Abstract
Genome Browsers are software that allow the user to view genome annotations in the context of a reference sequence, such as a chromosome, contig, scaffold, etc. The Generic Genome Browser (GBrowse) is an open source genome browser package developed as part of the Generic Model Database Project (see Unit 9.9; Stein et at., 2002). The increasing number of sequenced genomes has to a corresponding growth in the field of comparative genomics, which requires methods to view and compare multiple genomes. Using the same software framework as GBrowse, the Generic Synteny Browser (GBrowse_syn) allows the comparison of co-linear regions of multiple genomes using the familiar GBrowse-style web page. Like GBrowse, GBrowse_syn can be configured to display any organism and is currently the synteny browser used for model organisms such as C. elegans (WormBase; www.wormbase.org; see Unit 1.8) and Arabidopsis (TAIR; www.arabidopsis.org; see Unit 1.11). GBrowse_syn is part of the GBrowse software package and can be downloaded from the web and run on any unix-like operating system, such as Linux, Solaris, Mac OS X etc. GBrowse_syn is still under active development. This unit will cover installation and configuration as part of the current stable version of GBrowse (v1.71).
Introduction
GBrowse_syn was designed to be portable and configurable like its parent application GBrowse. It can be run on any unix-like operating system with the MySQL database management system installed. GBrowse_syn views multiple genomes by comparing co-linear regions of one or more genomes against a single reference sequence, with the ability to toggle between reference and target sequences. The Original use case was for comparison of three nematode genomes at WormBase but, as the number of sequenced nematode and other genomes continues to grow, more than three species can be compared with this software. GBrowse_syn is designed to use the same database adapters as GBrowse for displaying sequence annotations and uses a central joining database to link any number of GBrowse data sources and render them in the same screen.
This unit has two main protocols and one Alternate Protocol. Basic Protocol 1 shows how to configure GBrowse_syn to use the example data set of two aligned rice genomes with the alignments and sequence annotations in MySQL relational databases. In addition to multiple sequence alignment data, GBrowse_syn can use any kind of co-linearity data that has coordinate and strand information. Basic Protocol 2 shows how to configure OrthoCluster (see Unit 6.10) Synteny blocks to be loaded and browsed in GBrowse_syn. Whole genome alignment strategies for complex genomes usually involve hierarchical strategies where syntenic (or co-linear) regions are first identified and then aligned at the nucleotide sequence level. Alternate Protocol 2 shows how to load the GBrowse_syn alignment database from the relatively more complex output of the MERCATOR/MAVID whole genome alignment workflow (Dewey, 2007). Support Protocol 1 describes how to install GBrowse_syn and its dependencies from the most current stable source code (version 1.71 at time of writing).
Basic Protocol 1: Configuring and Using GBrowse_syn
GBrowse_syn is installed along with the GBrowse package. Sample alignment and configuration data are included with the installation. This protocol will describe the basic configuration and use of GBrowse_syn.
Example Data
Genome annotation files are provided in GFF3 for two rice species, referred to throughout the first part of the protocol as ‘rice’, and ‘wild_rice’, and blastz-derived (Schwartz et al., 2003) whole genome alignment data between the genomic DNA of the two species. The files are installed in the databases directory under the GBrowse document root, the HTDOCS option described in Unit 9.9, which is the location of GBrowse cascading style sheets, help files, tutorial, etc. The location of the document root will vary according to system architecture and user options selected at install time. The correct location of these files on the server will be displayed in the welcome screen shown when GBrowse_syn is used for the first time (See Support Protocol 1). In this example the location of the document root is
/var/www/html/gbrowse
Necessary Resources
Hardware
Unix (Linux, Solaris, or other variety) workstation or Macintosh with OS X 10.2.3 or higher
Internet connection
Software
No additional software is required if Support protocol 1 has been completed
Files
The data and configuration files needed for this protocol are pre-installed with the GBrowse package or its prerequisites, as described in Support Protocol 1.
This protocol assumes a unix-like operating system. The examples shown in this protocol are run on Linux (CentOS release 5.3) using MySQL server version 5.0.77. Many steps will require the sudo command for administrator level access to the system.
Obtaining example data
These are instructions for setting up and using GBrowse_syn with the examples that are installed along with the GBrowse package. The alignment data and genome annotation data were provided courtesy of Bonnie Hurtwitz.
-
1)
Go to the document root using the unix cd command. The $ symbol represents the Linux command prompt.
$ cd /var/www/html/gbrowse
-
2)
Examine the document tree of the databases directory using the ls -R command to examine the databases directory.
$ \ls -R databases
databases:
gbrowse_syn yeast_chr1+2
databases/gbrowse_syn:
alignments rice wild_rice
databases/gbrowse_syn/alignments:
rice.aln.gz
databases/gbrowse_syn/rice:
rice.gff3
databases/gbrowse_syn/wild_rice:
wild_rice.gff3
databases/yeast_chr1+2:
chr1.fa chr2.fa yeast_chr1+2.gff3
In some systems the ls command may be aliased to use other options by default. The backslash (\) before the ls command will invoke ls with no options other than the specified -R (recursive) argument. The files you will need are under the gbrowse_syn subdirectory.
-
3)
Change to the directory with the alignment data file and unpack the compressed file using the gunzip command.
$ cd databases/gbrowse_syn/alignments
$ sudo gunzip rice.aln.gz
Figure 9.12.1 shows the first few lines of the alignment file. The syntax of the sequence names in the alignment is critical because it contains meta-data required to get the coordinates and strand of each sequence.
The syntax is:
Species-seqid(strand)/start-end
The database loading script load_alignments_msa.pl (discussed below) will check the name format while parsing the alignments. Violations of the required syntax will cause a fatal exception and script will not execute.
-
4)
Create a database named ‘rice_synteny’ (you will need a MySQL account with CREATE and GRANT privileges). Substitute your own user name and password for ‘user’ and ‘pass’.
$ mysql -uuser -ppass
mysql> create database rice_synteny;
Query OK, 1 row affected (0.00 sec)
-
5)
Grant SELECT privileges to use ‘www-data’, the default web user name for mysql in this installation, then quit the mysql shell.
mysql> grant SELECT on rice_synteny.* to ‘www-data‘@’localhost’;
Query OK, 0 rows affected (0.02 sec)
mysql> quit Bye
-
6)
Load the database using the load_alignments_msa.pl script, which is pre-installed with GBrowse and can be run without specifying the location of the script. This will load the alignment file above into the database. The command is all on one line.
