Abstract
We previously reported that CD8+ T cells are directed predominantly toward the immunodominant Her-2/neu (neu) epitope RNEU420–429 in nontolerized FVB/N but not tolerized HER-2/neu (neu-N) mice. In this study, we screened overlapping peptides of the entire neu protein and identified six new epitopes recognized by vaccine-induced neu-N–derived T cells. Evaluation of individual nondominant responses by tetramer staining and IFN-γ secretion demonstrate that this repertoire is peripherally tolerized. To address the role that the complete CD8+ T cell repertoire plays in vaccine-induced antitumor immunity, we created a whole-cell vaccine-expressing neu cDNA that has been mutated at the RNEU420–429 anchor residue, thereby abrogating activation of immunodominant epitope responses. Studies comparing the mutated and nonmutated vaccines indicate that nondominant CD8+ T cells can induce antitumor immunity when combined with regulatory T cell-depleting agents in both neu-N and FVB/N mice. Collectively, these studies demonstrate that the neu-directed T cell repertoire is not intrinsically incapable of eradicating tumors. Rather, they are suppressed by mechanisms of peripheral tolerance. Thus, these studies provide new insights into the function of the complete T cell repertoire directed toward a clinically relevant tumor Ag in tumor-bearing hosts.
The success of cancer immunotherapy will depend on the development of vaccine approaches that specifically activate T cell responses against a panel of tumor-associated Ags (TAAs). Tumor-specific CD8+ T cells, in particular, recognize epitopes that derive from these Ags when bound to MHC class I molecules on the tumor cell surface. Early research in animal and human studies demonstrated the activation of CD8+ T cells against single epitopes contained within the amino acid sequence of one TAA (1–3). More recently, other nondominant CD8+ T cell responses have been identified as noted in Cancer Immunity’s peptide database. However, addressing the role that the complete CD8+ T cell repertoire plays in antitumor immunity is fundamental to the development of cellular-based vaccines and immunotherapy.
Immunodominance refers to the lymphocyte response that is predominantly directed toward one or a few epitopes in a protein rather than all of the potential determinants contained within that protein. Factors that lead to the development of immunodominance include Ag availability and processing, TAP specificity, peptide binding affinity to MHC class I, TCR affinity/avidity, the available precursor CD8+ T cell repertoire, and immunodomination (T cell–T cell competition) (4, 5). These factors ultimately generate CD8+ T cell responses that are larger and more robust than responses to other epitopes when evaluated by TCR avidity, effector function, and population size. The immunodominance of antigenic epitopes has been studied best in infectious disease models. Although immunodominant high-avidity CD8+ T cell responses have also been shown to be important in some cancer models (6–8), other models suggest that only low-avidity/nondominant TAA-specific T cells are available for activation in tolerized hosts. Often high-avidity immunodominant epitope-specific responses are tolerized due to expression of the Ag on nontumor tissues (9), whereas low-avidity/nondominant CD8+ T cell responses are thought to escape central and peripheral tolerance (10–12). Several reports have demonstrated that activation of low-avidity/nondominant CD8+ T cells are capable of expanding and lysing tumors (13–15). However, other studies suggest that there may be additional functional characteristics of the nondominant T cell repertoire that prevent effective T cell activation and function capable of causing tumor regression (6, 16, 17).
We have used the clinically relevant Her-2/neu transgenic (neu-N) mice to evaluate Her-2/neu (neu)–directed CD8+ T cell responses under tolerizing conditions. These mice overexpress the nontransforming rat neu cDNA under the mammary-specific mouse mammary tumor virus promoter and develop spontaneous neu-expressing mammary carcinomas (18). We previously reported that neu-targeted vaccination induces a predominantly high-avidity CD8+ T cell response to the immunodominant epitope RNEU420–429, which is capable of clearing neu-expressing tumors in the parental FVB/N nontolerized mice (8, 19, 20). In contrast, the RNEU420–429-specific CD8+ T cell response is tolerized in neu-N mice, which leads to the prevalence of a polyclonal but less functional T cell repertoire that is specific for other neu-derived epitopes (6, 19, 21). Interestingly, immune-modulating doses of cyclophosphamide (Cy) administered prior to vaccination to deplete T regulatory cells (Tregs) induces high-avidity RNEU420–429-specific CD8+ T cells that cause tumor regression in ~20% of neu-N mice (6).
In this report, we screened an overlapping 15-aa peptide library spanning the complete neu protein and identified six new epitopes that are recognized by both the FVB/N and neu-N CD8+ T cell repertoires. We now establish that T cells responses against this expanded panel of neu epitopes are indeed subdominant to RNEU420–429 in the nontolerized FVB/N mice. However, T cell responses against these epitopes in the tolerant neu-N mice form a new dominance hierarchy based on effector function. Additionally, we demonstrate that functional dominance does not always correspond with relative TCR avidity. Furthermore, we demonstrate that vaccination with a modified neu-targeted vaccine lacking expression of the RNEU420–429 immunodominant epitope allows for enhanced function of the nondominant CD8+ T cell repertoire and tumor recognition.
Materials and Methods
Cell lines and medium
The NT2.5 tumor cell line was originally derived from naturally arising spontaneous tumor from neu-N mice. These cells were created and cultured as previously described (16). The 3T3neuGM vaccine and 3T3GM mock vaccine cells lines were generated and cultured as previously reported (16). The cell lines used as T cell targets, that is, T2-Dq, NIH-3T3, and NT2.5B7-1, were also produced and maintained as previously described (21, 22).
Mice, tumor, and immunization protocols
Mice 7–12 wk of age were used in all experiments in accordance with protocols approved by the Animal Care and Use Committee of The Johns Hopkins University School of Medicine. FVB/N mice were purchased from Harlan Laboratories, The Jackson Laboratory, and Taconic. Neu-N mice (18) were originally provided by William Muller and The Jackson Laboratory and were bred and housed at The Johns Hopkins University. These mice were bred to homozygosity as verified by Southern blot analysis (16).
In all experiments, mice were treated with 100 mg/kg Cy (Bristol-Myers Squibb) and 50 μg PC61 (American Type Culture Collection) per mouse 1 d prior to vaccination. Cy and PC61 were diluted in PBS and individually administered i.p. in 500 μl doses. Mice were vaccinated with either a total of 3 × 106 irradiated (5000 rad from a [137Cs] source) 3T3neuGM or 3T3GM cells or 6 × 106 3T3neumut vaccine cells with 3 × 106 3T3GM cells. All vaccine cells were distributed into three 100 μl s.c. injections and admininstered in both arms and one leg to include immunization of three different lymph node regions as previously described (23). Tumor treatment experiments were conducted by injecting neu-N mice with 5 × 104 and FVB/N mice with 2 × 106 NT2.5 cells in a 100 μl volume in the mammary fat pad 3 d prior to vaccination.
Peptides
All peptides were synthesized (>95% purity) by either the Peptide Synthesis Facility or the Biosynthesis and Sequencing Facility at The Johns Hopkins University. Peptides were designed as either 15-mer overlapping by nine amino acids or 10-mer overlapping by one amino acid. The nucleoprotein (NP)118–126 (RPQASGVYM) peptide is from the lymphocytic choriomeningitis virus nucleoprotein (24) and was used as an irrelevant H2-Dq–binding peptide.
Ex vivo splenic stimulation and CD8+ T cell isolation
Splenocytes and lymphocytes were isolated from either neu-N or FVB/N mice 1 wk postvaccination. Spleens and draining lymph nodes were mashed, strained using a 100-μm mesh (BD Falcon), and RBCs were removed using ACK lysis buffer (Quality Biological). Splenocytes were cultured for 1 wk at 37°C and 5% CO2 with either 25 μg/ml peptide pool containing 10 overlapping peptides or 2.5 μg/ml individual peptides in CTL media consisting of RPMI 1640 (Invitrogen), 10% FBS (HyClone), 0.5% L-glutamine (Invitrogen), 1% penicillin/streptomycin (Invitrogen), and 0.1% 2-ME (Invitrogen). One week postpeptide stimulation, CD8+ T cells were separated from dead cell debris using a Ficoll (GE Healthcare) density gradient and isolated using a Dynal mouse CD8 cell negative isolation kit (Invitrogen).
GM-CSF release ELISA
Neu-specific peptides, RNEU420-429 and NP118-126, were pulsed onto T2-Dq cells at 26°C for 2 h in CTL cell media at a final concentration of 2.5 μg/ml. Pulsed targets (3 × 104) were then incubated with 3 × 105 isolated CD8+ T cells in 96-well flat bottom plates with a final volume of 150 μl CTL cell media at 37°C and 5% CO2. The supernatant was collected 24 h later and analyzed in a murine GM-CSF ELISA (R&D Systems). Graphed data depict peptide data minus background data (NP118–126).
Abs, tetramer, and flow cytometric analysis
The mouse anti–IL-2 Rα-chain (CD25) mAb, PC61, is isolated from the PC61.5.3 hybridoma (American Type Culture Collection) grown in protein-free hybridoma medium II (Invitrogen) supplemented with 0.1% penicillin/streptomycin (Invitrogen) at 37°C and 10% CO2. Expression of neu was analyzed using 7.16.4 (IgG2a), a neu mAb specific for the extracellular domain of neu. 7.16.4 was obtained from the M. Greene hybridoma (University of Pennsylvania, Philadelphia, PA) and purified from ascites prepared by Harlan Bioproducts for Science. The H-2Dq–specific Ab was isolated from the supernatant of the 30.5.7s hybridoma (American Type Culture Collection). Cells stained by either the neu or H-2Dq–specific Abs were labeled with a FITC-conjugated goat anti-mouse IgG (SouthernBiotech).
H-2Dq/neu–specific tetramer was assembled as previously described (25). PE-conjugated tetramer at varying dilutions was incubated with 5 × 105 isolated CD8+ T cells for 15 min at 4°C. CD8+ T cells were further incubated with anti–CD8-allophycocyanin and tetramer for an additional 15 min at 4°C. The percentage of cells that were CD8+tetramer+ was calculated from the total CD8+ population, and the percentage of Ag-specific cells was calculated by subtracting the H-2Dq/NP118–126 value from the neu-specific value.
