in regular PCR. After reverse transcription using the SuperScript First-strand Synthesis System for RT-PCR (Invitrogen), real-time PCR was performed in TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA, USA) using an ABI Prism 7000 Sequence Detection System (Applied Biosystems). To compare different gene expression levels, threshold cycle (CT) values were compared for relative gene expression analysis. The expression levels of target genes were presented as ratio to that of GAPDH. All samples were amplified four times.
Table 2.
Custom-made Primers List
*Primers and probe sequences for quantitative real-time RT-PCR.
Changes in mRNA Expression by rhBMP Treatment
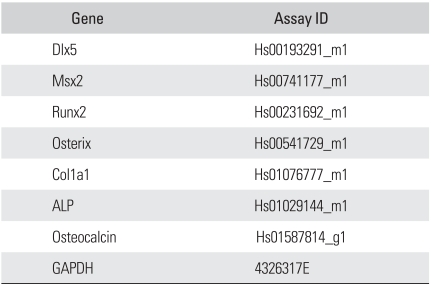
The plated cells were grown in osteogenic media (DMEM containing 10% FBS, 100 nM dexamethasone, 10 mM sodium-glycerophosphate, and 0.05 mM ascorbic acid-2-phosphate). They were divided into two groups. Group I was grown solely in osteogenic media. Group II was treated with 100 ng/mL rhBMP2 (R&D Systems) for 2 weeks. Total RNA was extracted following 2 weeks of treatment. The mRNA levels of osteoblast markers, such as col1a1, osteocalcin, and alkaline phosphatase, liver/bone/kidney type (ALPL), were quantitatively analyzed by real-time RT-PCR using ready-made primers. The mRNA levels of transcription factors involved in osteoblastic differentiation, such as Dlx5, Msx2, Runx2, and Osterix, were also quantitatively analyzed by real-time RT-PCR using ready-made primers (Table 3). Real-time PCR and quantification method was identical to that described previously. The expression level of rhBMP2 treated cell was divided by that of untreated cell to calculate the extent of up-regulation or down-regulation of the expression.
Table 3.
Ready-made Primer Set List for Real-time PCR*
*TaqMan® Gene Expression Assay.
RESULTS
Wnt Ligands and Receptors Expression
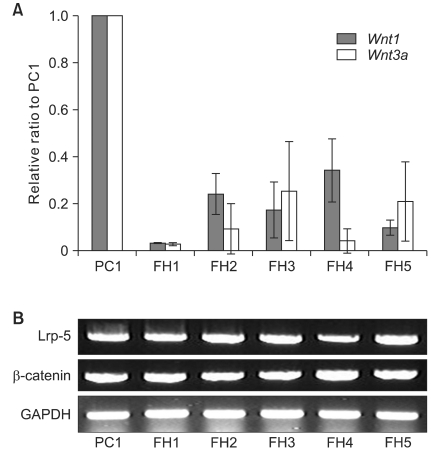
Control tissue (PC1) and fibrous hamartoma tissues from five patients (FH1-5) underwent quantitative real-time RT-PCR for Wnt1 and Wnt3a, and qualitative RT-PCR for Lrp-5 and β-catenin. In the fibrous hamartoma from all 5 CPT patients, the mRNA levels of Wnt ligands were lower than that of control tissue. Expression level of these ligands varied from patient to patient. The mRNA level of Wnt1 in the fibrous hamartoma ranged from 3.3% to 34.2% of that of normal control tissue, while Wnt3a mRNA expression level ranged from 0.14% to 13.5%. All tissues showed strong mRNA expression of both Lrp-5 and β-catenin in RT-PCR (Fig. 1).
Fig. 1.
The mRNA expression of Wnt ligands and their canonical receptors. (A) When the mRNA level of periosteal cell (PC1) was set as 1.0, the relative levels of mRNA expression of FH1-5 were less than 0.4 in both Wnt1 and Wnt3a, which are the most important factors in canonical signal pathway involved in osteoblastic differentiation. (B) PCR showed abundant expression of Lrp-5 and β-catenin mRNA in all tissues tested. PC: periosteal cell, FH: fibrous hamartoma, Lrp: low density lipoprotein receptor-related protein, Wnt: wingless-type MMTV intergration site.