$ load_alignments_msa.pl -u user -p pass -d rice_synteny -v -c rice.aln
where
-u username with CREATE, INSERT, GRANT privileges -p password (if required) -d database name -v verbose progress reporting (optional) -c start new database. This option overwrites any existing database of that name (recommended).
Now that we have loaded the alignment database, also referred to as the joining database because it links together the data sources for each of the species, turn to the species annotation data in GFF3 format (Figure 9.12.2). The GFF3 format is described in Table 9.9.1 of Unit 9.9. The location of the species annotation data relative to the document root is:
databases/gbrowse_syn/rice/rice.gff3
databases/gbrowse_syn/wild_rice/wild_rice.gff3
By default, the GFF files are used with a flat-file adapter that can access the GFF files directly. Due to the large number of gene models for the two species, using flat files as species databases may be slow and cause excessive latency in rendering the images for GBrowse_syn on some servers. It is a relatively simple process to convert the GFF3 to MySQL databases using the script bp_seqfeature_store.pl, which is installed with the bioperl-live distribution that will have been completed in Support protocol 1.
-
7)
Repeat steps 4-5 to create and set the permissions on two additional databases, named ‘rice’ and ‘wild_rice’.
-
8)
Load the rice.gff3 file into a Bio∷DB∷SeqFeature∷Store database using the bp_seqfeature_load.pl script.
$ bp_seqfeature_load.pl -u user -p pass -d rice -c -f rice.gff3
where
-u username with MySQL root-level privileges -p password (if required) -d database name -f specifies fast loading. This feature is a big time saver but the GFF3 file must be well formatted, so that all subfeatures with the same ID are situated together in the file. The example files in this protocol are compatible with this option. -c start new database. This option overwrites any existing database of that name. (recommended)
-
9)
Change to the wild_rice subdirectory and repeat step 8 for the wild_rice.gff3 file.
$ cd ../wild-rice
$ bp_seqfeature_load -u user -p pass -d wild_rice -c -f wild_rice.gff3
Note the database name is also changed to wild_rice.
Figure 9.12.1.

The first few lines of the rice.aln file, rice.aln is a CLUSTAL-formatted alignment file. Note, this is simply a formatting convention and does not imply that the CLUSTAL program was used to generate the data.
Figure 9.12.2.

A sample of the genome annotations for the ‘rice’ data source. These annotations are in GFF3 format, which is explained in detail in Unit 9.9. This sample contains three gene models in a three level containment hierarchy (gene > mRNA > CDS).
Configuration files
-
10)
Find the configuration files for the alignment data and the two rice species at the locations below, relative to the configuration root. The configuration root is the full system path to the configuration files. The actual path will vary by operating system and configuration options at the time of installation. In this example, the configuration root is
/var/www/conf/gbrowse.conf
/var/www/conf/gbrowse.conf/synteny/oryza.synconf.disabled
/var/www/conf/gbrowse.conf/synteny/rice_synteny.conf
/var/www/conf/gbrowse.conf/synteny/wild_rice_synteny.conf
The file oryza.synconf.disabled will have its name changed to oryza.synconf in a subsequence step. It has the ‘disabled’ extension so that GBrowse_syn will not try to load the data source until the configuration file is ready and the data source is fully configured.
The most important difference in the configuration between GBrowse_syn and GBrowse is that GBrowse_syn uses a joining database that links different species together via database features corresponding to alignment data, synteny blocks, gene orthology, etc. This example uses three configuration files, one for each of the rice species and one to link the species together via the joining database. The species configuration files have the same structure and options as GBrowse configuration files and specify track display options, etc. The example shown in Figure 9.12.3 is a minimal configuration file. For examples of the many configurable options for GBrowse, see Unit 9.9. The other configuration file is the GBrowse_syn and specifies the joining database that links the species, information about the species and their configuration files and display options. See Table 9.12.1 for configurable options for the GBrowse_syn configuration file.
When first installed, as shown in Support protocol 1, GBrowse_syn scans the configuration directory for files ending in .synconf to look for configured data sources. If none are found it prints a welcome screen, which is described in Support protocol 1.
Species configuration files have the same structure as GBrowse configuration files, though they tend to be less complex (see Figure 9.12.4). Note the data source is configured by default to use the memory adapter for flat files. Flat file databases are best used for small data sets. Because the rice example annotations contain many gene models, there can be excessive latency in rendering images on some system configurations. In order to speed up GBrowse_syn for the example data provided, you will use MySQL databases for the two species' genome annotations.
-
11)
In order to deploy the MySQL databases you have loaded for the rice data, use a text editor such as pico, emacs, vi, etc, and edit rice_synteny.conf so that the database arguments read (you will have to use sudo to edit the installed configuration files):
db_adapter = Bio∷DB∷SeqFeature∷Store db_args = dsn dbi:mysql:rice The Bio∷DB∷SeqFeature∷Store adapter and relational database schema are optimized for GFF3 and is the method of choice for loading this format into a relational database management system, in this case MySQL. This greatly accelerates data access and decreases the latency the user experiences when browsing multiple genomes with GBrowse_syn.
-
12)
Repeat step 11 for rice_synteny.conf, using the database name ‘wild_rice’.
-
13)
Reload the web page to see the display shown in figure 9.12.5. Click on the example rice 3:16050173..1606497. You should see the display shown in figure 9.12.6, which now has the alignment between the rice and wild rice genomes shown, with rice as the reference genome.
Figure 9.12.3.

Complete configuration file for the ‘oryza’ data source that is installed as an example with the GBrowse package. This file is similar in structure to a GBrowse configuration file, as described in Unit 9.9. In addition to the connection information for the joining database, this file specifies the location of the configuration files for the species to be compared in GBrowse_syn and the theme color and tracks to load for each species.
Table 9.12.1.