IFN-γ cytokine staining was performed using the BD Biosciences mouse intracellular cytokine stain kit. Isolated T cells were incubated with either NT2.5B7-1 or T2-Dq cells that were pulsed with 2.5 μg/ml peptide at an equal ratio in the presence of GolgiStop. After a 5-h incubation, cells were stained with CD8-CyChrome, fixed and permeabilized, and stained with IFN-γ-PE and CD8-CyChrome or an isotype control. The percentage of CD8+IFN-γ+ was calculated from the total CD8+ population, and the percentage of Ag-specific cells was calculated by subtracting the NP118–126 background value from the neu-specific value.
All samples were read on the BD FACSCalibur cytometer (BD Biosciences) and analyzed using FlowJo software (Tree Star).
Peptide binding studies
T2-Dq cells are washed twice in RPMI 1640 (Life Technologies) and resuspended at 2 × 105 cells in 250 μl RPMI 1640 supplemented with 3 μg/ml human β2-microglobulin (Sigma-Aldrich) and varying concentrations of peptide (0, 0.1, 1.0, 10, 100 μg). Cells are incubated for 18 h, shaking, at 26°C. Cells are washed with 1× PBS (Life Technologies) and stained for the MHC molecule H-2Dq using the mAb 30.5.7S (American Type Culture Collection). Samples were analyzed based on their mean fluorescence intensity.
Rat Her-2/neu mutagenesis and cloning
Rat Her-2/neu was mutated using the QuikChange II XL site-directed mutagenesis kit (Stratagene). The mutagenic primers used were designed to generate a leucine to alanine substitution at position four of RNEU420–429: forward, 5′-GCCAGACAGTGCCCGTGACCTCAGTGTCTTCCAGAACCTTC-3′ and reverse, 5′-GAAGGTTCTGGAAGACACTGAGGTCACGGGCACTGTCTGGC-3′. These primers were used to amplify the mutated pSV2/neu-N plasmid using the following conditions: an initial step of 95°C for 1 min, 18 cycles of 95°C for 50 s, 60°C for 50 s, 68°C for 10 min, and a final step of 68°C for 7 min. Mutated rat neu was excised from pSV2 using HindIII, EcroRI, and SalI (New England Biolabs) restriction enzymes and re-ligated into pcDNA3 expression vector (Invitrogen) using HindIII and XhoI (New England Biolabs).
Transfection and cloning of mutated Her-2/neu vaccine sorting
pcDNA3/neumut was linearized by PvuI (New England Biolabs) digestion and purified by phenol-chloroform extraction and ethanol precipitation. NIH-3T3 cells were transfected with linearized pcDNA3/neumut by electroporation (30 μg/1 × 107 cells). 3T3neumut cells were selected and cloned using 700 μg neomycin or G418 (Invitrogen).
Statistics
A Student t test was applied to compare statistical significance between peptide-specific responses and treatment groups of tumor-challenged mice, with p < 0.05 being significant. All analysis was performed using Excel software (Microsoft Office).
Results
Screening of an overlapping 15-aa peptide library of the entire neu protein identifies six new CD8+ T cell epitopes
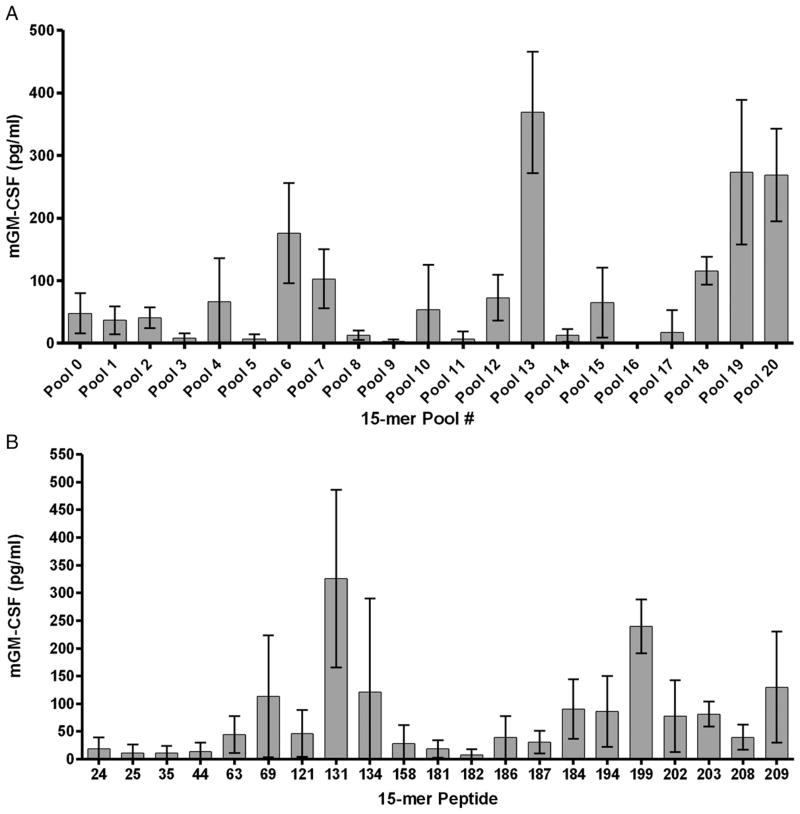
We previously reported that vaccine-induced neu-directed CD8+ T cells are predominantly directed at the immunodominant RNEU420–429 epitope in FVB/N mice (18). However, follow-up studies suggested that the neu-N CD8+ T cell repertoire is more polyclonal, less functional, and mainly specific for other alternate epitopes. To elucidate the differences between the different neu-directed CD8+ T cell repertoires that are predominant in tolerant versus nontolerant environments, we first identified the additional epitopes recognized by each T cell population within the neu-N CD8+ T cell repertoire. To accomplish this, we used immunized lymphocytes from neu-N mice treated with a vaccine consisting of 3T3 cells genetically modified to express neu and secrete GM-CSF (3T3neuGM) and immune-modulating doses of Cy to deplete Tregs to screen the entire neu protein for additional CD8+ T cell epitopes. Neu-N mice treated with this vaccine approach can mount CD8+ T cell responses specific for multiple epitopes that are associated with tumor regression ~20% of the time (6). A library containing two hundred nine 15-aa peptides that overlapped by 9 aa was synthesized to cover the entire sequence of neu and divided into 21 pools containing ten 15-aa peptides each for initial screening. Splenocytes were isolated 1 wk following vaccination and stimulated in vitro for 7 d with each of the 21 peptide pools. CD8+ T cells stimulated with the individual pools of peptide were subsequently evaluated by ELISA for GM-CSF secretion. Initial studies demonstrated activation of the neu-N CD8+ T cell repertoire by many peptide pools (Fig. 1A). The corresponding 15-aa peptides in positive pools were then tested individually using the same procedure. With this approach, 21 individual 15-aa peptides tested positive in multiple ELISA assays (Fig. 1B). Next, 10-aa peptides overlapping by 9 aa corresponding to each positive 15-aa peptide were tested using either a GM-CSF ELISA assay or an IFN-γ intracellular cytokine stain assay. After extensive screening, six 10-aa peptides and one 9-aa peptide were confirmed to be recognized by the neu-N CD8+ T cell repertoire (Table I). Analysis of the primary structure of the neu protein revealed that five of the identified epitopes are located within the intracellular domain, and one 10-mer and the 9-mer are located within the extracellular domain (Table I). A comparison of the rat neu epitope sequence homology to mouse neu demonstrated that five of the identified epitopes are 100% homologous to mouse neu (Table I). Thus, a full TAA peptide screen is feasible and can identify an extended panel of epitopes recognized by a polyclonal T cell repertoire. These data further support our hypothesis that an Ag-specific T cell repertoire is primarily specific for nondominant CD8+ T cell epitopes in a tolerized host.
FIGURE 1.
Screening of overlapping 15-aa peptides spanning the entire length of the Her-2/neu protein with neu-specific CD8+ T cells from vaccinated neu-N mice identifies six new epitopes. Splenocytes from five vaccinated neu-N mice were pulsed with each peptide pool or a single peptide for 1 wk. CD8+ T cells were isolated and incubated with T2-Dq cells pulsed with the corresponding peptide(s) or the irrelevant LCMV nucleoprotein peptide NP118–126. A, Detection of GM-CSF secretion by neu-specific CD8+ T cells after stimulation with peptide pools containing 10 overlapping 15-aa peptides. B, Individual 15-aa peptides elicit responses from neu-specific CD8+ T cell repertoire that correlate with the peptide pool data. Plotted are the mean GM-CSF ELISA values for five mice after background NP118–126 levels were subtracted.
Table I.
