Abstract
Murine embryonic stem cells have been shown to exist in two functionally distinct pluripotent states, embryonic stem cells (ES cell)- and epiblast stem cells (EpiSCs), which are defined by the culture growth factor conditions. Human ES cells appear to exist in an epiblast-like state, which in comparison to their murine counterparts, is relatively difficult to propagate and manipulate. As a result, gene targeting is difficult and to-date only a handful of human knock-in or knock-out cell lines exist. We explored whether an alternative stem cell state exists for human stem cells as well, and demonstrate that manipulation of the growth factor milieu allows the derivation of a novel human stem cell type that displays morphological, molecular and functional properties of murine ES cells and facilitates gene targeting. As such, the murine ES-like state provides a powerful tool for the generation of recombinant human pluripotent stem cell lines.
INTRODUCTION
Embryonic stem cells (ES cells) were first derived in 1981 from the inner cell mass (ICM) of murine preimplantation blastocyst embryos (Evans and Kaufman, 1981; Martin, 1981). ES cells are pluripotent, meaning they are able to expand indefinitely in vitro while retaining the capacity to generate derivatives of all three germ layers both in vitro and in vivo. The discovery of murine ES (mES) cells was a major breakthrough in developmental biology, since it enabled the study of mammalian gene function in vivo, using transgenic and knockout technologies. The subsequent derivation of human ES (hES) cells raised the expectation that these cells would similarly revolutionize our insights into human development and disease. Unfortunately, human pluripotent stem cells are remarkably resilient to non-viral genetic manipulation and to date only a handful of human knock-in or knock-out cell lines exist. As a result, the application of human pluripotent stem cells has been more limited than previously anticipated.
While both human and murine ES cells are derived from blastocyst-stage embryos, they demonstrate profound differences (Thomson et al., 1998). Murine ES cells grow in three-dimensional, tightly packed colonies with a population doubling time of approximately 16 hours and their maintenance is dependent on LIF and BMP4 growth factor signaling (Smith et al., 1988; Xu et al., 2005; Ying et al., 2003). In contrast, human ES cells form flattened two-dimensional colonies and are maintained in a bFGF and Activin A/TGFbeta signaling dependent manner (Thomson et al., 1998). HES cells proliferate slowly, with a population doubling time averaging 36 hours. Epigenetically, human and murine ES cells display a different X-chromosome inactivation pattern and promoter occupancy by pluripotency transcription factors (Boyer et al., 2005; Silva et al., 2008; Tesar et al., 2007). In addition, hES cells are passaged as small clumps of cells, and most hES cell lines cannot be passaged as single cells by trypsin digest. The inability of hES cell lines to grow from single cells greatly impedes genetic modification of these cells, since the introduction of transgenes is typically followed by clonal selection.
Two reports on the derivation of murine epiblast stem cells (EpiSCs) recently provided a new perspective on the nature of human ES cells (Brons et al., 2007; Tesar et al., 2007). EpiSCs are derived from post-implantation murine epiblast embryos under culture conditions similar to hES cell culture conditions. EpiSCs display many of the characteristics of human ES cells including their dependence on bFGF/Activin A signaling, their flattened colony morphology, their slower proliferation rate compared to murine ES cells, their X-inactivation status and their requirement to be passaged as small clumps of cells (Brons et al., 2007; Tesar et al., 2007).
The culture dynamics and the specific characteristics of murine ES cells and EpiSCs appear to be largely determined by the growth factor conditions under which these cell types are derived and maintained. Indeed, recent work from our group demonstrates that culture growth factor conditions play a critical role in defining the pluripotent stem cell state (Chou et al., 2008). Intriguingly, while pluripotent stem cells can be stably derived and propagated from multiple species in an epiblast-like state, including the rat and ‘non-permissive’ mouse strains, the LIF-dependent pluripotent state appears to be unstable in these species. (Buehr et al., 2008; Hanna et al., 2009; Li et al., 2009; Liao et al., 2009). However the LIF-dependent pluripotent state can be stabilized through the constitutive ectopic expression of one or more of the reprogramming factors (Oct4, Sox2, Klf4, cMyc), which induce the generation of induced pluripotent stem cells (iPS cells) from somatic cells (Takahashi et al., 2007; Takahashi and Yamanaka, 2006). In the non-permissive NOD mouse strain for example, the constitutive ectopic expression of either Klf4 or cMyc is sufficient to allow the derivation of ES-like cells from blastocyst embryos (Hanna et al., 2009). Small molecule inhibitors of glycogen synthase kinase 3 beta (GSK3β) and the mitogen-activated protein kinase (MAPK) signaling pathway can replace some of the reprogramming factors during iPS cell generation (Li and Ding, 2009). These inhibitors can similarly stabilize the LIF-dependent mES-like pluripotent stem cell state from both the non-permissive NOD mouse strain and the rat (Buehr et al., 2008; Hanna et al., 2009; Li et al., 2009; Liao et al., 2009). Thus, it appears that the LIF-dependent pluripotent state is metastable in these species, meaning it is dependent on either the constitutive expression of ectopic reprogramming factors or the continued inhibition of GSK3β and/or the MAPK signaling pathways.
While distinct pluripotent states are known to exist in mouse and rat, they have thus far not been described for human stem cells. A recent report demonstrates that stable human iPS cells (hiPS) can be derived in the presence of LIF and inhibitors of GSK3β and the TGFβ and MEK/ERK signaling pathways (Li et al., 2009). However, these cells appear to be molecularly identical to conventional hiPS cells. We used hiPS cell derivation as a tool to investigate the influence of growth factor signaling on human stem cell pluripotent state. Here we demonstrate the derivation of human cell lines that display many characteristics of murine ES cells including a dome-shaped colony morphology, the ability to be propagated by trypsin digest and to clonally grow from single cells, and the activation of LIF downstream signaling pathways. We demonstrate that in this state, the human cells are more amenable to the introduction of transgenes and allow homologous recombination-mediated gene targeting. The LIF-state is metastable, since it depends on the constitutive expression of ectopic reprogramming factors. Yet a combination of growth factors and inhibition of MEK-kinase signaling allows the conversion of the human LIF-iPS cells to a stable, pluripotent human iPS cell state.
Our findings support the idea that, analogous to mouse strains and the rat, human iPS cells adopt murine-ES cell properties, when the cells are derived in the presence of LIF and ectopic reprogramming factors. Importantly, this novel state facilitates homologous recombination-mediated gene targeting in human stem cells. As such the intermediate iPS state described here can be a useful tool in research and future cell therapies.