Configurable options for the GBrowse_syn configuration file. Options shown in bold face are required. Options shown in italics are recommended.
| Option | Description |
|---|---|
| Join | The data source name (DSN) for the joining database. Figure x.x.x shows a typical example. |
| source map | The mapping of symbolic source (name), configuration file name and description for each species. Figure 9.12.3 shows a typical example. |
| Tmpimages | The URL (location relative to the document root) where temporary image and cached data should be stored, eg: /gbrowse/tmp. |
| Buttons | The location for common GBrowse images of buttons, arrows, etc., eg: /gbrowse/images/buttons. Default images will be used unless otherwise specified. |
| Stylesheet | The URL for a cascading style sheet (CSS) file that specifies various configurable web display options. These can be customized. The default GBrowse stylesheet is used unless otherwise specified. |
| Examples | Example segments to display. These specify the reference species, sequence and coordinates. Some examples are shown in fig x.x.x. |
| zoom levels | which zoom levels will be available in the navigation menu default: zoom levels = 5000 10000 25000 50000 100000 200000 400000 |
| config_extension | The file extension (.syn or .conf) for species configuration files. Note this extension has to be used consistently throughout the GBrowse_syn configuration directory. default: syn |
| description | The description of the data source for public display. default: none |
| max_span | The gap between inset panels, expressed as the portion of the referebce panel with, to trigger merging of inset panels. default: 0.3 |
| max_segment | The maximum allowed sequence length to be displayed in the reference panel. default: 400Kb |
| min_alignment_size | The minimum alignment size, expressed as a fraction of the total reference sequence length, that will be used to create an inset panel. default: 0.01 |
| imagewidth | The default with, in pixels, of the reference panel. default: 5 |
| interimage_pad | The space between inset panels, in pixels. default: 5 |
| vertical_pad | The vertical space between panels, in pixels. default: 5 |
| align_height | The height of the alignment or syntenic block features, in pixels. default: 5 |
| max_gap | The maximum gap allowed between chained alignment features. default: 50Kb |
| overview_ratio | The relative width of the overview panel in relation to the with of the reference panel. default: 0.9 |
| overview bgcolor | The background color of the overview panel. Named web colors of hexidecimal codes are acceptable. default: gainsboro |
| grid coordinates | This option is for sparse grid coordinate data. If set to ‘exact’, all coordinates will be used. Otherwise, coordinates that are multiples of 10, 100, 1000, etc will be used depending on the size of the displayed segment. |
Figure 9.12.4.
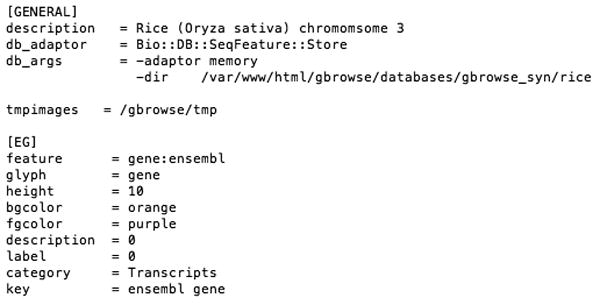
The rice_synteny.conf configuration file. Minimal information is required as this is not intended as a stand-alone genome browser. A Detailed list of configurable options for GBrowse configuration files can be found in Unit 9.9. Note that the [EG] track is referenced by the main configuration file oryza.synconf in figure 9.12.3.
Figure 9.12.5.
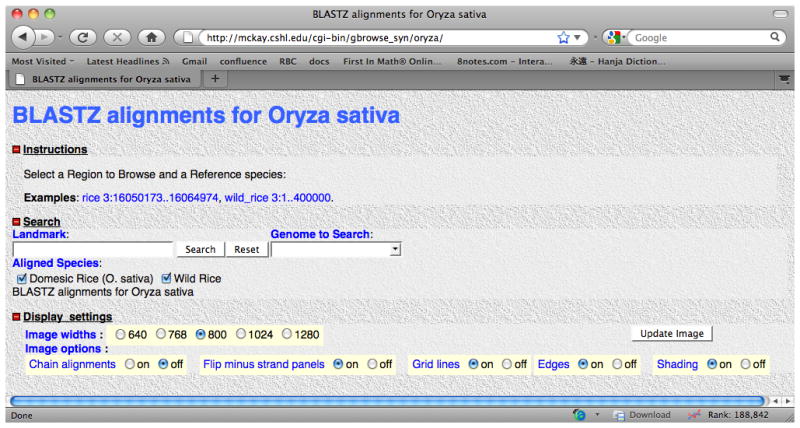
The startup screen for the Oryza sativa sample data source included with the GBrowse package. Clicking on one of the example segment links is a good way to get started browsing.
Figure 9.12.6.

Example segment rice 3:16050173..1606497. With the default options, shaded polygons with grid lines are shown. The grid lines correspond to mapped sequence coordinates in the aligned segments.
NOTE: The reference target relationship is stored reciprocally in the alignment database. Clicking on a part of the inset panel for other genome that does not have other behaviors, such as popup balloons or links, will reload the page with the reference/target relationship reversed.
Interpreting Results
-
14)
Examine the general layout which shows a central reference panel or a lower reference panel in cases where there are only two species. Inset panels for other species with matching regions appear above or below the reference sequence panel. Clicking on one of the inset panels, which will then become the reference sequence, facilitates rapid switching between reference sequences. The example used in this protocol uses two closely related species where, in most regions the whole segment is collinear and there is only one inset panel.
-
15)
Hover the mouse over the blue text web page features (figure 9.12.5) to display a popup balloon. Clicking these links take you to a help page describing that feature. A detailed list of features is described on a web-based help page (http://gmod.org/wiki/GBrowse_syn_Help). Figure 9.12.7 shows an excerpt from this help page, which is kept up to date as new features are added to GBrowse_syn.
-
16)
In the overview panel, click and drag on the scale-bar to activate the rubber band selection to aid in moving, re-centering, or resizing, the viewed region. Clicking anywhere on the overview panel will also move the detailed view to that region.
The overall look and feel are similar to GBrowse, though not all GBrowse features such as draggable tracks and rubber band selection in the detail panels are available.
-
17)
Compare the species. There is no upper limit on the number of species that can compared with GBrowse_syn. Because only a single reference sequence is shown at one time, the reference panel is repeated as many times as necessary to compare it to all species. An “all in one view” is also available, although it is not very informative if there are a large number of species being compared. Figure 9.12.8 shows a more complex example of a five species comparison from WormBase (http://www.wormbase.org). The lower section of the web page offers a number of image display options, such as width, shading and grid lines for aligned regions. The grid lines option is especially useful, as it tracks corresponding nucleotide residue positions at columns in the DNA sequence alignment, which highlights relatively large insertions and deletions. The example shown in figure 9.12.8 is of particular interest because it shows extensive insertions and deletions among the five genomic DNA sequences being compared.