Nondominant epitopes recognized by the neu-specific CD8+ T cell repertoire of vaccinated neu-N mice
| 15-mer | Protein Location | 9/10-mer | Sequence | Amino Acids | Homology to Mouse Her-2/neu (%) |
|---|---|---|---|---|---|
| 24 | ECD | 24F | LQLRSLTEIL | 144–153 | 10/10 (100) |
| 24 | ECD | 24F1 | LQLRSLTEI | 144–152 | 10/10 (100) |
| RNEU | ECD | — | PDSLRDLSVF | 420–429 | 7/10 (70) |
| 131 | ICD | 131E | YVSRLLGICL | 785–794 | 10/10 (100) |
| 134 | ICD | 134C | VTQLMPYGCL | 797–806 | 10/10 (100) |
| 158 | ICD | 158B | RLPQPPICTI | 944–953 | 9/10 (90) |
| 184 | ICD | 184E | QSLSPHDLSP | 1103–1112 | 10/10 (100) |
| 202 | ICD | 202F | HPSPAFSPAF | 1212–1221 | 10/10 (100) |
Nonimmunodominant CD8+ T cell epitopes bind H-2Dq
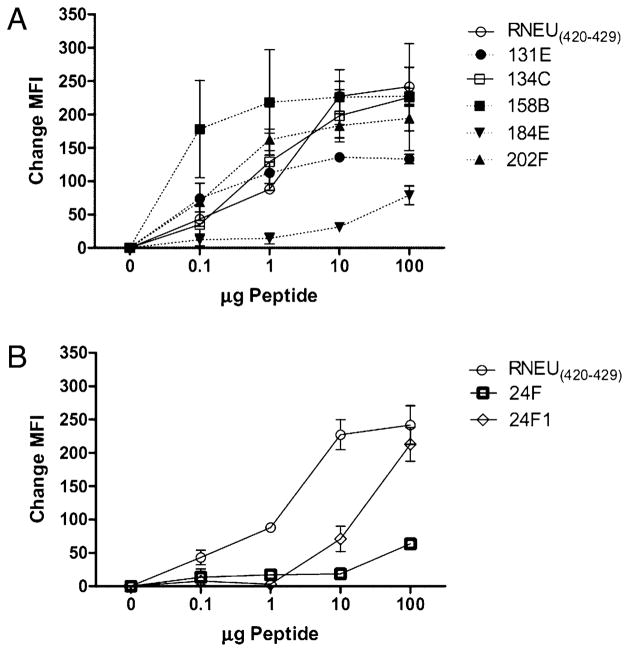
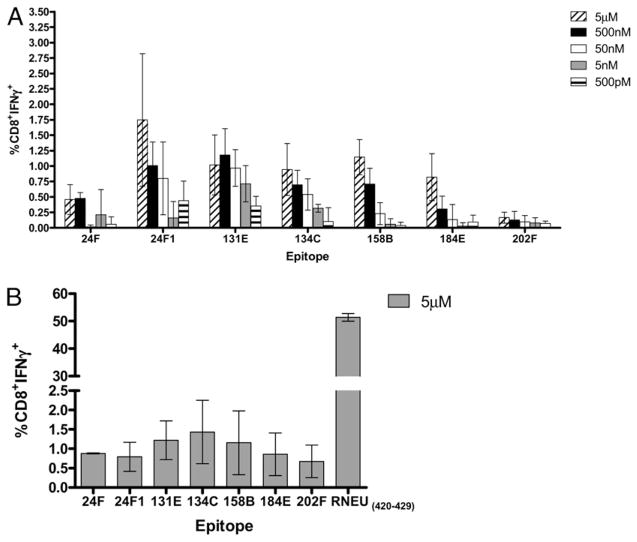
A series of additional analyses were performed on the six 10-mer and one 9-mer epitopes to confirm that these peptides induced neu-specific T cell responses rather than nonspecific activation of CD8+ T cells due to peptide cross-reactivity. Validation was carried out by first assessing the ability of each epitope to bind to H-2Dq. With these dilutional peptide binding assays, we observed that the epitopes 24F1 (9-aa), 131E, 134C, 158B, and 202F bind to H-2Dq with an affinity range similar to RNEU420–429. In contrast, 24F (10-aa) and 184E epitopes have weaker binding affinities for H-2Dq in comparison with RNEU420–429 (Fig. 2). Next, CD8+ T cells specific for each peptide were evaluated for IFN-γ production in a peptide titration assay. These functional T cell assays demonstrated that there are differences in the magnitude of CD8+ T cell responses to each epitope and identified a new hierarchy of functional dominance among the non–RNEU420–429-specific CD8+ T cell repertoire. The CD8+ T cell responses to 24F1, 131E, and 134C were the most robust based on the percentage of CD8+IFN-γ+ detected at nanomolar concentrations of peptide (Fig. 3A). These responses were followed by T cell responses specific for the 158B and 184E epitopes. Finally, the weakest CD8+ T cell responses were to the 24F and 202F epitopes (Fig. 3A). FVB/N CD8+ T cell responses specific for each epitope were also evaluated. Interestingly, CD8+ T cell responses directed at these non-dominant epitopes were observed, but the magnitude of these responses was 10-fold less than the immunodominant RNEU420–429 response (Fig. 3B). Thus, these experiments demonstrate the relative binding affinities of the nondominant CD8+ T cell epitopes. Importantly, these studies identify a functional hierarchy among the nondominant CD8+ T cell repertoire.
FIGURE 2.
Alternate epitopes bind to H-2Dq with varying affinities when compared with RNEU420–429. T2-Dq cells were pulsed with each 10-aa epitope at varying concentrations of peptide and β2-microglobulin for 18 h at room temperature. Samples were washed, stained with the H-2Dq mAb 30.5.7S, and analyzed based on the mean fluorescence intensity (MFI). The change in MFI from the no peptide control indicates stabilization of surface MHC class I by the peptide and relative strength of peptide binding to MHC class I. Shown is the average of triplicates after the no peptide control was subtracted. These experiments were repeated at least twice with similar results.
FIGURE 3.
Emergence of a functional hierarchy of neu-specific nondominant CD8+ T cell responses isolated from vaccinated neu-N and FVB/N mice. One week following vaccination, splenocytes and vaccine draining lymph nodes were collected from five neu-N (A) or FVB/N (B) mice and incubated with 2.5 μg/ml peptide for 7 d. Stimulated lymphocytes were collected and incubated for T2-Dq cells pulsed with the corresponding peptide or NP118–126 at varying concentrations. Cells were stained for CD8 and IFN-γ and analyzed by intracellular cytokine staining. A, CD8+ T cell responses directed against 24F1, 131E and 134C are the most prevalent in vaccinated neu-N mice as determined by peptide titration studies. B, Vaccinated FVB/N mice mount CD8+ T cell responses specific for nondominant epitopes that are significantly weaker than responses to RNEU420–429. Shown for both are the mean percentage of CD8+ T cells that are IFN-γ + after subtraction of background NP118–126 responses. These experiments were repeated at least twice with similar results.
The nondominant CD8+ T cell responses are less avid than RNEU420–429 T cell responses in both FVB/N and neu-N mice
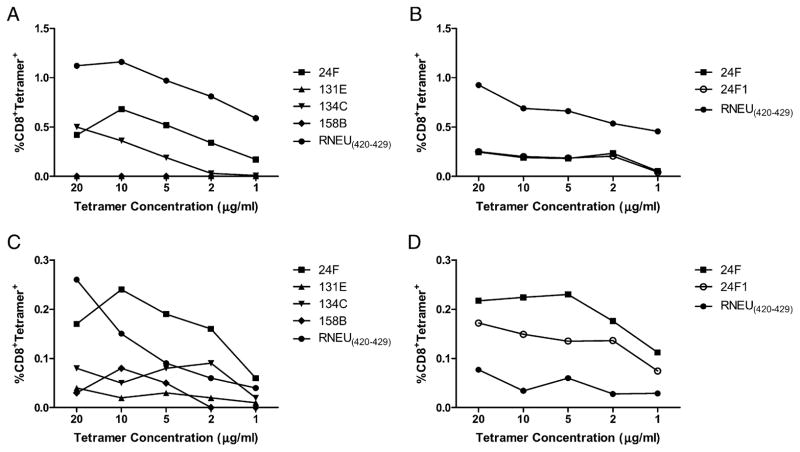
TCR avidity has been shown to be an important measure of the quality of a T cell response (26). Furthermore, we previously reported that Cy given as a Treg-depleting agent with the 3T3neuGM vaccine induces higher avidity T cell responses against the immunodominant RNEU420–429 epitope that are associated with tumor regression in neu-N mice (6). We therefore used dilutional tetramer staining to compare RNEU420–429-specific T cell avidity with the relative TCR avidities of the nondominant CD8+ T cell populations specific for the four epitopes against which the most robust responses are induced. We used our functional hierarchy data to limit our analysis to the 24F, 131E, 134C, and 158B 10-mer epitopes and the one 9-mer epitope because these four epitopes represent a range of functional avidity responses among the six 10-mer identified. We developed individual tetramers for each, and for the positive and negative control epitopes, RNEU420–429 and NP118–126, respectively, were used (for analysis, H-2Dq/NP118–126 was subtracted from the percentage CD8+tetramer+ as background). These tetramers were used to stain CD8+ T cell populations that were directly isolated from tumor-bearing FVB/N and neu-N mice treated with the 3T3neuGM vaccine (T cells populations were analyzed straight ex vivo without an in vitro stimulation as previously described by us; see Ref. 6). As expected, the RNEU420–429 response was the most avid and prevalent, and it could be detected down to a dilution of 1:1000 (1 μg/ml tetramer protein). Among the nondominant repertoire, the response to the 9-aa and 10-aa peptide 24F1 and 24F were the most avid and prevalent, followed by the response to peptide 134C. CD8+ T cell responses to 131E and 158B were too weak to be detected via tetramer staining in FVB/N mice (Fig. 4A, 4B, Supplemental Fig. 1). The CD8+ T cell responses detected in the neu-N mice were similar to those observed in FVB/N mice in that the hierarchy of epitope-specific responses based on relative avidities remained the same. Responses directed against the 24F1, 24F, and RNEU420–429 epitopes were the most avid, followed by the less avid responses toward the 134C, 158B, and 131E epitopes. However, the neu-N responses directed against 24F1, 24F, 134C, and RNEU420–429 were not as prevalent or avid in comparison with their counterparts in FVB/N mice. The exceptions to this observation were the responses to 131E and 158B. Unlike in FVB/N mice, these responses were detected in neu-N mice, but remained the least avid of the epitope-specific responses (Fig. 4C, 4D, Supplemental Fig. 1). Thus, the nondominant CD8+ T cell responses are functionally distinct and weaker than the immunodominant CD8+ T cell response against RNEU420–429. Additionally, both immunodominant and nondominant relative TCR avidities and T cell population sizes are affected by the presence of peripheral tolerance.
FIGURE 4.