RESULTS
Derivation of metastable human iPS cell lines with murine ES cell characteristics
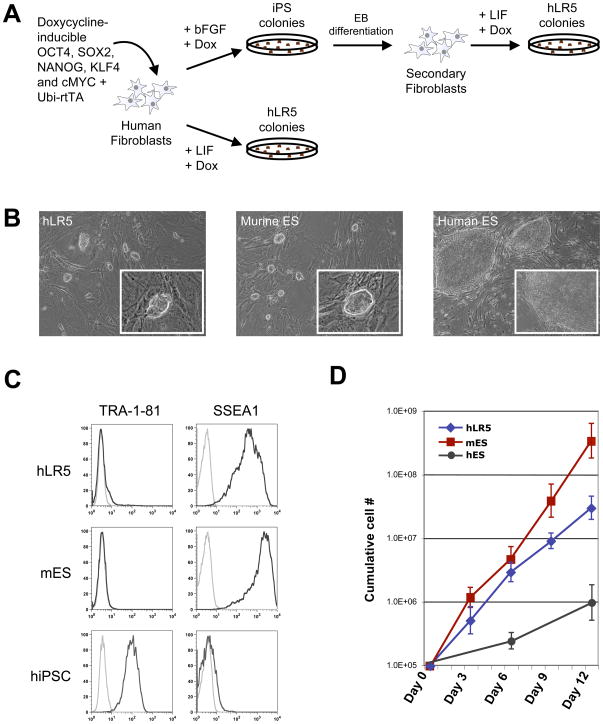
We used the recently reported induced pluripotent stem cell (iPS cell) strategy to explore the possibility of deriving human iPS cells in the presence of LIF. The five reprogramming factors OCT4, SOX2, NANOG, c-MYC and KLF4 were expressed in human fibroblast using a recently reported doxycyline inducible lentiviral system (Figure 1A) (Maherali et al., 2008). Fibroblasts were reprogrammed either directly from the primary fibroblasts or from so-called ‘secondary fibroblasts’, derived from differentiated ‘primary’ hiPS cells (Maherali et al., 2008). Using either approach, reprogramming of human fibroblasts in the presence of LIF, resulted in the formation of two types of colonies, transient, irregularly shaped colonies that deteriorated a few days after their first appearance (Supplemental figure 1A) and smaller, tightly packed colonies (Supplemental figure 1B). We picked individual colonies of the latter for further clonal analysis. These clones displayed the hallmark, tightly packed, bright, dome-shaped morphology of mES cells (Figure 1B), contrasting the flattened two-dimensional colony morphology of hES cells (Figure 1B). We designated these cells human LR5-iPS cells (hLR5) (human LIF + the constitutive expression of 5 reprogramming factors).
Figure 1. A metastable human iPS state with murine ES cell properties.
(A) Schematic representation of the used strategy.. Doxycycline-inducible lentiviral vectors were added either as individual vectors (Maherali et al., 2008) or the polycystronic human STEMCCA virus (Sommer et al., 2009) and inducible NANOG as indicated in the text
(B) Colony morphology of hLR5 cells (left panel), murine ES cells (middle panel) and human ES cells.
(C) FACS analysis of cell surface marker expression on hLR5 cells, murine ES cells and human iPS cells. Black lines: Cell surface marker using the indicated primary antibody. Grey line: no primary antibody control.
(D) Growth curve of hLR5 cells, mES and hES over a period of 12 days. Cumulative cell number is plotted against days (n=3, SD).
Next we investigated the cell surface marker profile of the hLR5 cells. Murine and human pluripotent stem cells express a mutually exclusive complement of cell surface markers. SSEA1 is expressed on undifferentiated murine pluripotent stem cells while human pluripotent stem cells express the SSEA3, SSEA4, TRA-1-81 and TRA-1-60 cell surface markers. Flow cytometry analysis of the hLR5 cells revealed a marker profile that resembles the surface marker profile of mES cells (Figure 1C). hLR5 cells do not express the TRA-1-81 cell surface marker (Figure 1C) but demonstrated high expression of the SSEA1 cell surface marker, which is typically expressed on murine pluripotent stem cells (Figure 1C). A fourth clone, which was derived through direct reprogramming of human fibroblasts from a different genetic background also lacked TRA-1-81 expression but expressed low SSEA1 levels, suggesting that the level of SSEA1 expression is heterogeneous between hLR5 clones of different genetic backgrounds. In addition to the general lack of TRA-1-81 expression, hLR5 cells also do not express SSEA3, SSEA4 and TRA-1-60, as tested by flow cytometry and immunohistochemistry (not shown). Unlike hES cells the hLR5 cells can be propagated by trypsin digest. This result suggested that similar to mES cells, hLR5 cells are tolerant to passaging as single cells. Indeed, upon single cell sorting of hLR5 cells into 96 well plates, hLR5 clones re-emerged in approximately 22% of the wells (n=10), similar to the efficiency of single-cell sorted mES cells (~30%) whereas upon single-cell sorting of the trypsin-adapted HUES3 hES cell line (Cowan et al., 2004), no colonies re-emerged (n=10). In addition, the hLR5 cells displayed a much higher proliferation rate than human ES- or iPS cells, with a cell doubling time of approximately 22 hours (Figure 1D). The hLR5 proliferation rate is close to the mES- or -miPS cell proliferation rate (doubling time ~16 hours) and much higher than the proliferation rate of hES- or hiPS cells (doubling time ~36 hours).
Activation of the JAK-STAT pathway and downstream target genes in hLR5 cells
The growth factor environment is known to be an important determinant of the stem cell pluripotent state (Brons et al., 2007; Chou et al., 2008; Tesar et al., 2007). In mES cells, LIF activates the JAK/STAT3 and the RAS/MEK/MAPK signaling pathways, which have opposing roles in mES cell maintenance and differentiation. Activation of the JAK-Stat3 signaling pathway has been shown to be important for long-term self-renewal of mES cells, whereas the RAS/MAPK pathway drives mES cell differentiation. Indeed, pharmacological inhibitors of the RAS/MEK/MAPK pathway have been shown to enhance mES cell self-renewal and in combination with inhibitors of GSK3β allow growth factor independent maintenance of pluripotent stem cells (Ying et al., 2008). Since hLR5 cells display many characteristics of murine ES cells, we investigated the effect of LIF and its signaling pathways on these cells.
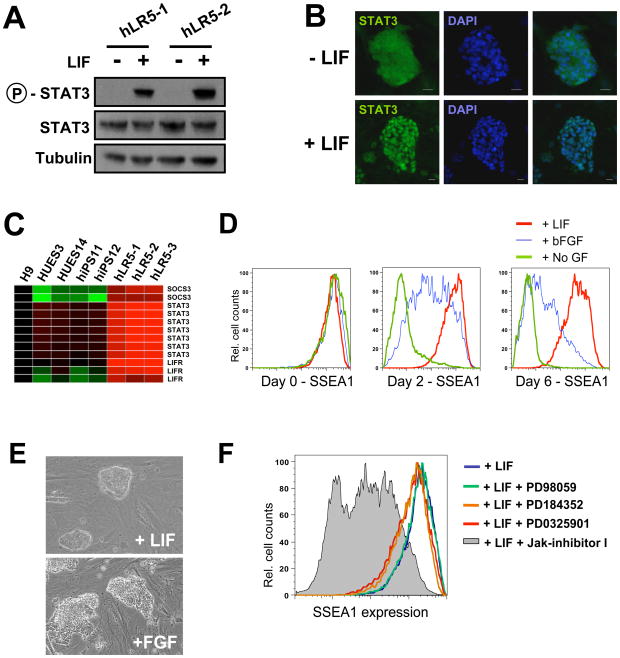
As shown in Figure 2A, STAT3 phosphorylation is robustly stimulated in hLR5 cells in a LIF-dependent manner. Upon LIF activation of the JAK-STAT signaling cascade, STAT3 translocates from the cytosol to the nucleus and directly activates downstream target genes. Immunofluorescence staining of STAT3 in hLR5 cells revealed nuclear translocation in response to LIF stimulation (Figure 2B). This translocation results in activation of STAT3 downstream target genes, including STAT3 itself as well as SOCS3 and the LIF-receptor, indicating that this pathway is functionally active in response to LIF stimulation of the hLR5 cells (Figure 2C).