Figure 9.12.7.
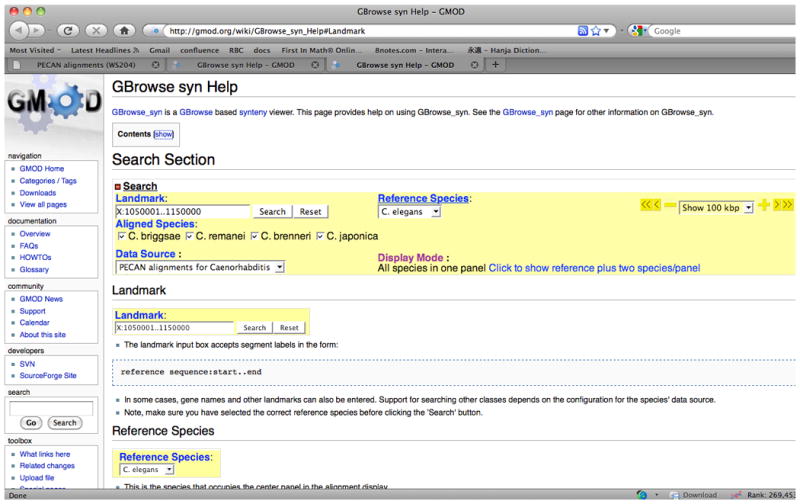
An excerpt from the GMOD (Generic Model Organism Database) Wiki pages that describes web page features for GBrowse_syn. These features continue to be updated and changes are posted to the Wiki.
Figure 9.12.8.

A five species whole genome DNA sequence alignment comparison from WormBase (http://www.wormbase.org), showing regions that are co-linear with Caenorhabditis elegans genomic segment X:1085001..1115000. The displayed region uses the default settings for the display options shown in the bottom panels of the image.
Alignment chaining
-
18)
Select the chain alignments option in the Display Setting part of the GBrowse_syn web page.. GBrowse_syn will perform an “on the fly” analysis of the alignments or co-linear regions from other sources, as well as merge or join parts that are within a configurable distance of each other (the default is 50kb), are on the same strand, and have either monotonically increasing or increasing coordinates depending on the orientation (see Figure 9.12.9). This method is analogous to the blastz chaining described in Kent et al., (2003).
Figure 9.12.9.

Alignment chaining. A) alignment of a segment of the rice and wild-rice genomes with the alignment data provided. B) the same region with the “chain alignments” option selected. Same-stand alignments with monotonically increasing (or decreasing) coordinates are merged or connected by dashed lines where there are gaps. This example allows gaps of up to 50kb between chained alignments. Note the loss of two genes in domestic vs. wild rice.
Basic Protocol 2: Browsing Orthocluster Synteny Blocks with GBrowse_syn
Although Gbrowse_syn was developed with whole genome DNA sequence alignments in mind, it can also be used to display syntenic or co-linear regions that are not based on DNA sequence alignments. For example, OrthoCluster (Ng et al. 2009; Zeng et al. 2008) is a tool that has been developed for the accurate detection of synteny blocks among multiple species. Briefly, OrthoCluster takes as input two types of files: (i) a genome file, which contains the list of all genes with its chromosome/contig, start position, end position and strand; and (ii) a correspondence file, which contains the orthologous relationships among genes in all genomes. A detailed protocol on generating these input files and on running OrthoCluster is available (see Unit 6.10). The following protocol illustrates how to generate the GBrowse_syn input files based on pair-wise synteny block detection using OrthoCluster for three nematode genomes: Caenorhabditis elegans (ele), Caenorhabditis briggsae (bri) and Pristionchus pacificus (ppa). The procedure shown here can be extended to any number and type of genomes.
Necessary Resources
Hardware
Unix (Linux, Solaris, or other variety) workstation or Macintosh with OS X 10.2.3 or higher
Internet connection
Software
All necessary software should be installed if Support Protocol 1 has been completed.
Files
Data and configuration files data are contained in the supplementary file orthocluster.tar.gz.
This protocol assumes that the user already executed OrthoCluster in a pair-wise manner for the three species and that the GBrowse package (Unit 9.9) has been installed. The examples shown in this protocol are run on Linux (CentOS release 5.3) using MySQL server version 5.0.77.
-
1)
Create a working directory called ORTHOCLUSTER by unpacking the supplemental file orthocluster.tar.gz with the tar command.
$ tar zxf orthocluster.tar.gz
where
z decompress the gzipped archive x extract the files f use the archive file orthocluster.tar
-
2)
Examine the contents of the directory using the ls -R command.
$ \ls -R ORTHOCLUSTER/
ORTHOCLUSTER/:
conf genome_files gff pairs scripts
ORTHOCLUSTER/conf:
bri.conf ele.conf orthocluster.synconf ppa.conf
ORTHOCLUSTER/genome_files:
genome_bri.txt genome_ele.txt genome_ppa.txt
ORTHOCLUSTER/gff:
bri.gff ele.gff ppa.gff
ORTHOCLUSTER/pairs:
bri_ppa ele_bri ele_ppa
ORTHOCLUSTER/pairs/bri_ppa:
perfect.cluster perfect.log
ORTHOCLUSTER/pairs/ele_bri:
perfect.cluster perfect.log
ORTHOCLUSTER/pairs/ele_ppa:
perfect.cluster perfect.log
ORTHOCLUSTER/scripts:
gff32gbrowse_syn.pl orthocluster2gff3.pl
The conf directory contains configuration files for GBrowse_syn (discussed below).
The genome_files folder contains the genome annotations for each species under analysis. For example, use the ‘head’ command to examine the first few lines of the genome file genome_bri.txt:
$ cd ORTHOCLUSTER $ head -5 genome_files/genome_bri.txt CBG14914 chrI 2983447 2985061 1 CBG08849 chrV 2000498 2001982 -1 CBG01738 chrIV 9847651 9848961 1 CBG03691 chrII 2283559 2284251 1 CBG26761 chrUn 4289107 4291920 -1 There are four tab-delimited fields: gene name, chromosome, start position, end position and strand. This information will be required later to generate data for internal grid-lines in the GBrowse_syn display.