Osteoblast Marker Gene Expression Change by BMP Treatment
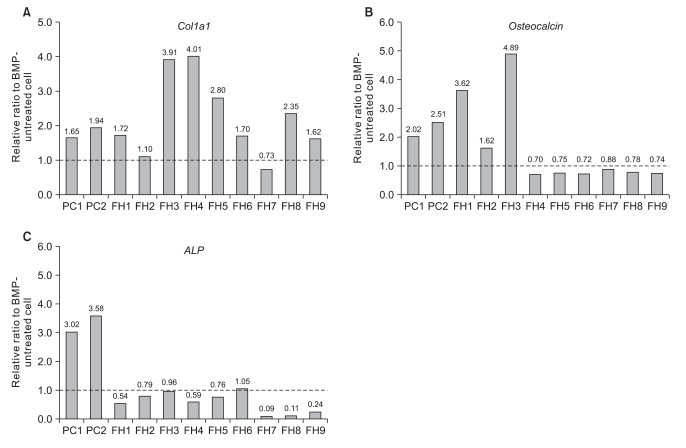
The mRNA level of osteoblast markers, including col1a1, osteocalcin, and ALPL were analyzed in 2 control tissues (PC1, PC2) and 9 fibrous hamartoma tissues (FH1-FH9). The control tissues (periosteal cells) showed up-regulation of mRNA expression of col1a1, osteocalcin and ALPL in response to rhBMP-2 treatment. Col1a1 mRNA level increased by 1.65- or 1.94-fold, osteocalcin by more than 2.0-fold, and ALPL by more than 3.0-fold. However, the response of fibrous hamartoma remained inconsistent. In 7 of 9 FH's, the mRNA expression of col1a1 increased by more than 10% of non-treatment control, while in 2 FH's, it increased by 10% or decreased by 21%, respectively (Fig. 2A). Osteocalcin mRNA expression was up-regulated by rhBMP in fewer FH's than col1a1. Only 2 FH's showed up-regulation of osteocalcin gene expression, and 6 FH's showed down-regulation of osteocalcin gene expression by rhBMP2 treatment (Fig. 2B). ALP gene expression was down-regulated by rhBPM2 treatment in 7 FH's, and that of the remaining 2 FH's remained within 10% of that of untreated control. This was in contrast to more than 3-fold up-regulation in the periosteal cells (PC1 and PC2) (Fig. 2C).
Fig. 2.
Response of mRNA expression of osteoblast markers to rhBMP2 treatment. The mRNA expression level of BMP-treated cell was divided by that of BMP-untreated cell. BMP: bone morphogenetic protein, PC: periosteal cell, FH: fibrous hamartoma, Col1a1: collagen type 1 alpha 1 chain, ALP: alkaline phosphatase, liver/bone/kidney type.
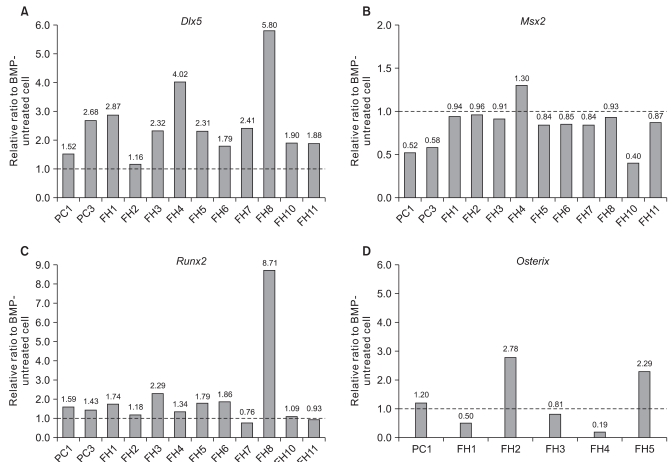
Change in the mRNA Expression of Transcription Factors Involved in Osteoblastogenesis by BMP Treatment (Fig. 3)
Fig. 3.