Figure 2. LIF-responsiveness of hLR5 cells.
(A) Western blot analysis of STAT3 phosphorylation in two independent hLR5 clones, with or without LIF-stimulation as indicated
(B) Immunostaining of STAT3 subcellular localization in hLR5 cells before (top panel) or after (bottom panel) LIF stimulation. Note the translocation of STAT3 from the cytoplasm (top panel) to the nucleus (bottom panel). Cell nuclei were visualized with DAPI.
(C) Gene expression analysis of downstream target genes of the JAK/STAT signaling pathway in three hLR5 clones as well as human ES cell lines and conventional human iPS cell lines.
(D) Flow cytometry analysis of SSEA1 surface marker on hLR5 cells upon LIF removal. Red lines: LIF control, Blue line: LIF substitution with bFGF, Green lines: no added growth factor.
(E) Colony morphology of the hLR5 cells before (top panel) and after LIF substitution with bFGF.
(F) Flow cytometry analysis of SSEA1 cell surface marker expression on hLR5 cells in the presence of small molecule inhibitors. hLR5 cells were maintained in hLR5 media for 1 week in the presence of inhibitors. Grey shaded area: Jak-inhibitor, colored lines: MEK inhibitors (PD9805, PD184352 and PD0325901, as indicated) Blue line: no inhibitor control
Upon removal or substitution of LIF from the hLR5 culture media, SSEA1 expression waned (Figure 2D). In addition, we noticed a change in colony morphology (Figure 2E). Next we used specific inhibitors of JAK/STAT signaling or the MAPK/MEK signaling pathway to examine the roles of these pathways in hLR5 cells. SSEA1 cell surface marker expression was used as a readout. As shown in Figure 2C, inhibition of the JAK/STAT3 pathway resulted in a marked decrease of SSEA1 on hLR5 cells (Jak-inhibitor I (0.6 μM), gray shaded area), whereas specific inhibition of the MEK/ERK1/ERK2 pathway did not affect cell surface marker expression (PD98059 (50 μM), PD184352 (0.8 μM) and PD0325901 (1 μM), colored lines). Together these results indicate that LIF stimulation of hLR5 cells results in activation of the JAK-STAT3 signaling cascade and upregulation of downstream target genes while LIF withdrawal results in changes in hLR5 colony phenotype. However, LIF withdrawal does not result in hLR5 differentiation, perhaps due to the persistent doxycyclin-induced ectopic expression of reprogramming factors.
The hLR5 state requires continued ectopic expression of five reprogramming factors
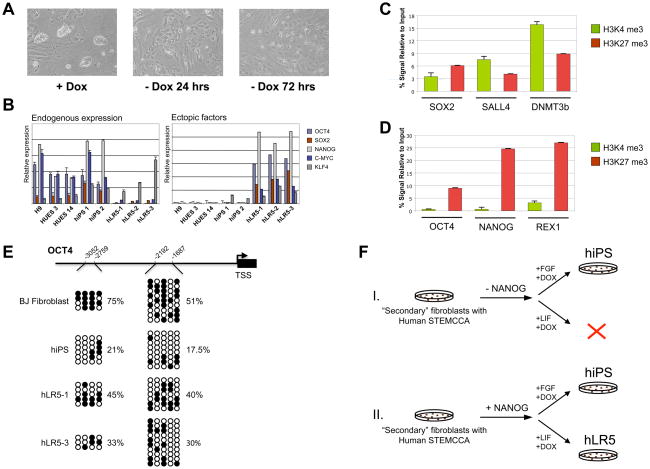
Next we examined whether hLR5 cells could be stably propagated in the absence of ectopic reprogramming factors. As shown in figure 3A, doxycycline withdrawal resulted in the rapid loss of hLR5 colony morphology, with all cells adopting a fibroblast-like appearance within 3 days. A similar dependence was found for rat iPS cells (Liao et al., 2009) and for iPS cells from the non-permissive NOD mouse strain (Hanna et al., 2009). The need for ectopic reprogramming factors suggested that in hLR5 cells, the endogenous pluripotency genes have not yet been fully activated. Q-PCR analysis of the expression of endogenous and ectopic pluripotency factors revealed that hLR5 cells fail to re-activate endogenous OCT4 and NANOG expression, while the expression of endogenous SOX2 and c-MYC are low (Figure 3B). Endogenous KLF4 was expressed at similar levels in hLR5 cells and human ES- or iPS cells.
Figure 3. The hLR5 state depends on ectopic pluripotency factors but is poised for re-activation of endogenous pluripotency genes.
(A) Ectopic factor dependence of hLR5 cells. Upon doxycycline withdrawal, hLR5 colony morphology is lost and cells adopt a fibroblast-like appearance. Days of differentiation are indicated
(B) Quantitative RT-PCR analysis of the expression of reprogramming factors used for the derivation and maintenance of hLR5 cells. Left panel: expression of endogenous genes. Right panel: expression of the Doxycyclin-inducible ectopic reprogramming factors. Human ES strains (H9, HUES3, HUES14) and human iPS strains (hiPS1, hiPS2) were used as controls. Color coding of the genes is indicated (n-3, SD).
(C) ChIP-qPCR analysis of the presence of Histone 3 lysine 4 (H3K4, green bars) marks and Histone 3 Lysine 27 (H3K27, red bars) marks at the promoter regions of the pluripotency genes SOX2, DNMT3b and SALL4 as indicated in hLR5 cells (n=3, SD).
(D) ChIP-qPCR analysis of the presence of Histone 3 lysine 4 (H3K4, green bars) marks and Histone 3 Lysine27 (H3K27, red bars) marks at the promoter regions of the pluripotency genes OCT4, NANOG and REX1 as indicated in hLR5 cells(n=3, SD).
(E) DNA methylation analysis of two CpG islands in the OCT4 promoter as indicated in the schematic of the OCT4 promoter region. Open circles indicate unmethylated and filled circles indicate methylated CpG dinucleotides. Shown are representative sequenced clones from BJ fibroblasts, human iPS cells and two independent clonal hLR5 cell lines. The percentage of CpG methylation at each CpG island in the respective cell lines is indicated. TSS: Transcription start site
(F) Schematic representation of the generation of hLR5 cells in the absence (Top panel, I.) or presence (Bottom panel, II.) of NANOG. While in the absence of NANOG expression traditional hiPS cell can be derived, no hLR5-like colonies form. Addition of ectopic NANOG results in the formation of hLR5 colonies.
We then analyzed the presence of activating and silencing histone marks at the promoter regions of critical regulators of pluripotency. Using chromatin-immunoprecipitation (ChIP) and Q-PCR, we tested the presence of two histone marks; Histone 3 Lysine 4-trimethylation (H3K4me3), a histone mark that activates transcription (Pray-Grant et al., 2005; Santos-Rosa et al., 2003; Sims et al., 2005; Wysocka et al., 2005) and Histone 3-Lysine 27-trimethylation (H3K27me3) which promotes stable transcriptional repression (Francis et al., 2004; Ringrose et al., 2004). Unexpectedly, ChIP-qPCR analysis revealed the presence of both marks at the promoter regions of SOX2, DNMT3b and SALL4 (Figure 3C). The H3K4 and H3K27 methylation marks are simultaneously present in so-called ‘bivalent domains’ which in ES cells are often found at promoters of important transcriptional regulators of development (Bernstein et al., 2006). Bivalent domains result in transcriptional repression, but leads to a ‘poised’ state which allows rapid activation or permanent repression by removing either one of the opposing histone marks and is consistent with the low-level endogenous expression of these genes in hLR5 cells (Figure 3B).