Lower relative avidities are detected among nondominant CD8+ T cell responses when compared with RNEU420–429 T cell responses in FVB/N and neu-N mice. A and B, RNEU420–429-specific responses are the most abundant and avid in FVB/N mice. C and D, 24F- and 24F1-specific responses are the most avid in neu-N mice. Lymphocytes were isolated 14 d after tumor challenge from 10 tumor-bearing mice treated with the 3T3neuGM vaccine and stained with increasing dilutions of H-2Dq/peptide tetramer and anti-CD8 straight ex vivo. Tetramer dilutions were made from 1 mg/ml tetramer stock solutions. Graphed data depict the percentage of CD8+ T cells that are tetramer positive after subtraction of background tetramer staining with H-2Dq/NP118–126. These experiments were repeated at least twice with similar results.
Generation of a neu-directed vaccine that does not induce RNEU420–429-specific responses allows further functional characterization of the nondominant neu-specific CD8+ T cell repertoire
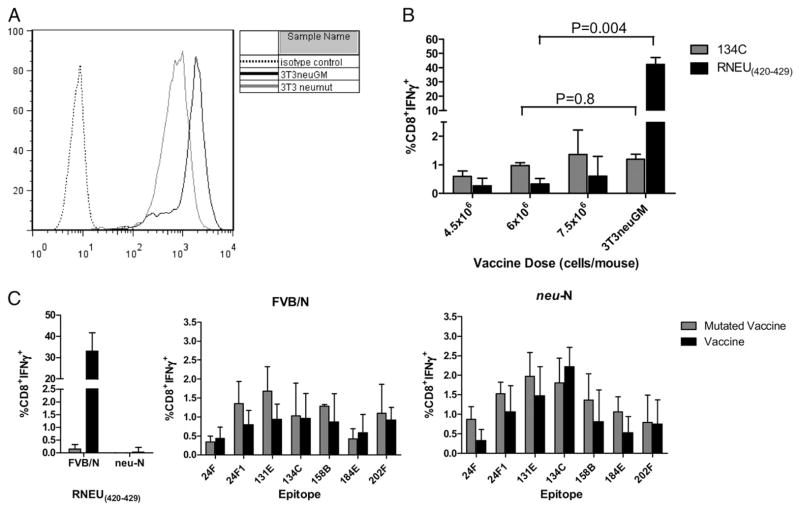
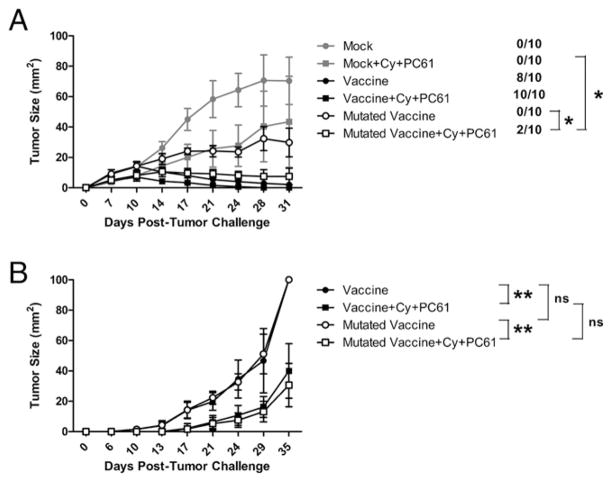
Next, we wanted to address whether the nondominant CD8+ T cell repertoire is capable of generating antitumor immunity in the absence of the dominant RNEU420–429 epitope in both FVB/N and neu-N mice. To accomplish this, we created a mutated vaccine, with an amino acid substitution at one of the anchor residues used to bind the immunodominant RNEU420–429 epitope to H-2Dq. This substitution should prevent binding of the epitope to H-2Dq, and thus presentation to and activation of RNEU420–429-specific T cells. We previously published that the substitution of an alanine residue into either position 4 or 10 of RNEU420–429 abrogates the ability of the peptide to bind H-2Dq (21). We therefore mutated the fourth anchor residue from leucine to alanine using PCR-based site-directed mutagenesis. The mutated rat neu cDNA was then electroporated into NIH-3T3 cells, and clones with the highest neu expression were sorted (Fig. 5A). To validate that the mutated vaccine (3T3neumut) was ineffective at inducing RNEU420–429 T cell responses, we compared lymphocytes isolated from vaccinated FVB/N mice for recognition of both the RNEU420–429 and the 134C epitopes, following a week of in vitro stimulation with the corresponding epitope. As expected, there was a significant decrease in detection of RNEU420–429-specific T cell responses in mice receiving the mutated vaccine (p = 0.004), as great as 100-fold less when compared with the wild-type vaccine. In contrast, responses directed at the 134C epitope were not significantly altered (p = 0.8) with the mutated vaccine (Fig. 5B). Next, immunized lymphocytes from both FVB/N and neu-N mice treated with either the wild-type or mutated vaccine were stimulated for 1 wk in vitro with each of the nondominant epitopes and RNEU420–429 and further assessed for their activation with and without the presence of RNEU420–429-specific T cells. Unexpectedly, we found that the nondominant T cell repertoire is not significantly altered in either FVB/N or neu-N mice (Fig. 5C). This finding led us to address whether the nondominant repertoire was functional enough to compensate for the loss of RNEU420–429-specific responses by eradicating neu-expressing tumors in tumor-bearing mice. In tumor treatment experiments, tumor-bearing FVB/N mice were treated with either the wild-type or mutated vaccine given alone or with Cy and PC61 (to more completely deplete Tregs) and assessed for tumor progression. Although all mice treated with the wild-type vaccine alone cleared tumor, mice treated with the mutated vaccine did not. However, mice treated with the mutated vaccine alone to induce nondominant CD8+ T cell responses demonstrated a stabilization of their tumor burden when compared with mice treated with mock vaccine that is statistically significant (p < 0.003). Interestingly, Cy administered with the mutated vaccine induced complete tumor clearance in 10–20% of FVB/N mice (Fig. 6A). Similar tumor treatment studies conducted in the neu-N mice demonstrated that the mutated vaccine given with Cy and PC61 can also induce significant tumor protection in tolerized mice when compared with mock vaccine (data not shown), neu vaccine, and mutated vaccine alone (p < 0.002) or mock vaccine given with Cy and PC61 (p < 0.01, data not shown) (Fig. 6B). Importantly, tumor outgrowth and survival kinetics remained similar in neu-N mice with or without RNEU420–429-specific CD8+ T cell responses, thus demonstrating that the nondominant CD8+ T cells do play a role in antitumor immunity in tolerized hosts (Fig. 6B).
FIGURE 5.
Alanine substitution at anchor residue four within RNEU420–429 eliminates RNEU420–429-specific but maintains nondominant epitope-specific T cell responses following treatment of neu-N and FVB/N mice with the mutated vaccine. A, The 3T3neumut cell line expresses half the level of neu as 3T3neuGM vaccine cells. Full-length rat neu cDNA was mutated using PCR mutagenic primers and transfected into NIH-3T3 cells. Transfected cells were screened and sorted based on neu expression. B, Vaccination with 3T3neumut generates responses specific for 134C but not RNEU420–429. Three FVB/N mice were vaccinated with varying doses of 3T3neumut along with 3 × 106 3T3GM cells or 3 × 106 3T3neuGM cells. One week later, splenocytes were pulsed with either RNEU420–429 or the 134C epitope for 7 d. Lymphoctyes were isolated and incubated with T2-Dq cells pulsed with the corresponding peptide or NP118–126. Plotted is the mean percentage of CD8+ T cells that are IFN-γ + after subtraction of NP118–126 values. C, Nondominant CD8+ T cell responses are not significantly altered when comparing FVB/N and neu-N mice vaccinated with either the mutated or nonmutated vaccines. Experiments were carried out as described in B using 3 × 106 vaccine or 6 × 106 3T3neumut with 3 × 106 3T3GM cells. These experiments were repeated at least twice with similar results.
FIGURE 6.
Nondominant CD8+ T cell repertoire induces antitumor immunity. A, Combining immune-modulating agents (Cy and PC61) with vaccination leads to tumor clearance in FVB/N mice treated with either 3T3neuGM or 3T3neumut/3T3GM. Listed next to each curve is the proportion of mice that were tumor free at the end of the experiment (day 31). *p < 0.002 for selected groups 14 d posttumor challenge. B, Significant protection in tumor-challenged neu-N mice when treated with Cy and PC61 in combination with the mutated or nonmutated vaccines. Groups of 10 FVB/N (A) and neu-N (B) mice were treated with or without 100 mg/kg Cy and 50 μg PC61/mouse 2 d after tumor injection (2 × 106 and 5 × 104 NT2.5 cells, respectively). Mice were vaccinated 3 d posttumor injection with either 3T3neuGM or 3T3neumut/3T3GM cells. All experiments were repeated three times with similar results. **p < 0.002.