Response of mRNA expression of transcription factors involved in osteoblastic differentiation. The mRNA expression level of BMP-treated cell was divided by that of BMP-untreated cell. BMP: bone morphogenetic protein, PC: periosteal cell, FH: fibrous hamartoma, Dlx: distal-less homeobox, Runx: runt-related transcription factor, Msx: muscle segment homeobox, drosophila homolog, Osx: osterix.
Transcription factors involved in osteoblastic differentiation, such as Dlx5, Msx2, Runx2, and Osterix, were analyzed in 10 FH's (FH1-FH8, FH10, FH11) and 2 periosteal cells. Periosteal cells showed responses compatible with osteoblastic differentiation. Dlx5, Runx2, and Osterix mRNA expressions were up-regulated with rhBMP2 treatment by 52%, 59, and 20%, respectively, and that of Msx2 was down-regulated by 48%. In FH's, changes in mRNA expression of these transcription factors were not consistent. Dlx5 expression was up-regulated in all FH's tested. The extent of up-regulation was similar to or larger than the two periosteal cells in 9 of 10 FH's tested. FH2 showed mild up-regulation, which was smaller than that of either PC1 or PC3. Msx2 was down-regulated as periosteal cells in 9 of 10 FH's tested. However, only one FH (FH10) showed down-regulation more than the PC's. In 8 of 10 FH's tested, the extent of down-regulation of Msx2 mRNA expression was less than 20% of the untreated cells, while that of PC's were more than 40%. One FH (FH4) even showed up-regulation of Msx2 expression. Runx2 mRNA expression was up-regulated in 8 FH's, and down-regulated in 2 FH's. FH8 showed 8.7-fold up-regulation, which also showed the largest up-regulation of Dlx5 expression. However, FH8 showed down-regulation of osteocalcin and ALPL mRNA expression. Osterix mRNA expression was diverse. Two HF's showed up-regulation larger than PC1, while three showed down-regulation by rhBMP2 treatment.
DISCUSSION
This study was limited by the fact that mRNA expressions of fibrous hamartoma from CPT associated with NF1 were compared to that of periosteal cells from non-NF1 patients. The findings might reflect the difference between Nf1 haploinsufficient cells and wild-type cells, but it does not explain why fibrous hamartoma develops on distal tibia specifically in systemically Nf1 haploinsufficient individual. In order to address this question, fibrous hamartoma cells should be compared with periosteal cells from normal-looking contralateral distal tibia of the same patient. However, it would be unethical to harvest it only for research purpose.
We investigated several aspects of the osteoblastic differentiation process in primary cultured cells from fibrous hamartoma believed to be the main pathologic tissue in CPT associated with NF1. Although the data were not consistent throughout all cases investigated in this study, fibrous hamartoma cells showed deficient expression of the Wnt ligands which are important factors in the regulation of bone mass, postnatal bone formation and fracture healing process.18) When stimulated by rhBMP2 treatment, most fibrous hamartoma cells up-regulated col1a1 expression. However, only a subset up-regulated osteocalcin expression, and all of them down-regulated ALP expression. These findings were consistent with a published report on a smaller number of cases.16)
Osteoblastogenesis is a complex process of cell differentiation from mesenchymal stem cell through osteoprogenitor and preosteoblast to osteoblast. Wnts and BMPs are the most important signal molecules that regulate this process. In this study, we investigated mRNA expression of the Wnt ligands (Wnt1 and Wnt3a), and their receptors of the canonical pathway (Lrp5 and β-catenin). Wnts are a large family of growth factors involved in a variety of biological processes. The canonical pathway of Wnt signal mediated through β-catenin/Lrp5 modulates most aspects of osteoblastic physiology, including proliferation, differentiation, bone matrix formation/mineralization and apoptosis.19) We showed that the mRNA expression of Wnt1 and Wnt3a, which are known to be involved in osteoblastogenesis, were significantly low in fibrous hamartoma cells, although the receptors' mRNA expression was comparable to control tissue. This means that deficiency of local Wnt ligands may contribute to disturbed osteoblastic differentiation in fibrous hamartoma in the pathogenesis of CPT. It appears worthwhile to further investigate Wnt signaling systematically in CPT associated with NF1.