ChIP-qPCR analysis of the H3K4me3 and H3K27me3 marks at the OCT4, NANOG and REX1 promoters revealed the presence of the repressive H3K27me3 mark in the hLR5 cells, corroborating the absence of endogenous expression of these genes (Figure 3D). In somatic cells, OCT4 and NANOG are silenced through additional epigenetic mechanisms including DNA methylation, which is thought to be a permanent transcriptional barrier. Indeed, DNA methylation has been shown to be a limiting step during iPS cell reprogramming, and inhibition of DNA methylation can complete the reprogramming of partially reprogrammed iPS cells (Mikkelsen et al., 2008). Bisulfite sequencing showed that unexpectedly, the OCT4 promoter regions are hypomethylated in the hLR5 cells as compared to the parental BJ fibroblasts (Figure 3E).
Together, these results suggest that hLR5 cells exist in a ‘poised’ state of near-pluripotency, in which some pluripotency genes, including SOX2, DNMT3b and SALL4 are in a bivalent histone methylation state, whereas others, such as OCT4, NANOG and REX1 still carry the transcriptionally repressive H3K27 methylation mark, but already display hypomethylation at the OCT4 promoter region.
Surprisingly, in addition to the four common reprogramming factors, ectopic expression of Nanog is also required for the maintenance of the hLR5 state. Using the ‘secondary fibroblasts’, we analyzed hLR5 derivation in the presence or absence of ectopic Nanog expression (Figure 3F). iPS reprogramming was induced with doxycycline using either conventional hES medium (with bFGF) or in hLR5 conditions (LIF). While in the presence of bFGF hiPS colonies formed with or without ectopic NANOG (Figure 3F, I.), under hLR5 culture conditions colony formation is dependent on ectopic NANOG expression (Figure 3F, II.), demonstrating that NANOG is required for the de-novo derivation of hLR5 cells. In addition, we explored the effect of ectopic NANOG expression when reactivating reprogramming in existing hiPS cells in hLR5 medium. Using hiPS cells derived with the STEMCCA lentivirus, we induced ectopic factor expression in the presence of human LIF with or without ectopic NANOG expression (Supplemental Figure 2A). In the presence of Nanog expression, colonies appeared after 2–3 passages that showed the typical hLR5 morphology (Supplemental Figure 2B, left panel), while without ectopic NANOG expression hiPS colony morphology rapidly deteriorated in hLR5 conditions (Supplemental Figure 2B, right panel). However, hLR5 cell cultures directly derived from hiPS cells remained heterogeneous, indicating that direct conversion of hiPS cells into hLR5 cells was incomplete and may require prolonged passaging and/or selection. Indeed, in similar manner, the conversion of murine EpiSCs into mES-like cells requires prolonged culture and passaging in combination with selection for mES-like cells (Bao et al., 2009)
Conversion of hLR5 cells to a stable pluripotent state
Previous reports have demonstrated that, similar to our hLR5 cells, rat iPS and miPS cells from the NOD background are unstable. However, NOD-derived iPS cells can be converted to a stable, epiblast-like pluripotent state by simultaneously removing the ectopic reprogramming factors and altering the culture growth factor conditions (Hanna et al., 2009). We examined whether changes in the growth factor environment could similarly induce the conversion of hLR5 cells into a stable pluripotent state.
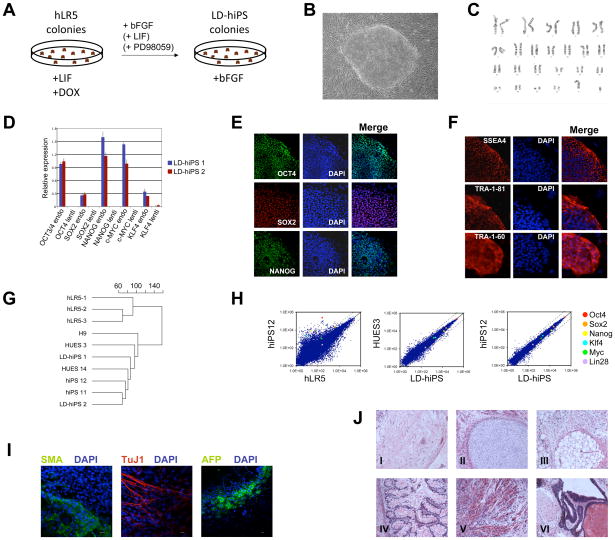
Figure 4A outlines the procedure of converting hLR5 cells into stable iPS cells. Clonal hLR5 cell lines were plated at a density of 5000 cells per cm2 in media containing human LIF and doxycycline. The next day, doxycycline was withdrawn from the hLR5 cultures and cells were further maintained in the presence of bFGF alone (Figure 4A). Withdrawal of ectopic reprogramming factors resulted in the rapid differentiation of most of the hLR5 colonies (Figure 1E). However, after 7–10 days stable colonies emerged that required mechanical passaging and displayed a typical hiPS-like colony morphology (Figure 4B). We termed these cells hLR5-derived human iPS cells (LD-hiPS). The conversion frequency of hLR5 cells into LD-hiPS cells was approximately 0.01%, similar to the conversion of murine metastable iPS cells into stable EpiSC-like iPS cells (Hanna et al., 2009). Pharmacological inhibitors of GSK3β and/or MAPK signaling were shown to stabilize the LIF-dependent pluripotent state in iPS and ICM-derived stem cell lines from NOD mice and rat. We analyzed whether LIF and/or small molecule inhibitors could positively influence the conversion of hLR5 cells (Li and Ding, 2009). Addition of LIF or the MEK inhibitor PD98059 (50 μM) alone resulted in a slight increase in hLR5 conversion rate (Supplemental figure 3), but in combination resulted in a near 8-fold increase in conversion frequency compared to FGF alone. Emerging converted colonies displayed the typical hiPS cell morphology and were subsequently maintained with bFGF alone, indicating that the resulting iPS cells are not LIF-dependent. Characterization of three independent LD-hiPS cell lines revealed a normal (2n=46, XY) karyotype of these cells (Figure 4C). ChIP analysis of the H3K4 and H3K27 histone methylation marks at the promoter regions of key pluripotency mediators suggested reactivation of pluripotency genes in LD-hiPS cells (Supplemental Figure 4). Indeed, Q-PCR analysis of pluripotency regulators demonstrated reactivation of endogenous OCT4, SOX2, NANOG, KLF4 and cMYC in LD-hiPS cells, and the absence of ectopic factors (Figure 4D). The expression and nuclear localization of OCT4, SOX2 and NANOG was further confirmed using immunofluorescence staining of LD-hiPS cells (Figure 4E). LD-hiPS cells displayed a cell surface marker profile characteristic of human pluripotent stem cells, with expression of TRA-1-60, TRA-1-81 and SSEA4 cell surface markers (Figure 4F), while the SSEA1 cell surface marker was absent (not shown). In addition, E-Cadherin, a cell-cell interaction protein that is important for pluripotent stem cell maintenance and differentiation, is induced upon conversion of hLR5 cells into LD-hiPS cells (Supplemental Figure 5). Hierarchical cluster analysis of the global gene expression profiles of hLR5 cells, LD-hiPS cells, human ES cells and human iPS cells revealed that LD-hiPS are highly similar to human ES- and iPS cells, whereas hLR5 cells form a separate cluster of unrelated cells (Figure 4G). Scatter plots of the microarray expression analysis of hLR5 cells and LD-hiPS further highlight the differences between hLR5 cells and the genetically identical hiPS#12 cell line. In contrast, there is high similarity between LD-hiPS and the genetically identical hiPS#12 cell line, but also between LD-hiPS and the genetically unrelated HUES3 human ES cell line (Figure 4H) (Maherali et al., 2008). Finally we examined the ability of LD-hiPS to generate derivatives of the three embryonic germ layers. Embryoid bodies (EBs) were generated from LD-hiPS cells and plated onto gelatin-coated coverslips. Immunoflurescence staining revealed the presence of cells expressing TuJ1, a neural marker, Smooth Muscle Actin, a mesoderm lineage marker and alpha-fetoprotein, a marker of endoderm differentiation (Figure 4I). In addition, we observed that some of the EBs started beating, indicating the development of cardiac tissue with pacemaker function (Supplemental Movie 1). Finally, subcutaneous injection of hLR5 cells into immunocompromised mice resulted in the formation of teratomas containing differentiated derivatives of the three embryonic germ layers (Figure 4J) demonstrating that the LD-hiPS cells are indeed pluripotent.