The pairs directory contains the pair-wise species comparisons, with an orthocluster output file perfect.cluster and log file perfect.log copied from the results of an orthocluster run. The reciprocal of each comparison is not required, as the reciprocal synteny blocks are calculated during the GBrowse_syn database loading process. The original location of these files will vary according to how orthocluster was run. The file perfect.log is necessary to access the sorting of the genome done by OrthoCluster when detecting synteny blocks. Note that, since these files are created automatically by OrthoCluster, it is expected that the log file and the cluster file have the same name prefix (e.g. perfect).
-
3)
The ‘scripts’ directory contains scripts necessary to process the orthocluster data into a format suitable for loading into GBrowse_syn. Make sure the scripts are executable.
$ chmod u+x scripts/*.pl
The script orthocluster2gff3.pl generates two gff3 files: one for the synteny blocks of each pair-wise comparison and one for the orthologous relations found for each co-linear synteny block of ortholog pairs.
- IMPORTANT: since gbrowse_syn requires the syntenic relationship to include orientation, the script orthocluster2gff3.pl only works for synteny blocks generated with the -rs parameter in OrthoCluster.
-
4)
Use the command below to generate the gff3 files given each synteny block file within your working directory. (Note, the long command may be very long, use the ‘\’ to indicate line breaks within the single command).
./scripts/orthocluster2gff3.pl <.cluster output \
> <reference_genome_file> <target_genome_file> <avoid_\
nested_blocks> <minimum_block_size>
where
<.cluster output>: path to the OrthoCluster output. <reference_genome_file>: path to the reference genome annotation. <target_genome_file>: path to the target genome annotation. <avoid_nested_blocks>: 1 for yes, 0 if no. <minimum_block_size>: minimum block size (in number of genes). For example, for the synteny blocks detected using C. elegans as reference and C. briggsae as target, the user may run the following command (all on one line):
$ ./scripts/orthocluster2gff3.pl pairs/ele_bri/perfect.cluster \
genome_files/genome_ele.txt genome_files/genome_bri.txt 0 2
where the last two values, 0 and 2, indicate that nested synteny blocks, and blocks containing two or more genes will be included in the parsed output, respectively. This will generate two output files within the working directory:
genome_ele_genome_bri.orthologs.gff3: contains all the orthologous relationships found within synteny blocks in GFF3 format.
genome_ele_genome_bri.cluster.gff3: contains the coordinates of synteny blocks in the reference and target genomes.
-
5)
Repeat step 4 for each species pair directory.
There should now be the following gff3 files:
genome_bri_genome_ppa.cluster.gff3
genome_bri_genome_ppa.orthologs.gff3
genome_ele_genome_bri.cluster.gff3
genome_ele_genome_bri.orthologs.gff3
genome_ele_genome_ppa.cluster.gff3
genome_ele_genome_ppa.orthologs.gff3
-
6)
Use the script gff32gbrowse_syn.pl to use the GFF3 file to create the generic tab delimited GBrowse_syn loading file. This script print to STDOUT, so the contents should be redirected to a file.
$./scripts/gff32gbrowse_syn.pl >gbrowse_syn_data.tsv
Examine the top line of the output file to see its structure. Note that this will be a very long line. Artificial line breaks are indicated with ‘\’ for readability.
$ head -1 gbrowse_syn_data.tsv bri chrI 176154 183558 + .\ ppa Ppa_Contig88 27212 30786 + .\ 176154 27212 177594 30786 182118 27212 183558\ 30786 | 30786 183558 27212 182118 30786 177594\ 27212 176154 -
This line describes a “synteny block” of colinear genes. The first 12 columns are two blocks of six fields, repeated for each of the reference and target sequences:
Species
Seqid
Start
End
Strand
Cigar-string (reserved for future use)
Reciprocal coordinate maps, delimited by a | symbol, are appended to the end of the line. Each coordinate map is composed of pair-wise positional matches. For multiple sequence alignments, they map changes in relative nucleotide residue positions due to insertions/deletions but, in this case, the maps are adapted to represent the positions of the start and end points of each gene in an orthologous pair. There are three genes in the block represented above, hence three sets of pairs in each coordinate map corresponding to the start and end of each gene.
-
7)
Create the GBrowse_syn joining database. You will need a MySQL account with root level privileges. Substitute your own user name and password in the command below.
$ mysql -u user -p pass
mysql> create database orthocluster;
Query OK, 1 row affected (0.00 sec)
-
8)
Grant SELECT privileges to user ‘www-data’ which is the default web user account in this example. This access level is secure and only allows read-only access to the database from the web.
mysql> grant SELECT on orthocluster.* to ‘www-data‘@’localhost’;
Query OK, 0 rows affected (0.00 sec)
-
9)
Repeat steps 7and 8 to create one new database for each species (‘ele’, ‘bri’, ‘ppa’ – the names specified in the configuration files).
-
10)
Exit the MySQL shell.
mysql> quit
-
11)
Load the database using the load_alignment_database.pl script that is pre-installed with the GBrowse package (the command is all one line). Substitute your own MySQL user name and password for user and pass, respectively.
-
$ load_alignment_database.pl -u user -p pass -d orthocluster \
-c [-v] gbrowse_syn_data.tsv
where
-u username with root-level MySQL privileges -p password (if required) -d database name -v verbose progress reporting (optional) -c start new database. This option overwrites any existing database of that name.
-
Genome annotations
-
12)
Locate the genome annotation data for the three nematode species in the gff directory. These annotations are derived from WormBase (http://www.wormbase.org) release WS204, which are located in the conf directory. GFF2 or GFF (http://biowiki.org/GffFormat) is an older version of GFF still used by WormBase at the time WS204was released. GFF2 is still well supported by GBrowse's Bio∷DB∷GFF adapter and database schema.
-
13)
Load the ‘ele’ database from the GFF file ele.gff using the script bp_fast_load.pl, which was installed along with bioperl-live in Support Protocol 1. Substitute your own MySQl user name and password in the command below
$ bp_fast_load_gff.pl -u user -p pass -d database -c gff/ele.gff
where
-u username with MySQL root-level privileges -p password (if required) -d database name -c start new database. This option overwrites any existing database of that name.
-
14)
Repeat step 13 for the ‘bri’ and ‘ppa’ GFF files, taking care to also change the database names accordingly.