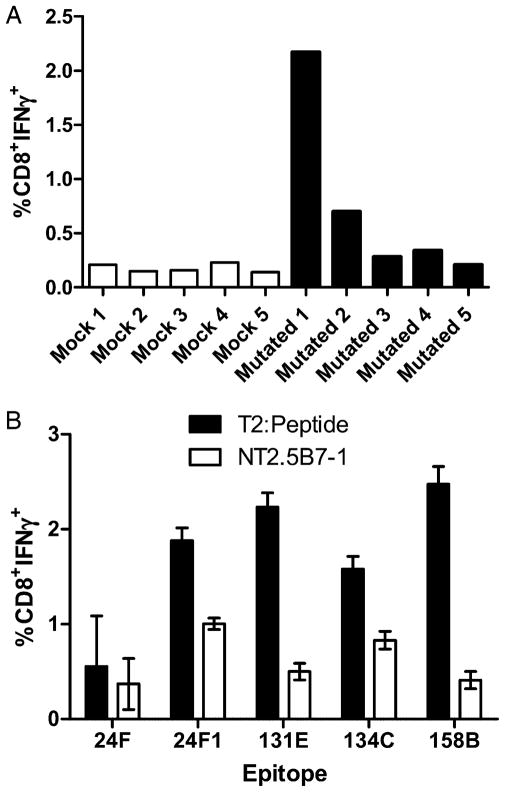
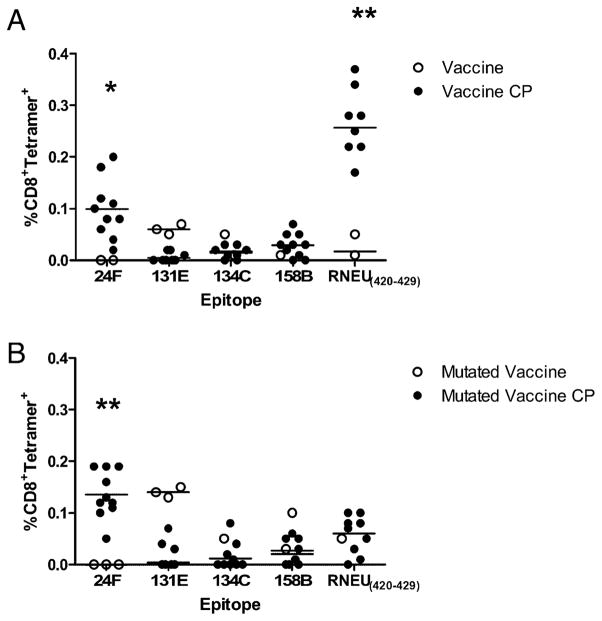
To better understand the function of the T cells directed at the nondominant epitopes, we analyzed the ability of this repertoire, in the absence of RNEU420–429-specific CD8+ T cell responses, to recognize neu-expressing mammary tumor cells straight ex vivo as well as after a 1-wk in vitro stimulation with each specific non-dominant epitope. Lymphocytes from tumor-bearing mice treated with Cy and PC61 and either mock or mutated vaccine were isolated and analyzed for IFN-γ secretion after incubation with NT2.5B7-1 tumor cells. The CD8+ T cell repertoire from the mutated vaccine-treated mice demonstrated enhanced recognition of tumor cells in three of the five mice in comparison with mock-treated mice (range, 0.14–0.23%) (Fig. 7A). To demonstrate that the nondominant CD8+ T cells we identified can recognize naturally processed neu epitopes on neu-expressing mammary tumors, we stimulated splenocytes from both the mock and mutated vaccine-treated mice in Fig. 7A with five of our identified epitopes. Results from these studies show that these nondominant CD8+ T cell responses do indeed recognize naturally processed epitopes on the mammary tumor cells (Fig. 7B). Additionally, we compared tetramer staining of four individual T cell populations from tumor bearing hosts straight ex vivo. Lymphocytes from tumor-bearing mice treated with or without Cy and PC61 and either the wild-type or mutated vaccine were also analyzed to dissect how Treg-mediated immune tolerance affects the neu-N CD8+ T cell repertoire. These studies revealed that 24F epitope-specific T cell responses are the only nondominant T cells enhanced after immune modulation with both the wild-type and mutated vaccines as measured by tetramer staining. All other nondominant responses tested were not significantly enhanced except for the RNEU420–429-specific responses in wild-type vaccine-treated mice (Fig. 8). Interestingly, the response to the 9-aa peptide 24F1 was not enhanced in comparison with the responses specific for the 10-aa peptide 24F (data not shown). These data suggest that only some epitope-specific T cell responses (likely the higher avidity responses such as RNEU420–429 and 24F targeted responses) are regulated by Tregs, whereas other immune regulatory mechanisms may alter the other epitope-specific T cell responses.
FIGURE 7.
Nondominant CD8+ T cell repertoire recognizes neu-expressing tumors. A, Enhanced tumor-specific CD8+ T cells responses are observed in tumor-challenged FVB/N mice that were depleted of Tregs and treated with the mutated vaccine. Tumor-bearing FVB/N mice were treated with 100 mg/kg Cy and 50 μg PC61/mouse 2 d after tumor injection. Mice were vaccinated 3 d posttumor injection with either 3T3GM or 3T3neu-mut/3T3GM cells. Fourteen days posttumor challenge, CD8+ T cells were isolated and stained with anti-CD8 and anti–IFN-γ after a 5-h incubation with NT2.5B7-1 tumor cells. B, Nondominant CD8+ T cell responses recognize neu-expressing tumors. Splenocytes isolated from mice in A were stimulated with peptide for 1 wk in vitro. Lymphoctyes were isolated and incubated with either T2-Dq cells pulsed with the corresponding peptide or NT2.5B7-1 tumor cells. Plotted is the mean percentage of CD8+ T cells that are IFN-γ + after subtraction of the mock values. These experiments were repeated at least twice with similar results.
FIGURE 8.
Nondominant CD8+ T cell repertoire is tolerized by Tregs in neu-N mice. A, Responses to 24F and RNEU420–429 epitopes are enhanced in tumor-bearing neu-N mice treated with Cy and PC61 in combination with 3T3neuGM vaccine. B, Responses to the 24F epitope are enhanced in tumor-bearing neu-N mice treated with Cy and PC61 in combination with the mutated vaccine (3T3neumut/3T3GM). Mice were tumor challenged and treated as described in Fig. 6. Fourteen days posttumor challenge, CD8+ T cells were isolated and stained with anti-CD8 and a 1:200 dilution (5 μg/ml of tetramer protein) of H-2Dq/peptide tetramer straight ex vivo. These experiments were repeated at least twice with similar results. *p = 0.014, **p < 0.0005. CP, Cy plus PC61.
Discussion
The studies presented in this work report four new findings. First, we identified six novel 10-mer and one novel 9-mer neu-specific epitopes using vaccine-induced lymphocytes to screen an overlapping 15-aa peptide library of the entire rat neu protein. Second, this nondominant T cell repertoire is capable of recognizing tumors in vitro and in vivo following vaccination in sequence with immune-modulating agents that alleviate Treg suppression. Third, the higher avidity nondominant CD8+ T cell responses are also subject to the Treg suppression that regulates the dominant RNEU420–429 T cell response in neu-N mice. Fourth, analysis of this T cell repertoire revealed a hierarchy of dominance among the neu-N CD8+ T cell repertoire based on both functional and relative TCR avidities.
The identification and validation of six 10-mer and one 9-mer neu-specific epitopes was the first step in understanding the role that the nondominant CD8+ T cell repertoire plays in the neu-directed antitumor response. It was also important to screen the entire neu sequence to obtain a complete epitope analysis to better comprehend how different CD8+ T cell populations respond to tumor-directed vaccination and therapy. Based on the size of neu (185 kDa) and the lack of previously identified H-2Dq binding epitopes in databases, the most effective strategy was to use an overlapping 15-aa peptide library. This methodology has been extensively used in viral research to fully characterize HIV- (27, 28), hepatitis C virus- (29), and human papillomavirus-specific (30) epitopes. To our knowledge, this is the first example employing this approach to identify the epitopes contained within a cancer Ag that are recognized by the complete CD8+ T cell repertoire. This screening process was effective in identifying epitopes located throughout all domains of the neu protein. Singh and Paterson (31, 32) previously reported the identification of eight new neu-specific CD8+ T cell epitopes recognized by lymphocytes isolated from FVB/N mice following vaccination with a Listeria monocytogenes bacterial delivery system, with each vector expressing an overlapping neu gene fragment. Five overlapping fragments were screened, and additional studies demonstrated a hierarchy of T cell responses based on a dilutional in vitro lytic assay. Interestingly, the epitopes identified using the L. monocytogenes vaccine approach were distinct from those identified with our vaccine platform (31–33). Importantly, in contrast to our findings, T cell responses induced by the L. mono-cytogenes vaccines against these epitopes did not correlate with tumor regression in the neu-N mice (32).
Several factors may account for the difference in epitopes identified by the two different vaccine approaches. First, it is possible that the two different vaccine approaches provide different mechanisms of Ag access to dendritic cells (DCs), the most likely APC involved in processing and presenting epitopes for priming of CD8+ T cells. In addition to the classical MHC class I presentation of Ag, at least two different intracellular pathways involved in cross-presentation of antigenic epitopes have already been described (34). There are several examples of differential epitope presentation based on which processing pathway the Ag enters (35–37). L. monocytogenes is known to initially gain entry through the lysosomal compartment and subsequently releases Ag directly into the cytosol via the enzyme listeriolysin O (38–40). Our group has shown that whole-cell GM-CSF–secreting vaccine cells are taken up exogenously and induce cross-priming by DCs in both animal (41) and human studies (42), although the intracellular mechanism by which Ags are processed for presentation to CD8+ T cells is still not understood. A second potential explanation is that the two different vaccine approaches predominantly access different subpopulations of DCs or other APCs. L. monocytogenes vaccines are known to access macrophages as well as DCs, and there is the potential for differential Ag processing and presentation among these two cell populations (43, 44). Finally, the cytokine milieu induced by the two approaches may activate the APCs differently, resulting in different mechanisms of Ag processing and presentation. L. monocytogenes vaccines are known to engage the innate immune response quite effectively through signals such as TLRs, but they do not induce B cell responses. In contrast, the GM-CSF–secreting vaccine cells produce a predominant adaptive response including B cell responses that even facilitate cross-priming of CD8+ Ag-specific immunity (45–47). Abs specific for neu have been shown to bind neu on the vaccinating cells, facilitating tumor cell uptake by CD8− DCs and resulting in enhanced in vivo tumor clearance (45). In fact, this mechanism might also explain why the GM-CSF–secreting vaccine cell approach induces more potent in vivo tumor regressions than observed with the L. monocytogenes approach (data not shown) (32). Additionally, our approach failed to induce T cell responses against the L. monocytogenes vaccine-derived epitopes (data not shown).