Figure 4. Conversion of hLR5 cells to a stable pluripotent state.
(A) Schematic representation of the conversion of hLR5 cells
(B) Representative image of the LD-hIPS colony morphology
(C) LD-hiPS cells have a normal 46 XY karyotype.
(D) Quantitative RT-PCR analysis of the expression of endogenous pluripotency factors and the silencing of the doxycycline-inducible ectopic reprogramming factors in two independent LD-hiPS clones (n=3, SD).
(E) Immunofluorescence analysis of OCT4 (top panels), SOX2 (middle panels) and NANOG (bottom panels) protein expression and nuclear localization in LD-hiPS cells. DAPI was used to visualize the cell nuclei.
(F) Immunofluorescence staining of characteristic cell surface markers of human pluripotent stem cells: SSEA4 (top panels), TRA-1-81 (middle panels) and TRA-1-60 (bottom panels). DAPI was used to visualize the cell nuclei.
(G) Unbiased cluster analysis of global gene expression profiles of three independent hLR5 clones, two independent LD-hiPS clones, three human ES cell lines and two human iPS cell lines.
(H) Scatter plots of microarray data on the global gene expression patterns of hLR5 cells, human iPS cells of the same genetic background (hiPS12, (Maherali et al., 2008)), Human ES cells (HUES3) and LD-hiPS cells as indicated. The position of individual pluripotency genes listed in the legend is indicated with colored circles.
(I) Immunostaining of differentiated LD-hiPS cell lines with markers for mesoderm (SMA, left panel), ectoderm (Tuj1, middle panel) and endoderm (AFP, right panel) as indicated. DAPI was used to visualize cell nuclei.
(J) H&E staining of teratomas generated from clonal LD-hiPS cells. Derivatives of all three germ layers are observed: I. Ganglion, II. Cartilage, III. Adipose tissue, IV. Gut, V. Muscle and VI. Respiratory epithelium and squamous epithelium
hLR5 cells facilitate transgenesis and gene targeting in human stem cells
A major obstacle for the application of human pluripotent stem cells in modeling human development and disease is the difficulty these cells have displayed in allowing the introduction of foreign genetic elements (Ptaszek and Cowan, 2007). While such basic molecular manipulations are mainstay in mES cells, generation of transgenic human stem cells is very inefficient and labor intensive.
Since the human hLR5 cells display many characteristics of murine ES cells, we examined whether these cells are more amenable for transgene insertion using standard electroporation procedures. We tested the transfection efficiency of hLR5 cells using either a 10 kb vector constitutively expressing a red fluorescent protein (tdTomato) and a puromycin selection casette or a 20 kb vector expressing tdTomato driven by the ISL1 promoter (Bu et al., 2009) and a hygromycin selection cassette. hLR5 cells, or control human ES cells, were electroporated with linearized constructs and after antibiotic-selection, the number of colonies was counted. Table 1 summarizes the result of 6 independent electroporations in two independent clonal hLR5 lines (hLR5-1 and hLR5-3) and 29 independent electroporations of hES cell lines (H9 and HUES3). Electroporation and selection of the same number of hLR5 cells with the same amount of vector yields over 200-fold more colonies that had incorporated the transgene compared to human ES cell electroporation. The high efficiency in which hLR5 cells incorporate transgenes is particularly important when large constructs such as BAC clones are used. Indeed, the same electroporation protocol allowed the introduction of a 250kb BAC clone with a puromycin selection cassette, albeit at a slightly lower efficiency approximately 1 colony per 106 electroporated cells.
Table 1.
Introduction of transgenes into human ES cells and hLR5 cells by electroporation
| Experiment | Cell type | # Cells per electroporation | # electroporations | Size of the construct | Drug selection | Average # colonies per 10 × 106 cells |
|---|---|---|---|---|---|---|
| #1 | Human ES | 1 × 107 | 10 | 20 kbase | Hygromycin | 6.8 |
| #2 | Human ES | 1 × 107 | 10 | 20 kbase | Hygromycin | 10.6 |
| #3 | Human ES | 1 × 107 | 6 | 20 kbase | Hygromycin | 12 |
| #4 | Human ES | 5 × 106 | 3 | 14 kbase | Puromycin | 5.7 |
| #5 | hLR5 | 1 × 107 | 2 | 20 kbase | Hygromycin | ≫1500 |
| #6 | hLR5 | 1 × 107 | 2 | 20 kbase | Hygromycin | ≫1500 |
| #7 | hLR5 | 1 × 107 | 1 | 14 kbase | Puromycin | 1300 |
| #8 | hLR5 | 1 × 107 | 1 | 14 kbase | Puromycin | 1100 |
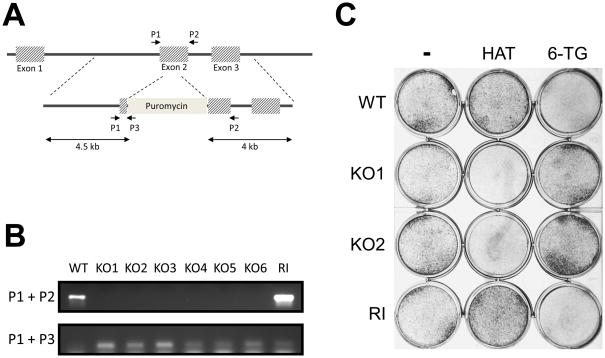
Finally, we tested the possibility of targeting specific loci in the hLR5 cell genome. We argued that since the hLR5 cells display many characteristics of mES cells, gene targeting may be facilitated in these cells. We chose to target the hypoxanthine phosphoribosyltransferase (HPRT) locus, as it offers the benefit of both positive and negative drug selection. Cells lacking HPRT are resistant to the drug 6-thioguanine (6-TG) but sensitive to HAT selection, while wild-type cells and cells with random integrations are HAT-resistant and 6-TG sensitive. Figure 5A shows a schematic representation of the human HPRT locus and the targeting construct, which disrupts the second exon by inserting a puromycin selection cassette. A combination of three primers (indicated, Fig 5A) can be used to distinguish between homologous recombination events and random integration of the targeting construct. Since the HPRT gene is located on the X-chromosome and therefore has only one copy in male hLR5 cells, a single targeting event can generate a knockout cell line. Upon electroporation of hLR5 cells with the HPRT targeting construct, cells were treated with Puromycin to select for positive clones and subsequently treated with 6-TG to select for homologous recombination events. Using this strategy, we determined the targeting efficiency to be approximately 1% (n=3). PCR analysis confirmed the disruption of the wild-type gene (Figure 5B, Upper panel). In addition, we tested the absence of functional HPRT expression in our targeted clones by culturing the cells in the presence of HAT supplement. As shown in Figure 5C, the HPRT knockout cells did not form colonies in the presence of HAT supplement while they were resistant to the positive-selection drug 6-TG.