In a previous study, BMP2, BMP4, BMPR1A, BMPR1B, and BMPR2 were reported to express in fibrous hamartoma of CPT.16) BMP signals induce receptor-mediated signaling pathways involving Smads, and subsequently transcriptions factors specific for osteoblastic differentiation. Runx 2 is the master osteogenic transcription factor, expresses in early skeletal development and persists through subsequent stages of bone formation.20,21) Dlx5 is a bone inducing homeodomain transcription factor, expressed in the later stages of osteoblastic differentiation, which is considered to be an upstream regulator of Runx2 and Osterix in the BMP-2 signaling pathway.22) Osterix is a zinc finger-containing transcription factor, specifically expressed in all developing bones, which is considered to act mainly during the terminal differentiation of osteoblasts.21) Msx2 is a mammalian homologue of the Drosophila muscle segment homeobox gene, which appears to play a negative role in osteoblast differentiation, stimulating cell proliferation and suppressing osteogenic differentiation.21) Hence, BMP signaling appears to be transduced via Smads to Dlx5, and then to Runx2 for early differentiation, and to Osterix for terminal differentiation and Msx2 antagonizes these pathways. In our control tissue - the periosteal cells - up-regulated Dlx5, Runx2 and Osterix were seen in response to BMP stimulation, whereas Msx2 was down-regulated. Hence, we attempted to delineate the response of these transcription factor expressions in fibrous hamartoma cells. It is noteworthy that both Dlx5 and Runx2 responded in a way similar to the periosteal cells in most cases of fibrous hamartoma. On the other hand, Msx2 expression was not suppressed by BMP stimulation as much as in the periosteal cells in most cases of fibrous hamartoma. In addition, Osterix expression was down-regulated in 3 of 5 FH's. These findings suggest that impairment of osteoblastic differentiation in fibrous hamartoma is the terminal step of the differentiation cascade and interference by Msx2 may work in this process. Another aspect of these findings that we need to pay attention is that the behavior of FH's varies from patient to patient.
In conclusion, the findings in this study support the hypothesis that fibrous hamartoma originates from aberrant growth of Nf1 haploinsufficient periosteal cells, which failed in terminal osteoblastic differentiation, arrested at a certain stage of this process. This pathomechanism of CPT should be targeted in the development of novel therapeutic biologic intervention.
ACKNOWLEDGEMENTS
This work was supported by the Korea Research Foundation Grant funded by the Korean Government (KRF-2008-313-E00352).
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.Ippolito E, Corsi A, Grill F, Wientroub S, Bianco P. Pathology of bone lesions associated with congenital pseudarthrosis of the leg. J Pediatr Orthop B. 2000;9(1):3–10. doi: 10.1097/01202412-200001000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Hefti F, Bollini G, Dungl P, et al. Congenital pseudarthrosis of the tibia: history, etiology, classification, and epidemiologic data. J Pediatr Orthop B. 2000;9(1):11–15. doi: 10.1097/01202412-200001000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Crawford AH, Schorry EK. Neurofibromatosis update. J Pediatr Orthop. 2006;26(3):413–423. doi: 10.1097/01.bpo.0000217719.10728.39. [DOI] [PubMed] [Google Scholar]
- 4.Friedman JM. Epidemiology of neurofibromatosis type 1. Am J Med Genet. 1999;89(1):1–6. [PubMed] [Google Scholar]