The use of an overlapping peptide library to screen an entire protein to identify a more complete panel of CD8+ T cell epitopes proved both effective and timely, and it identified additional epitopes that play a role in the in vivo eradication of tumors. Interestingly, T cell responses against these epitopes are regulated by tolerance mechanisms, including Tregs, even in the FVB/N mice in which the neu protein should be viewed as foreign. Additionally, the T cell responses against the alternative epitopes are less avid than the T cell responses to the immunodominant epitope, RNEU420–429, which are only subject to significant regulation by Tregs in the tolerized neu-N mice (6). These findings suggest that other T cell parameters, in addition to avidity, provide signals to induce mechanisms of T cell suppression in tumor-bearing hosts. Additional support for this concept comes from the fact that the T cell populations specific for the epitopes identified with the L. monocytogenes approach were also subject to Treg suppression in the nontolerant mice even though these populations were of higher avidity than the T cell populations identified by our approach. In contrast to our findings, the inhibition of Tregs did not result in enhanced in vivo tumor eradication with the L. monocytogenes vaccine approach (32). Additional studies are underway to better understand the in vivo mechanisms regulating these alternative T cell populations.
One potential technical issue that occurred was the observation of nonspecific cross-reactivity between positively identified 15-aa peptides that bound to MHC class I and were recognized by neu-specific T cells. However, pulsing micromolar concentrations of peptide onto APCs bypasses the endogenous processing and presentation machinery of APCs, which has been shown to lead to false-positives (48). This problem is easily addressed by a peptide validation scheme that eliminates epitopes that are lost to detection in assays that aim to determine the limits of detection by measures of avidity and function. Thus, peptides identified through this screening process must be further validated functionally as previously reported (49).
Analyses of this more complete CD8+ T cell repertoire also revealed a hierarchy of dominance among the neu-N CD8+ T cell repertoire based on both functional and relative TCR avidity. Specifically, we demonstrated that three of the six 10-aa epitopes were more dominant based on their binding affinities to H-2Dq and activation by low concentrations of Ag. Interestingly, one of the 10-aa peptide responses identified (24F) has a corresponding 9-aa peptide that shows enhanced IFN-γ secretion and MHC class I binding. This finding was surprising to us since we have previously shown that the amino acids at positions 4 and 10 are the critical amino acids for peptide binding to H-2Dq using an alanine scan that replaced each amino acid within RNEU420–429 (21). These data indicate that the T cell responses specific for the 9-aa peptide may have a proliferative advantage over the 10-aa–specific responses even though both responses have similar relative TCR avidities. A hierarchy of CD8+ T cell populations based on functional avidity has been shown in several other models, including an influenza viral model and a hepatocellular carcinoma model (49, 50). Interestingly, although we originally hypothesized that the relative TCR avidities of the nondominant CD8+ T cell responses would correlate with this functional avidity hierarchy (6, 51–53), tetramer staining analyses revealed that functional dominance may not always correlate with relative TCR avidities for all responses. These findings are supported by several other studies (54–58). In particular, Derby et al. (54) suggested that these discrepancies may be due to variable TCR expression and TCR signaling efficiency. However, this would not account for our findings because expression of CD8a and TCR remains similar among the nondominant neu epitope-specific T cell populations (data not shown). However, this does not rule out the possibility that TCR signaling efficiency may be diminished for each of these T cell responses based on changes in the localization of accessory molecules at the immunological synapse or other internal signaling molecules. Thus, the explanation for these differences remains unclear and warrants further investigation. Importantly, these data also highlight the need to better characterize more complete Ag-specific CD8+ T cell populations based on multiple functional parameters. These studies also caution against vaccination with a single peptide based on our current understanding of the function of different epitope-specific CD8+ T cell populations within the complete T cell repertoire.
Further evaluation of the nondominant CD8+ T cell hierarchies in the absence of the immunodominant T cell response were carried out to uncover functionally important T cell populations that might be suppressed by the dominant response. We theorized that the elimination of the immunodominant CD8+ T cell response would create a new hierarchy of CD8+ T cell responses, where the nondominant T cell responses would not be outcompeted for either immunologic resources or space. Evaluation of CD8+ T cell responses in both the neu-N and FVB/N mice demonstrate that regardless of whether the immunodominant response is present, the relative functional hierarchy and magnitude of T cell responses directed against nondominant epitopes remain unchanged. Similar observations were made by both Andreansky et al. (59) and Kedzierska et al. (60) in influenza A viral infection models. These finding may be explained by the correlations between TCR avidity and the T cell population burst size and duration of the expansion period (61). However, this does not rule out the possibility that the mutated vaccine may uncover other, unidentified responses that may shape and alter the nondominant CD8+ T cell repertoire. Additionally, it is possible that repeated boosting vaccinations might further alter the T cell repertoire toward a larger, more avid and functional overall response. The identification of these additional epitopes will allow further exploration of this question.
There is also evidence supporting the concept that TCR avidity directly affects the in vivo efficacy of CD8+ T cell responses, where more avid responses correlate with enhanced antitumor activity (62, 63). However, many of the Ags that are expressed on tumors are also expressed at low levels on normal tissue, which results in either thymic depletion or peripheral suppression of the most avid T cell responses in cancer patients, leaving only a blunted T cell response for activation by immunotherapies (64). Along these lines, both our previously published and unpublished data suggest that the nondominant neu-N CD8+ T cell repertoire is dysfunctional, and that only uncovering high-avidity RNEU420–429-specific responses confers tumor protection (6). However, these new findings evaluating vaccination with a mutated vaccine lacking the dominant T cell epitope suggest that the nondominant CD8+ T cell repertoire does indeed generate antitumor immune responses when Tregs are first inhibited. These data are further supported by the ability of the nondominant CD8+ T cell repertoire to recognize tumor in our in vitro assays. These observations are in contrast to data from Dobaño et al. (65), who showed that subdominant CD8+ and CD4+ T cell responses cannot compensate for the loss of the immunodominant response after Plasmodium yoelii circum-sporozoite protein DNA vaccination against P. yoelii. In their studies, the subdominant repertoire could only partially compensate against parasitic challenge after vaccination. However, in the neu-N model, the kinetics of tumor outgrowth and tumor-free survival of mice treated with either the mutated or nonmutated vaccine overlap. These data are supported by other published studies demonstrating the efficacy of low-avidity T cell responses against developing tumors (13–15). It is still not clear why tumor-free survival is not enhanced either with or without the presence of the immunodominant T cell response. We suspect that this may be partially due to the available CD8+ T cell precursor frequency. Another possibility may be the need to prolong the length of time that Tregs are suppressed, a question difficult to address without more specific Treg-depleting agents that will not also deplete activated T cells following vaccination. Treg populations recover 2 wk after administration of Cy (6) and the IL-2 receptor Ab PC61 (66).
It is also possible that additional peripheral tolerance mechanisms are providing inactivation signals to the nondominant T cell populations. Several published studies suggest that the non-dominant, or low-avidity, T cell responses escape the effects of central and peripheral tolerance (10–12). However, our tumor survival curves and tetramer data propose that the nondominant CD8+ T cell repertoire is regulated by peripheral tolerization mechanisms even in the absence of RNEU420–429-specific responses. Additional support for our findings come from Lyman et al. (17), where they demonstrated that low-avidity HA-specific CD8+ T cells that are adoptively transferred into InsHA mice are also tolerized. These data further emphasize the importance of addressing and blocking immune regulatory pathways to generate more effective cancer vaccines. Studies are currently underway to better elucidate the differences in T cell function between neu-N mice that demonstrate tumor regression versus progression.
In summary, these data provide a detailed characterization of a CD8+ T cell repertoire specific for a novel panel of neu-derived nondominant antigenic epitopes. The findings presented identify a functional hierarchy of T cell subpopulations when defined by both functional capability and TCR avidity measures. These findings also demonstrate the complex interactions that exist within an expanded T cell repertoire specific for just one Ag. These data disprove the prior dogma that high-avidity T cell responses for dominant tumor Ags are irreversibly tolerized, leaving in place for vaccine induction only the nontolerized, low-avidity, and nondominant antigenic T cell responses. Finally, these data provide strong support for the requirement of immune-modulating agents to enhance vaccine induction of all components of the T cell repertoire. These findings also caution against the use of immunization approaches targeting only one antigenic epitope.
Supplementary Material
Acknowledgments
We thank Dr. Barry Kobrin and Dr. Leisha Emens for scientific and technical assistance on this project.
This work was supported by National Cooperative Drug Discovery Groups National Institutes of Health/National Cancer Institute Grant 519CA113341, National Institutes of Health/National Cancer Institute Grant RO1 1R01CA122081, Specialized Program of Research Excellence in Breast Cancer Grant P50CA088843, and National Institutes of Health Training Grant 5-T32 AI007247. E.M.J. is the first recipient of the Dana and Albert “Cubby” Broccoli Professorship in Oncology.
Abbreviations used in this article
- Cy
cyclophosphamide
- DC
dendritic cell
- neu
Her-2/neu
- neu-N
Her-2/neu transgenic
- NP
nucleoprotein
- TAA
tumor-associated Ag
- Treg
T regulatory cell
Footnotes
The online version of this article contains supplemental material.
Disclosures
This work describes the use of a GM-CSF–secreting vaccine. Although none of the investigators on this manuscript has a financial interest, The Johns Hopkins University has the potential to receive royalties on the human vaccine equivalent described in this article through a licensing agreement with BioSante. This potential conflict of interest is being managed by the Conflict of Interest Committee.