Figure 5. Homologous recombination mediated gene targeting in hLR5 cells.
A: Schematic representation of the human HPRT locus and the targeting construct. The PCR primers used to detect the wild-type locus and the targeting construct (P1, P2, P3) are indicated.
B: PCR detection of 6 independent clones in which the HPRT locus was successfully targeted (KO1-6) and one clone with random integration of the targeting construct (RI). Wild-type cells (WT) were used as control. Upper panel, presence of the wild-type allele. Lower panel, detection of the targeting construct.
C: Confirmation of functional knockout of the HPRT gene in targeted hLR5 cells. WT: Wild-type, KO1 and KO2: knockout clones; RI: Clone with random integration, 6-TG: 6-thioguanine, HAT: HAT supplement
DISCUSSION
Stable and metastable pluripotent states
Recent reports have demonstrated that stem cells can exist in several distinct pluripotent states, which are defined, in part, by the culture growth factor conditions (Brons et al., 2007; Chou et al., 2008; Tesar et al., 2007). bFGF and Activin A allow the derivation of EpiSCs from murine post-implantation embryos (Brons et al., 2007; Tesar et al., 2007). EpiSCs display many characteristics of human ES- and iPS cells, which are derived and maintained under similar growth factor conditions. In contrast, LIF or a combination of GSK3β and MEK inhibitors (2i inhibitors) allows the derivation of murine ES cells from preimplantation blastocysts, which display a characteristic, dome-shaped colony morphology and differ from EpiSCs in their culture dynamics, molecular and epigenetic characteristics and notably, the ability to generate chimeric mice (Evans and Kaufman, 1981; Martin, 1981; Ying et al., 2008).
Yet, only a few inbred mouse strains can spontaneously give rise to stable ES cell lines, while other mouse strains and other species such as rat and primates only give rise to pluripotent stem cells with epiblast-like properties (Brons et al., 2007; Thomson et al., 1998; Thomson and Marshall, 1998). However, the combined use of LIF and 2i inhibitors recently allowed the derivation of true LIF-dependent ES cells from rat blastocysts and rat iPS cells (Buehr et al., 2008). Remarkably, in the absence of 2i inhibitors, these rat ES cells convert to an EpiSC-like pluripotent state (Buehr et al., 2008). In the mouse, EpiSCs can be converted into ES cells through the overexpression of Klf4 in the presence of 2i inhibitors (Guo et al., 2009). The derivation of ES-like iPS cells from the “non-permissive” NOD mouse strain is similarly dependent on either the constitutive expression of Klf4 or c-Myc or the addition of 2i inhibitors to the culture media (Hanna et al., 2009). It appears therefore that the EpiSC pluripotent state is the common stable pluripotent state for most strains of mice as well as other species, whereas the unique murine ES-like pluripotent state is ‘metastable’ in these genetic backgrounds.
We found that upon iPS reprogramming of human fibroblasts in the presence of human LIF colonies appear that display hallmark characteristics of mES cells, including the dome-shaped tightly packed mES cell morphology, the high proliferation rate, the activation of downstream targets of STAT signaling, and the tolerance of trypsin-passaging and single cell cloning. These hLR5 cells could be derived either through direct reprogramming of primary human fibroblasts, or using a more efficient ‘secondary’ fibroblast system (Schematic Figure 1A). hLR5 cells are metastable, since they depend on the constitutive expression of ectopic reprogramming factors. Upon removal of these factors, hLR5 cells convert to a stable pluripotent state that is indistinguishable from previously described hiPS cell lines. The conversion frequency of hLR5 cells into LD-hiPS cells was similar to the frequencies of the conversion of murine metastable iPS cells of the NOD strain into stable EpiSC-like iPS cells (Hanna et al., 2009), about 0.01% and is improved to almost 0.1% when LIF and the MEK inhibitor PD98059 are added during the conversion process. Several arguments support the notion that the emerging LD-hiPS cells are the result of conversion of hLR5 cells into a stable hiPS cell state rather than selection of pre-existing hiPS cells in the hLR5 population. First, the hLR5 cells were clonally derived and maintained for over 30 passages before conversion. Second, the hLR5 cells were continually passaged by trypsinization and third, LD-hiPS cells can be derived from hLR5 cells generated directly from primary fibroblasts, which have never before existed in a hiPS cell state. In hLR5 cells, the JAK/STAT3 signalling pathway is activated in a LIF-dependent manner resulting in the expression of STAT3 downstream target genes. In addition, hLR5 cells respond to LIF withdrawal with changes in cell morphology and surface marker expression. Since hLR5 cells in themselves do not form differentiated derivatives upon LIF-withdrawal, probably due to the forced ectopic expression of the five reprogramming factors, the cells are not LIF dependent to the same degree as mES cells are. However, continued maintenance of hLR5 in the presence of LIF is critical for the efficient conversion of hLR5 cells into pluripotent LD-hiPS cells, in particular in combination with the MEK inhibitor PD98059.
The conversion of hLR5 cells into hiPS cells is accompanied by epigenetic changes at the promoter regions of critical pluripotency regulators. Unexpectedly, these pluripotency factors, while transcriptionally silent, appear to be in a ‘poised’ state in hLR5 cells, from which they can be rapidly activated. SOX2, DNMT3b and SALL4 display the bivalent H3K4me3 and H3K27me3 histone methylation marks, which allow rapid conversion to the transcriptionally active H3K4me3 methylation state. OCT4, NANOG and REX1 are silenced by H3K27me3, yet the OCT4 promoter region is hypomethylated in the hLR5 cells, which greatly facilitates OCT4 reactivation. Similarly, the metastable iPS cells derived from the NOD mouse strain display hypomethylation at the Oct4 promoter (Hanna et al., 2009), suggesting that demethylation of promoter regions of critical pluripotency regulators is an essential property of the metastable state that allows gene reactivation and stable conversion to the epiblast-like pluripotent state.
Interestingly, the number of ectopic factors that is required to stabilize the mES-like state differs between the murine NOD strain, rat and human. While murine metastable NOD-iPS cells can be maintained with the constitutive expression of a single factor (either cMyc or Klf4) (Hanna et al., 2009), rat metastable iPS cell lines require the full complement of reprogramming factors (Liao et al., 2009), and in the case of the hLR5 cells this repertoire is expanded with the addition of NANOG. Genetic background has been shown to be a critical determinant in defining murine (meta)stable pluripotent states (Hanna et al., 2009), and it is possible that genetic background affects the reactivation of pluripotency genes in human hLR5 cells in a similar manner.