Configuration files
-
15)
Locate the conf directory which contains the necessary configuration file for the new GBrowse_syn data source. The file orthocluster.synconf (see Figure 9.12.10), contains all of the information necessary to set up the browser and link the species data sources ( ele.conf, bri.conf, ppa.conf). For an example of a species configuration file, see figure 9.12.11. The structure of orthocluster.synconf file is similar to the oryza.synconf file described in Basic Protocol 1 except that the sparse grid line data from the orthologous gene boundaries requires the following option.
grid coordinates = exact
This option configures GBrowse_syn to use all available coordinate data for drawing grid lines.
For alignment data, there is usually more coordinate information and every nearest 10th, 100th or 1000th coordinate pair is used, depending on the zoom level. With the ‘exact’ value, all grid coordinates are used at any zoom level.The structure of each species configuration file is similar to the rice examples in Basic protocol 1, except that the Bio∷DB∷GFF database adapter is configured to use GFF2 rather than GFF3 data. The file ele.conf is shown as an example (Figure 9.12.11).
-
16)
Copy the configuration files from the conf directory to the GBrowse_syn configuration root directory. The configuration root is the full system path to the configuration files. The actual path will vary by operating system and configuration options at the time of installation. In this example, the configuration root is
/var/www/conf/gbrowse.conf
$ copy conf/*conf /var/www/conf/gbrowse.conf/synteny
where the part in bold may vary by your specific system configuration.
This should complete the installation of the OrthoCluster data source. To test it, point your web browser to http://hostname/cgi-bin/gbrowse_syn/orthocluster, where hostname would be ‘localhost’ if you are running the browser on the same machine, or the server name if you are browsing remotely. You should see the startup screen shown in figure 9.12.12.
-
17)
Select the example bri chrX:255000..275000, if configured correctly, you should see the image shown in figure 9.12.13.
Figure 9.12.10.

The configuration file orthologs.synconf. Note that the coordinate sparse data require the use of the grid “coordinates = exact” option.
Figure 9.12.11.

The ele.conf species configuration file. Note that the Bio∷DB∷GFF adapter is used for the GFF2 gene annotation data.
Figure 9.12.12.

The starting page for the ‘orthocluster’ data source.
Figure 9.12.13.
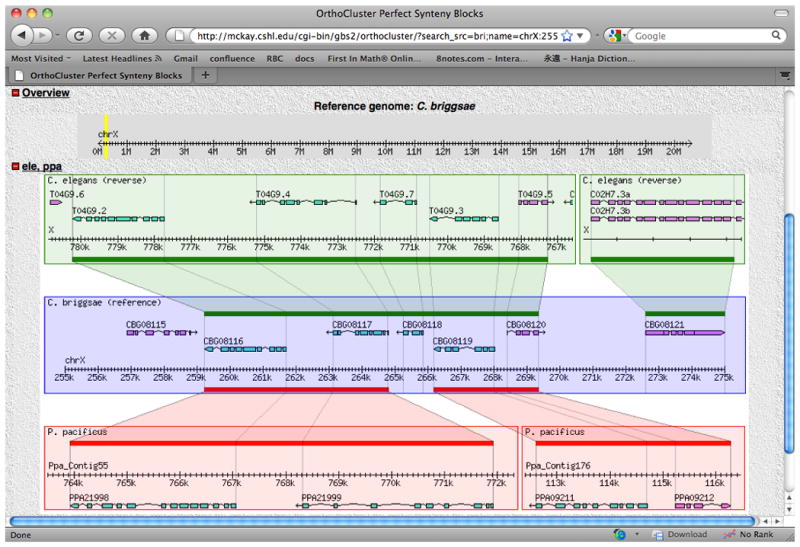
Example segment chrX:255000..275000.With the default options, shaded polygons with grid lines are shown. Note that the grid lines correspond to orthologous gene boundaries.
Alternate Protocol 2: Loading Mercator into the GBrowse_syn Database
MERCATOR (Dewey 2007) is a tool for whole genome alignment using protein coding exons as anchors in the alignment procedure. MERCATOR produces a map of the synteny blocks among the genomes compared and can be used for pairwise or multi-way alignments.
This protocol assumes that the user has already run the MERCATOR pipeline to produce DNA sequence alignments, that the GBrowse package (see Unit 9.9) has been installed, and that. The examples shown in this protocol are run on Linux (CentOS release 5.3) using MySQL server version 5.0.77.
Steps for running MERCATOR are outlined in (Dewey 2007) and in the appendix of (Dewey 2006). The results of MERCATOR include several files and directories; however, the necessary folder for this procedure is the ‘alignments’ directory. Running MERCATOR requires generating a gene annotation and genome files (not shown).
The typical directory structure, if following the MERCATOR instructions, includes an ‘input’ and ‘output’ directory. Within the ‘output’ directory there is a directory called ‘alignments’, this contains all the data necessary for transformation to GBrowse_syn.
Example data are taken from pairwise alignments for the species Drosophila yakuba and D. erecta from the web site http://www.biostat.wisc.edu/∼cdewey/fly_CAF1. These data are the result of a MERCATOR and MAVID alignment (Bray and Pachter, 2004) between these two fly species. Although MAVID is used for the example data, other DNA sequence alignment software could be used on the synteny blocks identified by MERCATOR.
Necessary Resources
Hardware
Unix (Linux, Solaris, or other variety) workstation or Macintosh with OS X 10.2.3 or higher
Internet connection
Software
All necessary software should be installed if Support Protocol 1 has been completed.
Files
Example data were taken from the Drosophila yakuba and D. erector alignments available at http://www.biostat.wisc.edu/∼cdewey/fly_CAF1/.
-
1)
Download the example MERCATOR/MAVID data for D. erecta and D. yakuba pair-wise alignments from http://www.biostat.wisc.edu/∼cdewey/fly_CAF1/ (note that the long line of this command is wrapped; a ‘\’ indicates a line break inside a single command).
-
2)
Unpack the compressed archive
$ tar zxf DroYak_CAF1-DroEre_CAF1.tar.gz
where
z decompress the gzipped archive x extract the files f use the archive file DroYak_CAF1-DroEre_CAF1.tar
-
3)
The directory DroYak_CAF1-DroEre_CAF1 is equivalent to the ‘alignments’ directory described above. Examine the directory with ls.