References
- 1.Kawakami Y, Eliyahu S, Sakaguchi K, Robbins PF, Rivoltini L, Yannelli JR, Appella E, Rosenberg SA. Identification of the immunodominant peptides of the MART-1 human melanoma antigen recognized by the majority of HLA-A2-restricted tumor infiltrating lymphocytes. J Exp Med. 1994;180:347–352. doi: 10.1084/jem.180.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fisk B, Blevins TL, Wharton JT, Ioannides CG. Identification of an immunodominant peptide of HER-2/neu protooncogene recognized by ovarian tumor-specific cytotoxic T lymphocyte lines. J Exp Med. 1995;181:2109–2117. doi: 10.1084/jem.181.6.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, McCluskey J. Immunodominance and immunodomination: critical factors in developing effective CD8+ T-cell-based cancer vaccines. Adv Cancer Res. 2006;95:203–247. doi: 10.1016/S0065-230X(06)95006-4. [DOI] [PubMed] [Google Scholar]
- 4.Sercarz EE, Lehmann PV, Ametani A, Benichou G, Miller A, Moudgil K. Dominance and crypticity of T cell antigenic determinants. Annu Rev Immunol. 1993;11:729–766. doi: 10.1146/annurev.iy.11.040193.003501. [DOI] [PubMed] [Google Scholar]
- 5.Yewdell JW. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity. 2006;25:533–543. doi: 10.1016/j.immuni.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Ercolini AM, Ladle BH, Manning EA, Pfannenstiel LW, Armstrong TD, Machiels JP, Bieler JG, Emens LA, Reilly RT, Jaffee EM. Recruitment of latent pools of high-avidity CD8+ T cells to the antitumor immune response. J Exp Med. 2005;201:1591–1602. doi: 10.1084/jem.20042167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutoit V, Rubio-Godoy V, Dietrich PY, Quiqueres AL, Schnuriger V, Rimoldi D, Liénard D, Speiser D, Guillaume P, Batard P, et al. Heterogeneous T-cell response to MAGE-A10254–262: high avidity-specific cytolytic T lymphocytes show superior antitumor activity. Cancer Res. 2001;61:5850–5856. [PubMed] [Google Scholar]
- 8.Zeh HJ, III, Perry-Lalley D, Dudley ME, Rosenberg SA, Yang JC. High avidity CTLs for two self-antigens demonstrate superior in vitro and in vivo antitumor efficacy. J Immunol. 1999;162:989–994. [PubMed] [Google Scholar]
- 9.Lewis JD, Reilly BD, Bright RK. Tumor-associated antigens: from discovery to immunity. Int Rev Immunol. 2003;22:81–112. doi: 10.1080/08830180305221. [DOI] [PubMed] [Google Scholar]
- 10.Cibotti R, Kanellopoulos JM, Cabaniols JP, Halle-Panenko O, Kosmatopoulos K, Sercarz E, Kourilsky P. Tolerance to a self-protein involves its immunodominant but does not involve its subdominant determinants. Proc Natl Acad Sci USA. 1992;89:416–420. doi: 10.1073/pnas.89.1.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grossmann ME, Davila T, Celis T. Avoiding tolerance against prostatic antigens with subdominant peptide epitopes. J Immunother. 2001;24:237–241. [PubMed] [Google Scholar]
- 12.Zehn D, Bevan MJ. T cells with low avidity for a tissue-restricted antigen routinely evade central and peripheral tolerance and cause autoimmunity. Immunity. 2006;25:261–270. doi: 10.1016/j.immuni.2006.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgan DJ, Kreuwel HT, Fleck S, Levitsky HI, Pardoll DM, Sherman LA. Activation of low avidity CTL specific for a self epitope results in tumor rejection but not autoimmunity. J Immunol. 1998;160:643–651. [PubMed] [Google Scholar]
- 14.Scardino A, Gross DA, Alves P, Schultze JL, Graff-Dubois S, Faure O, Tourdot S, Chouaib S, Nadler LM, Lemonnier FA, et al. HER-2/neu and hTERT cryptic epitopes as novel targets for broad spectrum tumor immunotherapy. J Immunol. 2002;168:5900–5906. doi: 10.4049/jimmunol.168.11.5900. [DOI] [PubMed] [Google Scholar]
- 15.Cordaro TA, de Visser KE, Tirion FH, Schumacher TN, Kruisbeek AM. Can the low-avidity self-specific T cell repertoire be exploited for tumor rejection? J Immunol. 2002;168:651–660. doi: 10.4049/jimmunol.168.2.651. [DOI] [PubMed] [Google Scholar]
- 16.Reilly RT, Gottlieb MB, Ercolini AM, Machiels JP, Kane CE, Okoye FI, Muller WJ, Dixon KH, Jaffee EM. HER-2/neu is a tumor rejection target in tolerized HER-2/neu transgenic mice. Cancer Res. 2000;60:3569–3576. [PubMed] [Google Scholar]
- 17.Lyman MA, Nugent CT, Marquardt KL, Biggs JA, Pamer EG, Sherman LA. The fate of low affinity tumor-specific CD8+ T cells in tumor-bearing mice. J Immunol. 2005;174:2563–2572. doi: 10.4049/jimmunol.174.5.2563. [DOI] [PubMed] [Google Scholar]
- 18.Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci USA. 1992;89:10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murata S, Ladle BH, Kim PS, Lutz ER, Wolpoe ME, Ivie SE, Smith HM, Armstrong TD, Emens LA, Jaffee EM, Reilly RT. OX40 costimulation synergizes with GM-CSF whole-cell vaccination to overcome established CD8+ T cell tolerance to an endogenous tumor antigen. J Immunol. 2006;176:974–983. doi: 10.4049/jimmunol.176.2.974. [DOI] [PubMed] [Google Scholar]
- 20.Machiels JP, Reilly RT, Emens LA, Ercolini AM, Lei RY, Weintraub D, Okoye FI, Jaffee EM. Cyclophosphamide, doxorubicin, and paclitaxel enhance the antitumor immune response of granulocyte/macrophage-colony stimulating factor-secreting whole-cell vaccines in HER-2/neu tolerized mice. Cancer Res. 2001;61:3689–3697. [PubMed] [Google Scholar]
- 21.Ercolini AM, Machiels JPH, Chen YC, Slansky JE, Giedlen M, Reilly RT, Jaffee EM. Identification and characterization of the immunodominant rat HER-2/neu MHC class I epitope presented by spontaneous mammary tumors from HER-2/neu-transgenic mice. J Immunol. 2003;170:4273–4280. doi: 10.4049/jimmunol.170.8.4273. [DOI] [PubMed] [Google Scholar]
- 22.Manning EA, Ullman JG, Leatherman JM, Asquith JM, Hansen TR, Armstrong TD, Hicklin DJ, Jaffee EM, Emens LA. A vascular endothelial growth factor receptor-2 inhibitor enhances antitumor immunity through an immune-based mechanism. Clin Cancer Res. 2007;13:3951–3959. doi: 10.1158/1078-0432.CCR-07-0374. [DOI] [PubMed] [Google Scholar]
- 23.Jaffee EM, Thomas MC, Huang AY, Hauda KM, Levitsky HI, Pardoll DM. Enhanced immune priming with spatial distribution of paracrine cytokine vaccines. J Immunother Emphasis Tumor Immunol. 1996;19:176–183. doi: 10.1097/00002371-199605000-00002. [DOI] [PubMed] [Google Scholar]
- 24.Schulz M, Aichele P, Schneider R, Hansen TH, Zinkernagel RM, Hengartner H. Major histocompatibility complex binding and T cell recognition of a viral nonapeptide containing a minimal tetrapeptide. Eur J Immunol. 1991;21:1181–1185. doi: 10.1002/eji.1830210513. [DOI] [PubMed] [Google Scholar]
- 25.Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- 26.McMahan RH, Slansky JE. Mobilizing the low-avidity T cell repertoire to kill tumors. Semin Cancer Biol. 2007;17:317–329. doi: 10.1016/j.semcancer.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beattie T, Kaul R, Rostron T, Dong T, Easterbrook P, Jaoko W, Kimani J, Plummer F, McMichael A, Rowland-Jones S. Screening for HIV-specific T-cell responses using overlapping 15-mer peptide pools or optimized epitopes. AIDS. 2004;18:1595–1598. doi: 10.1097/01.aids.0000131362.82951.b2. [DOI] [PubMed] [Google Scholar]
- 28.Kitano M, Kobayashi N, Kawashima Y, Akahoshi T, Nokihara K, Oka S, Takighuchi M. Identification and characterization of HLA-B*5401-restricted HIV-1-Nef and Pol-specific CTL epitopes. Microbes Infect. 2008;10:764–772. doi: 10.1016/j.micinf.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 29.Martin P, Parroche P, Chatel L, Barretto C, Beck A, Trépo C, Bain C, Lone YC, Inchauspé G, Fournillier A. Genetic immunization and comprehensive screening approaches in HLA-A2 transgenic mice lead to the identification of three novel epitopes in hepatitis C virus NS3 antigen. J Med Virol. 2004;74:397–405. doi: 10.1002/jmv.20189. [DOI] [PubMed] [Google Scholar]
- 30.Todd RW, Steele JC, Etherington I, Luesley DM. Detection of CD8+ T cell responses to human papillomavirus type 16 antigens in women using imiquimod as a treatment for high-grade vulval intraepithelial neoplasia. Gynecol Oncol. 2004;92:167–174. doi: 10.1016/j.ygyno.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 31.Singh R, Paterson Y. Vaccination strategy determines the emergence and dominance of CD8+ T-cell epitopes in a FVB/N rat HER-2/neu mouse model of breast cancer. Cancer Res. 2006;66:7748–7757. doi: 10.1158/0008-5472.CAN-05-4469. [DOI] [PubMed] [Google Scholar]
- 32.Singh R, Paterson Y. In the FVB/N HER-2/neu transgenic mouse both peripheral and central tolerance limit the immune response targeting HER-2/neu induced by Listeria monocytogenes-based vaccines. Cancer Immunol Immunother. 2007;56:927–938. doi: 10.1007/s00262-006-0237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh R, Paterson Y. Immunoediting sculpts tumor epitopes during immunotherapy. Cancer Res. 2007;67:1887–1892. doi: 10.1158/0008-5472.CAN-06-3960. [DOI] [PubMed] [Google Scholar]
- 34.Rock KL, Shen L. Cross-presentation: underlying mechanisms and role in immune surveillance. Immunol Rev. 2005;207:166–183. doi: 10.1111/j.0105-2896.2005.00301.x. [DOI] [PubMed] [Google Scholar]