- 5.Shen MH, Harper PS, Upadhyaya M. Molecular genetics of neurofibromatosis type 1 (NF1) J Med Genet. 1996;33(1):2–17. doi: 10.1136/jmg.33.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutmann DH, Wood DL, Collins FS. Identification of the neurofibromatosis type 1 gene product. Proc Natl Acad Sci U S A. 1991;88(21):9658–9662. doi: 10.1073/pnas.88.21.9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuorilehto T, Nissinen M, Koivunen J, Benson MD, Peltonen J. NF1 tumor suppressor protein and mRNA in skeletal tissues of developing and adult normal mouse and NF1-deficient embryos. J Bone Miner Res. 2004;19(6):983–989. doi: 10.1359/JBMR.040130. [DOI] [PubMed] [Google Scholar]
- 8.Leskela HV, Kuorilehto T, Risteli J, et al. Congenital pseudarthrosis of neurofibromatosis type 1: impaired osteoblast differentiation and function and altered NF1 gene expression. Bone. 2009;44(2):243–250. doi: 10.1016/j.bone.2008.10.050. [DOI] [PubMed] [Google Scholar]
- 9.Bollag G, Clapp DW, Shih S, et al. Loss of NF1 results in activation of the Ras signaling pathway and leads to aberrant growth in haematopoietic cells. Nat Genet. 1996;12(2):144–148. doi: 10.1038/ng0296-144. [DOI] [PubMed] [Google Scholar]
- 10.Cichowski K, Jacks T. NF1 tumor suppressor gene function: narrowing the GAP. Cell. 2001;104(4):593–604. doi: 10.1016/s0092-8674(01)00245-8. [DOI] [PubMed] [Google Scholar]
- 11.Schindeler A, Little DG. Recent insights into bone development, homeostasis, and repair in type 1 neurofibromatosis (NF1) Bone. 2008;42(4):616–622. doi: 10.1016/j.bone.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Jacks T, Shih TS, Schmitt EM, Bronson RT, Bernards A, Weinberg RA. Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nat Genet. 1994;7(3):353–361. doi: 10.1038/ng0794-353. [DOI] [PubMed] [Google Scholar]
- 13.Cichowski K, Shih TS, Schmitt E, et al. Mouse models of tumor development in neurofibromatosis type 1. Science. 1999;286(5447):2172–2176. doi: 10.1126/science.286.5447.2172. [DOI] [PubMed] [Google Scholar]
- 14.Upadhyaya M, Han S, Consoli C, et al. Characterization of the somatic mutational spectrum of the neurofibromatosis type 1 (NF1) gene in neurofibromatosis patients with benign and malignant tumors. Hum Mutat. 2004;23(2):134–146. doi: 10.1002/humu.10305. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Bollag G, Clark R, et al. Somatic mutations in the neurofibromatosis 1 gene in human tumors. Cell. 1992;69(2):275–281. doi: 10.1016/0092-8674(92)90408-5. [DOI] [PubMed] [Google Scholar]
- 16.Cho TJ, Seo JB, Lee HR, Yoo WJ, Chung CY, Choi IH. Biologic characteristics of fibrous hamartoma from congenital pseudarthrosis of the tibia associated with neurofibromatosis type 1. J Bone Joint Surg Am. 2008;90(12):2735–2744. doi: 10.2106/JBJS.H.00014. [DOI] [PubMed] [Google Scholar]
- 17.Mariaud-Schmidt RP, Rosales-Quintana S, Bitar E, et al. Hamartoma involving the pseudarthrosis site in patients with neurofibromatosis type 1. Pediatr Dev Pathol. 2005;8(2):190–196. doi: 10.1007/s10024-004-1004-1. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, Alman BA. Wnt pathway, an essential role in bone regeneration. J Cell Biochem. 2009;106(3):353–362. doi: 10.1002/jcb.22020. [DOI] [PubMed] [Google Scholar]
- 19.Bodine PV, Komm BS. Wnt signaling and osteoblastogenesis. Rev Endocr Metab Disord. 2006;7(1-2):33–39. doi: 10.1007/s11154-006-9002-4. [DOI] [PubMed] [Google Scholar]
- 20.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89(5):747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 21.Ryoo HM, Lee MH, Kim YJ. Critical molecular switches involved in BMP-2-induced osteogenic differentiation of mesenchymal cells. Gene. 2006;366(1):51–57. doi: 10.1016/j.gene.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 22.Ryoo HM, Hoffmann HM, Beumer T, et al. Stage-specific expression of Dlx-5 during osteoblast differentiation: involvement in regulation of osteocalcin gene expression. Mol Endocrinol. 1997;11(11):1681–1694. doi: 10.1210/mend.11.11.0011. [DOI] [PubMed] [Google Scholar]