$ \ls -1 DroYak_CAF1-DroEre_CAF1
1
10
100
--- truncated ---
98
99
DroEre_CAF1.agp
genomes
map
treefile
- NOTE: the −1 for ls option lists one file/line. There are a total of 116 numbered directories. The list has been truncated for display purposes. Each numbered directory contains a single file, mavid.mfa. The key files for conversion to GBrowse_syn are
x/mavid.mfa multiple sequence alignment produced by MAVID genomes lists the prefixed named used when Mercator alignments were run. map encodes the chromosome, start, stop, and strand locations of each synteny block in each of the genomes aligned in the order listed.
-
4)
Convert the data to GBrowse_syn loading format using the mercatoraln_to_synhits.pl script. If Support Protocol 1 has been completed and the current stable GBrowse is installed, this script will be pre-installed in the executable path, typically /usr/bin (may vary by operating system) and can be run without specifying the path to the script. The program prints to STDOUT, so redirect the output to a file. The command is all on one line.
$ mercatoraln_to_synhits.pl -d DroYak_CAF1-DroEre_CAF1 \
-a mavid.mfa > mercator.tab
where
-d the path to the folder with the necessary input files -a the name of the alignment file in each of the numbered subdirectories
The file mercator.tab is in a tab delimited format designed for direct loading into the GBrowse_syn alignment (or joining) database. The format has one tab-delimited record/line. Each line represents a synteny block, or alignment, with 13 fields:
Reference Species
Reference Seqid
Reference Start
Reference End
Reference Strand
Reference Cigar-string (not used; reserved for future use)
Target Species
Target Seqid
Target Start
Target End
Target Strand
Target Cigar-string (not used; reserved for future use)
Coordinate map (optional)
The coordinate map is used to save pair-wise nucleotide residue coordinates for columns in the aligned sequences. It is not necessary to store coordinates for every column. GBrowse_syn usually uses multiples of 10, typically 100. The purpose of storing the coordinate information is to position grid lines in the graphical display that will make large insertions and deletions in the sequences visible and intuitive. The grid lines are equidistant on the reference sequence but can show insertions or deletions by increasing or decreasing the distance between the lines, respectively, on the target sequence. The format of field 13 (with spaces, not tabs)
rcoord1 tcoord1 rcoord2 tcoord2 | tcoorda rcoorda tcoordb rcoordb
where
rcoordn reference nucleotide residue number n tcoordn target nucleotide residue number n n column in the alignment | Symbol delimiting reciprocal coordinate maps - NOTE: calculating the coordinate map is computationally intensive and the script may take a long time to run.
-
5)
Load the GBrowse_syn alignment database with the script load_alignment_database.pl. If Support Protocol 1 has been completed, the script is pre-installed and can be run without specifying the path. Substitute your MySQL user name and password in the command below.
$ load_alignment_database.pl -u user -p pass -d database -v -c \
mercator.tab
where
-u username with root-level MySQL privileges -p password (if required) -d database name -v verbose progress reporting (optional) -c start new database. This option overwrites any existing database of that name. (recommended)
Support Protocol 1: Installing GBrowse_syn in the Unix/Linux Environment
GBrowse_syn has been included in the GBrowse since version 1.69 and improved in version 1.7. Recent development of this software component requires updating from GBrowse 1.70 to the most recent stable version of the 1.7× series. GBrowse_syn and example data are also included in the GBrowse 2.0× series.
Necessary Resources
Hardware
Unix (Linux, Solaris, or other variety) workstation or Macintosh with OS X 10.2.3 or higher
Internet connection
Software
Most necessary pre-requisite software should be installed if GBrowse 1.70 is installed and Support Protocol 1 in Unit 9.9 has been completed. Subversion (SVN) and the CPAN shell are also required.
Files
The most recent GBrowse source code from the ‘stable’ branch of the SVN source code repository (http://gmod.org/wiki/GBrowse#SVN), the current version of bioperl-live (http://www.bioperl.org/wiki/Using_Subversion) and the Bio∷Graphics library from the Comprehensive Perl Archive Network (CPAN: http://cpan.org).
This protocol assumes a unix-like operating system with subversion installed. The examples shown in this protocol are run on Linux (CentOS release 5.3) using MySQL server version 5.0.77, subversion (svn) version 1.4.2 (r22196) and CPAN v1.9402. Details of files, permissions, etc. are described in Support Protocol 1 in Unit 9.9. There are many new features in the current development version, which have not yet been released. To get the latest version, it is best to use svn to get the most recent bioperl-live and GBrowse 1.7×. The Bio∷Graphics libraries have split from GBrowse and BioPerl and need to be installed or updated via the CPAN shell. You will need administrative access via the sudo command for many of these steps.
Check out and install the latest version of the source code
-
1)
Check out the latest build of bioperl-live. The $ symbol represents the Linux command prompt. The command is all on one line.
$ svn co svn://code.open-bio.org/bioperl/bioperl-live/trunk bioperl-live
This will check out the bioperl-live source code into a working directory bioperl-live
-
2)
Install bioperl-live from source.
$ cd bioperl-live
$ perl ./Build.pl
This script may ask about optional prerequisites and tests. Assuming you have GBrowse 1.7 installed and working, you can likely select the default options (usually [n]) whenever prompted. However, be sure to select the default option [a] when prompted to install all scripts, as some will be needed in Basic Protocol 1.
$ ./Build test (optional - there will be a lot of warnings)
$ sudo ./Build install
-
3)
Install or update Bio∷Graphics
$ sudo cpan
cpan shell -- CPAN exploration and modules installation (v1.9402)
Enter ‘h’ for help.
cpan[1]> install Bio∷Graphics
It is assumed that you have all or most Bio∷Graphics prerequisites already. Follow prerequisites if prompted. Installing via the CPAN shell requires that all tests pass. If there is a failure and it looks minor, try installing with force.
cpan[2]> force install Bio∷Graphics
-
4)
Check out the GBrowse stable branch. This will download version controlled source code for GBrowse 1.7× into a working directory Generic-Genome-Browser. The command is all on one line.
$ svn co https://gmod.svn.sourceforge.net/svnroot/gmod/Generic-Genome-Browser/branches/stable Generic-Genome-Browser
-
5)
Install GBrowse from the source code.