Gene targeting in human pluripotent cells
Murine ES cells have been instrumental in the discovery of gene function in the context of mammalian development and disease. The standard techniques that readily allow the introduction of transgene and reporter gene constructs in mES cells work poorly in human pluripotent stem cells. As a result, our ability to introduce foreign genetic elements into human cells is largely limited to the use of virus, or site-specific zinc-finger nucleases which are expensive and of which off-target effects are suspected but not well characterized. Recent studies in metastable pluripotent stem cells in the NOD mouse strain and the rat demonstrate that the LIF-dependent, mES-like pluripotent state allows the genetic manipulation of these cells using standard electroporation-based techniques. We demonstrate that in similar fashion, large reporter constructs and even BAC clones can be introduced into hLR5 cells. hLR5 cells even allow homologous recombination-based gene targeting. Until now, the (targeted) introduction of genetic elements into human pluripotent stem cells was highly inefficient and largely impractical. The intolerance of hES cells to grow from single cells resulted in very low yields upon antibiotic selection and the low proliferation rate made the process time consuming and labor intensive. Recently, Song et al reported a recombinant BAC-based strategy for gene targeting in hES cells (Song et al.). While a BAC-based system has the advantage of high homologous recombination efficiency, the system still suffers from the same practical difficulties associated with introducing a BAC clone into human pluripotent stem cells, the low numbers of clones and the added technical difficulties in identifying homologous recombination events. In contrast, hLR5 cells are tolerant to clonal selection and they have a high proliferation rate, which further facilitates clonal outgrowth and selection. Finally, hLR5 cells allow gene targeting with small (4kb) homologous arms using standard electroporation procedures that have been well established for the targeting of mES cells. Combined with the ability of hLR5 cells to convert into a stable iPS state we demonstrate here that the hLR5 intermediate provides an efficient platform for targeted gene modification and/or correction in human pluripotent stem cells (Figure 6). As such, it may find use in the generation of recombinant human cell lines for biomedical research, drug development and perhaps in future cell- or gene correction therapies.
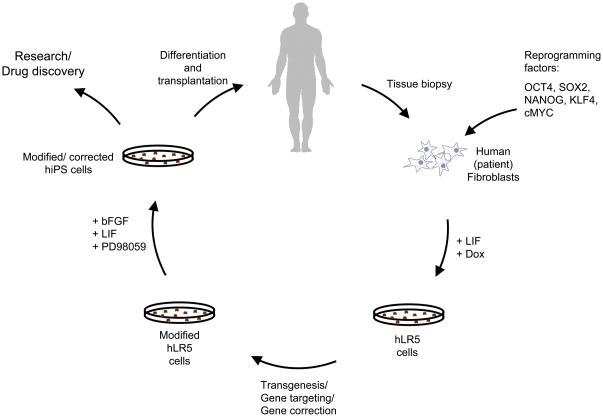
Figure 6. Application of the intermediate hLR5 state to generate recombinant human pluripotent stem cells for research, drug development and potential gene correction therapy.
Schematic model summarizing the procedure for gene targeting in human (patient) cells via the metastable hLR5 intermediate state. Human primary fibroblasts are collected and transduced with the five reprogramming factors OCT4, SOX2, NANOG, KLF4 and cMYC. Upon induction of reprogramming in the presence of human LIF, hLR5 colonies emerge. hLR5 cells are genetically modified using standard. The modified hLR5 cells are subsequently converted into pluripotent LD-hiPS cells, which in turn can be used in research or differentiated for use in future cell/gene correction therapies.
EXPERIMENTAL PROCEDURES
Culture of human ES and iPS cells
Human ES cells or iPS cells were maintained on γ-irradiated MEFs in hES cell medium (DMEM/F12 containing 20% knockout serum replacement (KOSR), 2mM L-Glutamine, 1% NEAA, 100 U of penicillin, 100 μg of streptomycin (all from Invitrogen), 0.1mM β-mercaptoethanol (Sigma), and 5 ng/ml bFGF(R&D systems). Cells were routinely passaged every 5–7 days. For EB derivation colonies were picked and collected in EB medium (IMDM containing 15% FBS, 2mM L-Glutamine, 1% NEAA, 1 mM Sodium Pyruvate, 100 U of penicillin, 100 μg of streptomycin (all from Invitrogen) 200 μg/mL iron-saturated transferrin (Sigma), 4.5 mM monothioglycerol (Sigma), 50 μg/mL ascorbic acid (Sigma)). Colonies were cultured with gentle agitation. After 7–9 days colonies were transferred to gelatin coated chamber slides, allowed to adhere and incubated for 3–5 more days. For teratoma formation LD-hiPS were injected subcutaneously into NOG mice (Jackson Laboratories, Bar Harbor, ME). After ~10–12 weeks teratomas were dissected, washed and fixed.. Parafin sections were stained with H&E.
Derivation and Maintenance of hLR5 cells
Clone#12 hiPSC colonies were differentiated as described (Maherali et al., 2008). The fibroblasts were induced to generate hLR5 cells by passaging the cells onto MEF feeders in hLR5 media (DMEM/F12 containing 20% KOSR, 100 U of penicillin, 100 μg of streptomycin, 2mM L-Glutamine, 1% NEAA (all from Invitrogen), 0.1mM β-mercaptoethanol (Sigma), 10 ng/ml human LIF (Sigma) and 2 ng/ml Doxycycline (Sigma). Emerging colonies were individually picked and subcultured by trypsin digest in hLR5 media on MEF feeders. For the direct reprogramming of human fibroblasts (HS27, ATCC) into hLR5 cells 105 cells per 1cm2 were transduced with the STEMCCA lentivirus (containing doxycycline-inducible human OCT4, SOX2, KLF4 and cMYC) and rtTA with or without a doxycycline-inducible lentivirus for NANOG (Maherali et al., 2008; Sommer et al., 2009; Stadtfeld et al., 2008). After approximately 30 days, emerging colonies were individually picked and expanded further by trypsin digest in hLR5 media with MEF feeders. hLR5 cells are passaged every 2–3 days, depending on culture density.
Microarray and Q-PCR analysis
Total RNA was extracted using Trizol (Invitrogen) following the manufacturer s protocol. cDNA synthesis (Superscript III First-Strand synthesis system, Invitrogen) was performed using random primers. qRT-PCRs were carried out using Brilliant II SYBR Green mix (Stratagene) and a Stratagene MXPro4000 real-time thermocycler. Primer sequences for the analysis of endogenous and ectopic pluripotency gene expression were reported previously (Maherali et al., 2008).
For genome-wide expression analysis total RNA was labeled and hybridized to Agilent Whole Human Genome Oligo 4X44K Microarrays (one-color platform) according to the manufacturer’s protocols. The gene expression results were analyzed using GeneSifter microarray analysis software.
FACS analysis
Cells were incubated with the antibodies against the indicated surface antigens for 30 min at 4°C in RPMI + 0.5%FBS. The following antibodies were used for cell surface marker profiling: SSEA1 (BD Biosciences), TRA-1-81, TRA-1-60, SSEA3 and SSEA4 (Millipore). Cells were washed twice and incubated with the relevant fluorphore-conjugated secondary antibody (BD Biosciences) for 30 min at 4°C. Cells were washed, resuspended in RPMI/0.5% FBS and analyzed on a Becton-Dickinson FACSCalibur cell analyzer.