- 35.Wolkers MC, Brouwenstijn N, Bakker AH, Toebes M, Schumacher TN. Antigen bias in T cell cross-priming. Science. 2004;304:1314–1317. doi: 10.1126/science.1096268. [DOI] [PubMed] [Google Scholar]
- 36.Shen X, Wong SB, Buck CB, Zhang J, Siliciano RF. Direct priming and cross-priming contribute differentially to the induction of CD8+ CTL following exposure to vaccinia virus via different routes. J Immunol. 2002;169:4222–4229. doi: 10.4049/jimmunol.169.8.4222. [DOI] [PubMed] [Google Scholar]
- 37.Donohue KB, Grant JM, Tewalt EF, Palmer DC, Theoret MR, Restifo NP, Norbury CC. Cross-priming utilizes antigen not available to the direct presentation pathway. Immunology. 2006;119:63–73. doi: 10.1111/j.1365-2567.2006.02406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berche P, Gaillard JL, Sansonetti PJ. Intracellular growth of Listeria monocytogenes as a prerequisite for in vivo induction of T cell-mediated immunity. J Immunol. 1987;138:2266–2271. [PubMed] [Google Scholar]
- 39.Brunt LM, Portnoy DA, Unanue ER. Presentation of Listeria monocytogenes to CD8+ T cells requires secretion of hemolysin and intracellular bacterial growth. J Immunol. 1990;145:3540–3546. [PubMed] [Google Scholar]
- 40.Schnupf P, Portnoy DA. Listeriolysin O: a phagosome-specific lysin. Microbes Infect. 2007;9:1176–1187. doi: 10.1016/j.micinf.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 41.Huang AY, Golumbek P, Ahmadzadeh M, Jaffee E, Pardoll D, Levitsky H. Role of bone marrow-derived cells in presenting MHC class I-restricted tumor antigens. Science. 1994;264:961–965. doi: 10.1126/science.7513904. [DOI] [PubMed] [Google Scholar]
- 42.Thomas AM, Santarsiero LM, Lutz ER, Armstrong TD, Chen YC, Huang LQ, Laheru DA, Goggins M, Hruban RH, Jaffee EM. Mesothelin-specific CD8+ T cell responses provide evidence of in vivo cross-priming by antigen-presenting cells in vaccinated pancreatic cancer patients. J Exp Med. 2004;200:297–306. doi: 10.1084/jem.20031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stier EM, Mandal M, Lee KD. Differential cytosolic delivery and presentation of antigen by listeriolysin O-liposomes to macrophages and dendritic cells. Mol Pharm. 2005;2:74–82. doi: 10.1021/mp049896v. [DOI] [PubMed] [Google Scholar]
- 44.Wick MJ, Ljunggren HG. Processing of bacterial antigens for peptide presentation on MHC class I molecules. Immunol Rev. 1999;172:153–162. doi: 10.1111/j.1600-065x.1999.tb01363.x. [DOI] [PubMed] [Google Scholar]
- 45.Kim PS, Armstrong TD, Song H, Wolpoe ME, Weiss V, Manning EA, Huang LQ, Murata S, Sgouros G, Emens LA, et al. Antibody association with HER-2/neu-targeted vaccine enhances CD8 T cell responses in mice through Fc-mediated activation of DCs. J Clin Invest. 2008;118:1700–1711. doi: 10.1172/JCI34333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dhodapkar KM, Krasovsky J, Williamson B, Dhodapkar MV. Antitumor monoclonal antibodies enhance cross-presentation of cellular antigens and the generation of myeloma-specific killer T cells by dendritic cells. J Exp Med. 2002;195:125–133. doi: 10.1084/jem.20011097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.den Haan JM, Bevan MJ. Constitutive versus activation-dependent cross-presentation of immune complexes by CD8+ and CD8− dendritic cells in vivo. J Exp Med. 2002;196:817–827. doi: 10.1084/jem.20020295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yewdell JW. The seven dirty little secrets of major histocompatibility complex class I antigen processing. Immunol Rev. 2005;207:8–18. doi: 10.1111/j.0105-2896.2005.00309.x. [DOI] [PubMed] [Google Scholar]
- 49.Belz GT, Xie W, Doherty PC. Diversity of epitope and cytokine profiles for primary and secondary influenza a virus-specific CD8+ T cell responses. J Immunol. 2001;166:4627–4633. doi: 10.4049/jimmunol.166.7.4627. [DOI] [PubMed] [Google Scholar]
- 50.Liu Y, Daley S, Evdokimova VN, Zdobinski DD, Potter DM, Butterfield LH. Hierarchy of alpha fetoprotein (AFP)-specific T cell responses in subjects with AFP-positive hepatocellular cancer. J Immunol. 2006;177:712–721. doi: 10.4049/jimmunol.177.1.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yee C, Savage PA, Lee PP, Davis MM, Greenberg PD. Isolation of high avidity melanoma-reactive CTL from heterogeneous populations using peptide-MHC tetramers. J Immunol. 1999;162:2227–2234. [PubMed] [Google Scholar]
- 52.Hernández J, Lee PP, Davis MM, Sherman LA. The use of HLA A2.1/p53 peptide tetramers to visualize the impact of self tolerance on the TCR repertoire. J Immunol. 2000;164:596–602. doi: 10.4049/jimmunol.164.2.596. [DOI] [PubMed] [Google Scholar]
- 53.Dutoit V, Rubio-Godoy V, Doucey MA, Batard P, Liénard D, Rimoldi D, Speiser D, Guillaume P, Cerottini JC, Romero P, Valmori D. Functional avidity of tumor antigen-specific CTL recognition directly correlates with the stability of MHC/peptide multimer binding to TCR. J Immunol. 2002;168:1167–1171. doi: 10.4049/jimmunol.168.3.1167. [DOI] [PubMed] [Google Scholar]
- 54.Derby MA, Wang J, Margulies DH, Berzofsky JA. Two intermediate-avidity cytotoxic T lymphocyte clones with a disparity between functional avidity and MHC tetramer staining. Int Immunol. 2001;13:817–824. doi: 10.1093/intimm/13.6.817. [DOI] [PubMed] [Google Scholar]
- 55.Rubio-Godoy V, Dutoit V, Rimoldi D, Lienard D, Lejeune F, Speiser D, Guillaume P, Cerottini JC, Romero P, Valmori D. Discrepancy between ELISPOT IFN-γ secretion and binding of A2/peptide multimers to TCR reveals interclonal dissociation of CTL effector function from TCR-peptide/MHC complexes half-life. Proc Natl Acad Sci USA. 2001;98:10302–10307. doi: 10.1073/pnas.181348898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Villacres MC, Lacey SF, Auge C, Longmate J, Leedom JM, Diamond DJ. Relevance of peptide avidity to the T cell receptor for cytomegalovirus-specific ex vivo CD8 T cell cytotoxicity. J Infect Dis. 2003;188:908–918. doi: 10.1086/377582. [DOI] [PubMed] [Google Scholar]
- 57.La Gruta NL, Turner SJ, Doherty PC. Hierarchies in cytokine expression profiles for acute and resolving influenza virus-specific CD8+ T cell responses: correlation of cytokine profile and TCR avidity. J Immunol. 2004;172:5553–5560. doi: 10.4049/jimmunol.172.9.5553. [DOI] [PubMed] [Google Scholar]
- 58.Moore TV, Lyons GE, Brasic N, Roszkowski JJ, Voelkl S, Mackensen A, Kast WM, Le Poole IC, Nishimura MI. Relationship between CD8-dependent antigen recognition, T cell functional avidity, and tumor cell recognition. Cancer Immunol Immunother. 2009;58:719–728. doi: 10.1007/s00262-008-0594-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andreansky SS, Stambas J, Thomas PG, Xie W, Webby RJ, Doherty PC. Consequences of immunodominant epitope deletion for minor influenza virus-specific CD8+-T-cell responses. J Virol. 2005;79:4329–4339. doi: 10.1128/JVI.79.7.4329-4339.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kedzierska K, Guillonneau C, Gras S, Hatton LA, Webby R, Purcell AW, Rossjohn J, Doherty PC, Turner SJ. Complete modification of TCR specificity and repertoire selection does not perturb a CD8+ T cell immunodominance hierarchy. Proc Natl Acad Sci USA. 2008;105:19408–19413. doi: 10.1073/pnas.0810274105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zehn D, Lee SY, Bevan MJ. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 2009;458:211–214. doi: 10.1038/nature07657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alexander-Miller MA. High-avidity CD8+ T cells: optimal soldiers in the war against viruses and tumors. Immunol Res. 2005;31:13–24. doi: 10.1385/IR:31:1:13. [DOI] [PubMed] [Google Scholar]
- 63.Laheru D, Lutz E, Burke J, Biedrzycki B, Solt S, Onners B, Tartakovsky I, Nemunaitis J, Le D, Sugar E, et al. Allogeneic granulocyte macrophage colony-stimulating factor-secreting tumor immunotherapy alone or in sequence with cyclophosphamide for metastatic pancreatic cancer: a pilot study of safety, feasibility, and immune activation. Clin Cancer Res. 2008;14:1455–1463. doi: 10.1158/1078-0432.CCR-07-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Disis ML, Cheever MA. HER-2/neu protein: a target for antigen-specific immunotherapy of human cancer. Adv Cancer Res. 1997;71:343–371. doi: 10.1016/s0065-230x(08)60103-7. [DOI] [PubMed] [Google Scholar]
- 65.Dobaño C, McTague A, Sette A, Hoffman SL, Rogers WO, Doolan DL. Mutating the anchor residues associated with MHC binding inhibits and deviates CD8+ T cell mediated protective immunity against malaria. Mol Immunol. 2007;44:2235–2248. doi: 10.1016/j.molimm.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 66.Onizuka S, Tawara I, Shimizu J, Sakaguchi S, Fujita T, Nakayama E. Tumor rejection by in vivo administration of anti-CD25 (interleukin-2 receptor α) monoclonal antibody. Cancer Res. 1999;59:3128–3133. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.