$ cd Generic-Genome-Browser
$ perl Makefile.PL
$ make
$ make test (optional)
$ sudo make install
- NOTE: If you already have an SVN working directory from a previous checkout:
$ sudo make realclean
$ svn update
$ perl Makefile.PL
$ make
$ make test (optional)
$ sudo make install
This should complete the installation of the most recent stable GBrowse. The locations of many of the GBrowse-related files vary by operating system, system architecture or options chosen, as described in Unit 9.9. Resaonable default choices are provided with each option when running Makefile.PL. It is recommended to use the default options. In the example configuration shown here, the necessary files for GBrowse_syn were installed in:
htdocs: /var/www/html/gbrowse databases: /var/www/htmn/databases/gbrowse_syn cgi-script: /var/cgi-bin/gbrowse_syn conf: /var/www/conf/gbrowse.conf/synteny
-
6)
Test the installation by pointing your web browser to http:hostname/cgi-bin/gbrowse_syn, where hostname would be ‘localhost’, if you are running the browser on the same machine, or the server name if you are browsing remotely. If GBrowse_syn is installed correctly and has never been configured before, you should see something similar to the image below in Figure 9.12.14. If you are updating, you should see one of your configured data sources.
Figure 9.12.14.
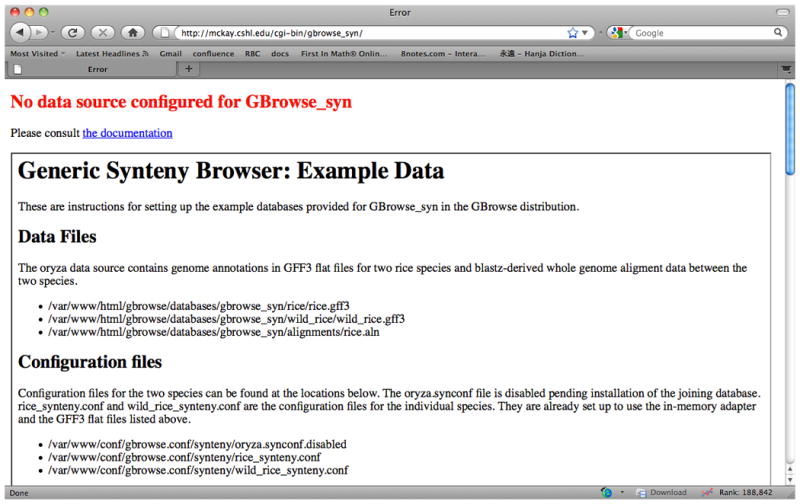
The welcome screen for a new, unconfigured gbrowse_syn installation.
Commentary
Background Information
GBrowse_syn has been part of the GBrowse package since version 1.69 but has undergone the most active development and debugging since version 1.70, which is why Support Protocol 1 emphasizes using the most current version of the 1.71 source code. GBrowse 2, which has many new features (see Unit 9.9 for an overview), was recently released but represents a major re-architecture of GBrowse, and both the 1.71 and 2.0 branches will both be maintained for some time as GBrowse 2.0 becomes more widely tested and stable. If you have installed a recent update of GBrowse 2, GBrowse_syn is also installed by default. Configuration and set-up are virtually identical in both versions 1.71 and 2.0 but this may change in future releases. GBrowse_syn 1.71 is still maintained and is recommended at the time of writing due to the stability and amount of testing that this branch has undergone. Note, however, that new features and future development will be on GBrowse version 2 branch.
Critical Parameters and Troubleshooting
As with GBrowse, the most useful resource for sorting out issues with GBrowse is the GMOD- GBrowse mailing list (see https://lists.sourceforge.net/lists/listinfo/gmod-gbrowse). Developers responsible for GBrowse and GBrowse_syn, as well as other users monitor the list and usually provide feedback or troubleshooting tips for setting up or configuring GBrowse. The list is also monitored by the GMOD help desk, which ensures that questions do not go unanswered. Archives of this list can be searched at http://www.nabble.com/gmod-gbrowse-f3500.html.
GBrowse_syn will send fatal error messages to the browser window. When asking for help, please be sure to record the text of the message. Also check the Web server error log file for warnings or error messages, which can be critical to understanding the problem. The error log file, error_log, or error.log, is located in the server log directory. Some common locations for the log files are: /usr/local/apache/logs etc/httpd/logs and /var/log/httpd. This may be a large file unsuitable for browsing in text editors. Try the unix command tail -50 error_log to look at the last 50 lines. Log entries are usually time-stamped. You may want to vary the number of lines (the −xx flag) depending on how many errors there are. Include the text of the errors in the email sent to contact the GBrowse mailing list for help.
Supplementary Material
Acknowledgments
Development of GBrowse_syn was funded in part from the National Institutes of Health grants P41-HG002223 (WormBase), 7U41HG004269-03 (The modENCODE Data Coordinating Center) and the National Science Foundation grant DBI 0735191 (The iPlant Collaborative: A Cyberinfrastructure Centered Community for A New Plant Biology). Ismael Vergara's work was partly supported by a Discovery Grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada (to Nansheng Chen).
Footnotes
Supplemental Data File Legend
Title: orthocluster.tar.gz
This file is a compressed archive containing the OrthoCluster Synteny data and genome annotation data for the three nematode species used in Basic Protocol 2. It also includes the required configuration files and processing scriptsUse of this file is described in the text of protocol 2.
Literature Cited
- Bray N, Pachter L. MAVID: Constrained ancestral alignment of multiple sequences. Genome Research. 2004;14:693–699. doi: 10.1101/gr.1960404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey CN. Whole-Genome Alignments and Polytopes for Comparative Genomics. EECS Department, University of California; Berkeley: 2006. [Google Scholar]
- Dewey CN. Aligning multiple whole genomes with Mercator and MAVID. Methods Mol Biol. 2007;395:221–236. doi: 10.1007/978-1-59745-514-5_14. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. Evolution's cauldron: Duplication, deletion, and rearrangement in the mouse and human genomes. PNAS. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng MP, et al. OrthoClusterDB: an online platform for synteny blocks. BMC Bioinformatics. 2009;10:192. doi: 10.1186/1471-2105-10-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, Lewis S. The generic genome browser: A building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, et al. OrthoCluster: a new tool for mining synteny blocks and applications in comparative genomics. 11th International Conference on Extending Technology (EDBT); March 25-30, 2008; Nantes, France. 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