Electroporation and gene targeting
Cells were trypsinized, strained, resuspended in 700 μl PBS containing 15–30 μg linearized DNA, and transferred to a 0.4cm gap electroporation cuvette (Biorad). The electroporation was carried out using a single 320V, 200uF pulse. Upon electroporation, cells were replated onto gelatinized dishes with DR4 MEFs. Antibiotic selection was started 48–72 h later using either 25 μg/ml Hygromycin (Invitrogen) or 0.25 μg/ml Puromycin (Invivogen) as indicated. For HPRT KO, cells were selected with Puromycin followed by treatment with 6-Thioguanine (6-TG, Sigma) to select for homologous recombinants. HAT-selection was carried out by adding 1x HAT supplement (Invitrogen) directly to the hLR5 medium.
ChIP
Cells were fixed in 1% formaldehyde for 10 minutes, quenched with glycine and washed 3 times with PBS. Cells were then resuspended in lysis buffer and sonicated 10 × 30 sec in a Bioruptor (Diagenode, Philadelphia, PA) to shear the chromatin to an average length of 600 bp.
Supernatants were precleared using protein-A agarose beads (Roche, Mannheim, Germany) and 10% input was collected. Immunoprecipitations were performed using polyclonal antibodies to H3K4trimethylated, H3K27trimethylated and normal rabbit serum (Upstate, Temucula, CA). DNA-protein complexes were pulled-down using Protein-A agarose beads and washed. DNA was recoverd by overnight incubation at 65°C to reverse cross-links and purified using QIAquick PCR purification columns (Qiagen, Maryland). Enrichment of the modified histones in different genes was detected by quantitative real time PCR using the primers in the supplemental methods.
Immunostaining
Primary antibodies used were: α-TRA-1-60, α-SSEA-3, α-SSEA-4, α-TRA-1-81, α-Sox2 (all from Millipore), α-SSEA1, α-Stat3 (both from Cell Signaling), α-Oct4, α-Nanog (both from Abcam), α-TuJ1 (Covenance), α-SMA (Sigma) and α-AFP (Santa Cruz). All secondary antibodies were from Invitrogen. The nuclei were visualized with DAPI.
Western blotting
Cells were lysed using RIPA buffer containing proteinase inhibitors. The protein concentration was estimated using Bradford reagents and equal amounts of protein were run on 4–12% Bis-Tris Gels (Invitrogen) and transferred to PVDF membranes (Millipore). Primary antibodies used were: Phospho-Stat3, Stat3, E-Cadherin (all Cell signaling) and Tubulin (Sigma). HRP coupled secondary antibody was from Cell Signalling.
ACCESSION NUMBERS
Microarray data have been deposited in the GEO database with the following accession numbers: XXXX, XXXX, XXXX, XXXX, XXXX, XXXX, XXXX
Highlights.
We report a metastable human stem cell state with mES cell properties
The murine-ES like human stem cell state is LIF-responsive
Reprogramming factor withdrawal results in reversion to standard hiPS cells
hLR5 cells facilitate gene targeting in human pluripotent stem cells
Supplementary Material
Acknowledgments
The authors thank Tim Ahfelt and Chad Cowan for providing human iPS cells, Laura Prickett-Rice and Kat Folz-Donahue at the HSCI/MGH Flow Cytometry Core facility cell sorting; Jason West and Meredith Bryden for technical support. This work was supported by grants from the NIH and a grant from the Dutch Science Organization (NWO). C.B. was supported by the Gottlieb Daimler- and Karl Benz-Foundation. H.H.C. was supported by the National Science Council (Taiwan, ROC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bao S, Tang F, Li X, Hayashi K, Gillich A, Lao K, Surani MA. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature. 2009;461:1292–1295. doi: 10.1038/nature08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Bu L, Jiang X, Martin-Puig S, Caron L, Zhu S, Shao Y, Roberts DJ, Huang PL, Domian IJ, Chien KR. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460:113–117. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- Buehr M, Meek S, Blair K, Yang J, Ure J, Silva J, McLay R, Hall J, Ying QL, Smith A. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135:1287–1298. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Chou YF, Chen HH, Eijpe M, Yabuuchi A, Chenoweth JG, Tesar P, Lu J, McKay RD, Geijsen N. The growth factor environment defines distinct pluripotent ground states in novel blastocyst-derived stem cells. Cell. 2008;135:449–461. doi: 10.1016/j.cell.2008.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan CA, Klimanskaya I, McMahon J, Atienza J, Witmyer J, Zucker JP, Wang S, Morton CC, McMahon AP, Powers D, et al. Derivation of embryonic stem-cell lines from human blastocysts. N Engl J Med. 2004;350:1353–1356. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Markoulaki S, Mitalipova M, Cheng AW, Cassady JP, Staerk J, Carey BW, Lengner CJ, Foreman R, Love J, et al. Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell. 2009;4:513–524. doi: 10.1016/j.stem.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ding S. Small molecules that modulate embryonic stem cell fate and somatic cell reprogramming. Trends Pharmacol Sci. 2009;31:36–45. doi: 10.1016/j.tips.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Li W, Wei W, Zhu S, Zhu J, Shi Y, Lin T, Hao E, Hayek A, Deng H, Ding S. Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell. 2009;4:16–19. doi: 10.1016/j.stem.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Liao J, Cui C, Chen S, Ren J, Chen J, Gao Y, Li H, Jia N, Cheng L, Xiao H, et al. Generation of induced pluripotent stem cell lines from adult rat cells. Cell Stem Cell. 2009;4:11–15. doi: 10.1016/j.stem.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Maherali N, Ahfeldt T, Rigamonti A, Utikal J, Cowan C, Hochedlinger K. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3:340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang H, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008 doi: 10.1038/nature07056. Advance online publication, May 28 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, 3rd, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- Ptaszek LM, Cowan CA. New tools for genome modification in human embryonic stem cells. Cell Stem Cell. 2007;1:600–602. doi: 10.1016/j.stem.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Ehret H, Paro R. Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol Cell. 2004;16:641–653. doi: 10.1016/j.molcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bernstein BE, Karabetsou N, Morillon A, Weise C, Schreiber SL, Mellor J, Kouzarides T. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol Cell. 2003;12:1325–1332. doi: 10.1016/s1097-2765(03)00438-6. [DOI] [PubMed] [Google Scholar]
- Silva SS, Rowntree RK, Mekhoubad S, Lee JT. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:4820–4825. doi: 10.1073/pnas.0712136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M, Rogers D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Chung SK, Xu Y. Modeling disease in human ESCs using an efficient BAC-based homologous recombination system. Cell Stem Cell. 6:80–89. doi: 10.1016/j.stem.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Marshall VS. Primate embryonic stem cells. Curr Top Dev Biol. 1998;38:133–165. doi: 10.1016/s0070-2153(08)60246-x. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Xu RH, Peck RM, Li DS, Feng X, Ludwig T, Thomson JA. Basic FGF and suppression of BMP signaling sustain undifferentiated proliferation of human ES cells. Nat Methods. 2005;2:185–190. doi: 10.1038/nmeth744. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.